Abstract
CpG dinucleotides are known to play a crucial role in regulatory domains, affecting gene expression in their natural context. Here, we demonstrate that intragenic CpG frequency and distribution impacts transgene and genomic gene expression levels in mammalian cells. As shown for the Macrophage Inflammatory Protein 1α, de novo RNA synthesis correlates with the number of CpG dinucleotides, whereas RNA splicing, stability, nuclear export and translation are not affected by the sequence modification. Differences in chromatin accessibility in vivo and altered nucleosome positioning in vitro suggest that increased CpG levels destabilize the chromatin structure. Moreover, enriched CpG levels correlate with increased RNA polymerase II elongation rates in vivo. Interestingly, elevated CpG levels particularly at the 5′ end of the gene promote efficient transcription. We show that this is a genome-wide feature of highly expressed genes, by identifying a domain of ∼700 bp with high CpG content downstream of the transcription start site, correlating with high levels of transcription. We suggest that these 5′ CpG domains are required to distort the chromatin structure and to increase gene activity.
INTRODUCTION
Due to the degeneracy of the genetic code, an individual protein can be encoded by a large number of different DNA sequences. This degeneracy can be utilized to generate optimized gene sequences, thereby increasing transgene expression efficiency in heterologous systems (1,2). The commonly applied gene optimizing algorithms combine known parameters, such as the adaptation of the coding sequences toward most frequently used tRNAs, the avoidance of strong 5′ RNA secondary structures and inverted repeats, as well as low A + T and high G + C sequence stretches. Moreover, the generation of sequence elements such as TATA-box like motifs, cryptic splice sites, destabilizing UA-rich RNA stretches or internal polyadenylation signals is excluded (3,4). In addition to these parameters, the nucleotide composition of a given gene also influences its potential to form stable nucleosomes in vivo (5). For example, AA, TT and TA dinucleotides are favored approximately every 10 bp where both DNA strands face toward the nucleosome core; GC dinucleotides tend to follow the same spacing where both phosphodiester backbones face outward (6). Yet, the critical contribution of various dinucleotides on gene expression is still barely understood. Among all dinucleotide combinations, CpG and TpA seem to be the most interesting ones: genome-wide analysis revealed more than 20 years ago that while CpG frequency has fallen to its lowest levels in transcriptionally silent DNA, TpA is most stringently excluded in DNA destined to function as RNA in the cytosol, which corresponds with short mRNA half-lives (7). A TpA dominance has been found in 5′ untranslated regions and gene bodies of cytokine and chemokine genes (8), accounting for their mRNA instability. In contrast, the abundance of overall CpG levels in humans is much lower than expected based on the GC content (9). The selective pressure leading to CpG-scarcity is provided by the inherent mutability of methylated cytosines. The deamination of 5-methylcytosines (m5C) results in thymine, which is not easily recognized as foreign and therefore leads to a transition mutation in the following replication (10). Although CpG dinucleotides in non-coding regions such as SINEs (short interspersed nuclear elements), LINEs (long interspersed nuclear elements) and endogenous retroviruses are generally subject to intensive methylation (11,12), there are regulatory regions of CpG clusters that stay unmethylated. These so-called CpG islands, first defined by Bird in 1986, are on average 400–500 bp of length, have a C + G content of 0.5 or higher and an observed to expected CpG ratio of 0.6 or higher within a range of 200 bp or greater (13,14). CpG islands are mostly found within the promoter and the first exon of several genes, particularly housekeeping genes (15,16). The methylation status of regulatory DNA sequences directly influences their transcriptional activity. Although DNA methylation in the promoter is exclusively correlated with gene repression (10), gene-body methylation was also found to be associated with enhanced gene expression levels (17). Methylated cytosines can influence transcription either via hindering transcription factors (TFs) to bind to DNA or by the recruitment of regulatory proteins such as histone-modifying and chromatin-remodeling enzymes (18). Chromatin structure and nucleosomal coverage are key regulators of gene expression and generally limit the accessibility of DNA sequences for DNA-binding factors (19). Therefore, chromatin-remodeling mechanisms are required to enable factor binding and transcriptionally active genes to adopt an open chromatin structure, whereas silent genes are highly compacted (20).
The diverse functions and opposing effects of CpG dinucleotides on gene expression make the usage of CpG dinucleotides in transgenes a critical parameter. We recently reported a drastic loss of reporter activity upon CpG removal by means of the reporter gene GFP, and could attribute this effect to decreased de novo transcriptional activity (21). Based on these observations, we sought to identify the role of intragenic CpG content, its distribution and the associated mechanism leading to increased gene expression by using cytokine genes as model systems. Cytokine expression is relevant for medical applications, such as cancer and immunotherapy (22), wound healing (23), allergy relief (24), treatment of autoimmune disorders (25), anti-viral therapy (26) and disease diagnosis (27).
By means of the model genes coding for the Macrophage Inflammatory Protein 1α (MIP-1α) and Granulocyte Macrophage-Colony Stimulating Factor (GM-CSF), we could show for the first time that the accumulation of CpG dinucleotides within the open reading frame (ORF) of transgenes leads to increased gene expression in mammalian cells. Our data provide convincing evidence that a high CpG dinucleotide content in the 5′ coding region of murine mip-1α increases the efficiency of transcription elongation by rearrangement of nucleosome positions, whereas intragenic CpG-depletion causes chromatin compaction, thereby impeding gene transcription. This hypothesis is supported by our genome-wide analysis demonstrating that efficiently expressed human genes contain a CpG-rich domain of ∼700 bp in length that does not classify as CpG island, directly downstream of the transcription start site (TSS). Our study revealed a novel role for these 5′ CpG domains in directly increasing transcription rates of endogenous genes.
MATERIALS AND METHODS
Plasmid construction
The cytokine genes human mip-1α (humip-1α), murine mip-1α (mmip-1α), human gm-csf (hugm-csf) and murine mgm-csf (mgm-csf) were modified in silico with respect to codon usage and CpG amount, synthesized via stepwise PCR from oligonucleotides (GeneArt/life technologies) and inserted into the eukaryotic expression vector pcDNA3.1 (+) (Invitrogen). Based on each wild type (wt) gene sequence, three gene variants with varying CpG content were generated. The resulting plasmids were named phuMIP-wt/-11/-0/-43, pmMIP-wt/-13/-0/-42, phuGM-CSF-wt/-12/-0/-63 and pmGM-CSF-wt/-21/-0/-61. The codon adaptation index (CAI) of the gene variants was calculated as described previously (21). For infection/transfection experiments, mmip-1α gene variants were cloned into the plasmid pPCR-Script Amp SK (+) (Stratagene) using HindIII and EcoRI, resulting in plasmids pT7-mMIP-wt, −0, −13 and −42. For the generation of stable cell lines, mmip-1α variants were inserted into pcDNA5/FRT (Invitrogen) via the restriction sites HindIII and BamHI, resulting in the plasmids pFRT-mMIP-wt, pFRT-mMIP-0, pFRT-mMIP-13 and pFRT-mMIP-42.
Cell culture, transient/stable transfections and infections
H1299 and human embryonic kidney (HEK) 293 derivate cell lines 293T/17 and 293 Flp-In cells (Invitrogen) were cultivated in D-MEM (Invitrogen) supplemented with 10% FCS, 1% penicillin/streptomycin and 2 mM L-glutamine. Flp-In chinese hamster ovary (CHO) cells (Invitrogen) were cultured in HAM-F12 (Invitrogen) supplemented with 10% heat inactivated FCS, 1% penicillin/streptomycin and 2 mM L-glutamine. Non-transfected Flp-In HEK 293 and Flp-In CHO cells were further supplemented with 100 µg/ml Zeocin. For transient and stable transfections, the calcium phosphate co-precipitation technique was applied (28). For the generation of stable cell lines, Flp-In 293 and CHO cells, respectively, were co-transfected with the Flp-recombinase encoding plasmid pOG44 (Invitrogen) and pFRT-mMIP-wt/-0/-13/-42 each at a ratio of 9:1. Positively transfected cells were selected by hygromycin B (Invitrogen) at a concentration of 100 µg/ml (293) or 500 µg/ml (CHO), respectively. For infection studies, cells were infected with an MVA strain expressing the T7-polymerase (MVA-T7), kindly provided by Prof. Gerd Sutter (Paul Ehrlich Institute, Langen, Germany) at an MOI of 50. One hour after infection, cells were washed with PBS and transfected with the respective plasmids pT7-mMIP-wt/-0/-13/-42 using Fugene 6 (Roche). Protein expression under control of the T7 promoter was quantified 12 h after infection by ELISA of cell culture supernatants.
ELISA
ELISA kits were purchased from R&D Systems for MIP-1α detection or BD Biosciences Pharmingen for GM-CSF detection, respectively, and experiments were conducted according to the manufacturer’s instructions using 1 µg total protein of culture supernatants.
Isolation of RNA
Nuclear and cytoplasmic RNA of 1 × 107 stably transfected CHO Flp-In cells was isolated using the RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol.
Northern blot analysis
An antisense RNA probe was generated by in vitro transcription using the Riboprobe in vitro transcription Kit (Promega) according to the manufacturer’s protocol. The sequences of primer used to generate the template for in vitro transcription are available on request.
Equal amounts of isolated RNA (10 µg) were used for northern blot analysis and blotted on a positive charged nylon membrane (Biodyne Plus) for 16 h. After hybridization with the denaturated DIG-probe for 16 h at 40°C, washing and blocking, the membrane was incubated with the Anti-DIG-antibody-AP solution for 30 min at RT. The membrane was covered by CDP-Star RTU-solution (1:100 in detection buffer, TROPIX, Bredford, USA) and detected by chemoluminescence (KODAK Biomax MR, Amersham).
PCR-techniques
First-strand cDNA was synthesized from DNA-free RNA using the DyNAmo™ cDNA Synthesis Kit (Finnzymes) with Oligo(dT)15 primers following the manufacturer’s instructions. Sequences of primers used to amplify the complete ORF to detect alternative splice products are available on request. Quantitative PCR (qPCR) was carried out by the StepOnePlus real-time PCR system using the DyNAmo™ Flash SYBR® Green qPCR Kit (Finnzymes) according to the manufacturer’s protocol. In relative quantification analyses of transcript levels, the amount of mmip-1α-specific transcripts was related to hph transcripts which served as an internal control. Product specificity was assessed based on melting curves. Data were analysed using the 2−ΔΔCT method or the Pfaffl method (29).
Nuclear run-on analysis
Approximately 3 × 107 stably transfected CHO Flp-In cells were used for nuclear run-on assays according to a previously described method (30). Briefly, nuclei of 3 × 107 stably transfected cells were prepared on ice, supplemented with biotin-16-UTP (Roche) for 30 min at 29°C, and labeled transcripts were bound to streptavidin-coated magnetic beads (Invitrogen). Total cDNA was synthesized by Oligo(dT)15 primed reverse transcription of captured molecules. mmip-1α- and hph-specific transcripts were quantified via qPCR using external standards as described above.
Analysis of mRNA half-live
Decay of respective mRNA transcripts was measured as described previously (31). 2.5 µM Actinomycin D (Roth) was added to cell culture supernatants of CHO Flp-In cells stably expressing mMIP-1α, followed by incubation for different time periods (0 h, 1.5 h, 3 h, 6 h, 12 h, 24 h) prior to cell harvest. Total RNA was isolated, reverse transcribed using Oligo(dT)15 and quantified via qPCR using hph as internal control gene as described above (Primer sequences available on request). The decay constant k is determined by plotting the amounts of RNA molecules of each gene variant exponentially as a function of time. This function reflects the decay constant k. After determination of the decay constant k, the respective half-live was calculated t1/2 = ln2/k.
Nucleosome reconstitution by salt dialysis
PCR fragments of mmip-wt, mmip-0 and mmip-42 were incorporated into mononucleosomes using the salt dialysis technique as previously described (32). Core histones of drosophila embryos were kindly provided Prof. Dr Längst (University of Regensburg). To distinguish between gene variants, PCR fragments were uniquely labeled using reverse primers with different fluorescent dyes at the 5′ end (DY 550 and DY647, respectively; sequences available on request). Assembly reactions (50 µl) contained 1.2 µg PCR product of each gene variant, varying amounts of histone octamers, 200 ng BSA/µl and 250 ng competitor DNA in high-salt buffer (10 mM Tris, pH 7.6, 2.0 M NaCl, 1.0 mM EDTA, 0.05% NP-40, 1.0 mM ß-mercaptoethanol). The NaCl concentration was reduced to 50 mM NaCl during 12–16 h. Analysis of mononucleosome positions was performed by polyacrylamide gelelectrophoresis (PAGE) on a 5.0% PAA in 0.4% TBE buffer followed by detection by the fluorescence imager FLA5000 (Fujifilm).
FAIRE
Formaldehyde-assisted isolation of regulatory elements (FAIRE) analysis was essentially done according to a published protocol (33). Approximately 3 × 107 exponentially growing Flp-In 293 cells stably expressing the mMIP-1α variants were cross-linked for 7 min at RT with 1% formaldehyde and subsequently quenched by glycine (125 mM). Cells were scraped off, washed twice with PBS and collected by centrifugation (700 × g; 5 min; 4°C). The cell pellet was resuspended in buffer IA (10 mM HEPES/KOH pH 7.9, 85 mM KCl, 1 mM EDTA, 1 × protease inhibitor cocktail (Roche)) and lysed on ice for 10 min in buffer IB (10 mM HEPES/KOH pH 7.9, 85 mM KCl, 1 mM EDTA, 10% Nonidet P-40, 1× protease inhibitor cocktail (Roche)). Cell lysate was centrifuged at 700 × g for 5 min and cell nuclei lysed in buffer II (50 mM Tris/HCl pH 7.4, 1% SDS, 0.5% Empigen BB, 10 mM EDTA pH 8.0, 1× protease inhibitor cocktail (Roche)). Samples were sonicated using a Bioruptor sonicator (Diagenode) to yield ∼200–500 bp DNA fragments. Cell debris was spun at 16 100 g for 5 min and the clarified supernatant was treated with RNAse A at a final concentration of 0.33 µg/µl for 1–2 h at 37°C. 25% of the sheared chromatin was isolated, treated with proteinase K (0.5 µg/µl) at 56°C for 1 h and reverse cross-linked o/n at 65°C. Released DNA was isolated by adding an equal volume of phenol–chloroform/isoamyl alcohol (25:24:1) in Phase Lock Gel Light Tubes. The remaining 75% of sheared chromatin was directly extracted by phenol–chloroform in the same way without prior proteinase K treatment and reverse cross-link. DNA from the aqueous phase of both chromatin fractions (with and without reverse cross-link) was subsequently precipitated by the addition of ammonium acetate (pH 7.5) to a final concentration of 2.5 M and an equal volume of isopropanol followed by o/n incubation at −20°C. The precipitate was collected the next day by centrifugation (30 min; 16 000 × g; 4°C), washed with 70% ethanol, air-dried and resuspended in 200 µl double-distilled water. Quantification of purified DNA was carried out by qPCR on the StepOnePlus™ instrument by Applied Biosystems using the DyNAmo Flash SYBR® Green qPCR Kit from Finnzymes according to the manufacturer's instructions. Primer were designed to cover the TSS and the 3′ UTR of mmip-1α as well as genes of the rdna as internal control. All results were normalized to rdna and referred to mmip-wt. They are presented as the ratio of DNA recovered from cross-linked cells divided by the amounts of the same DNA in the corresponding non-cross-linked samples. Data were analysed using the 2−ΔΔCT method.
ChIP assay
Approximately 3 × 107 recombinant Flp-In 293 cells stably expressing mMIP-1α variants were cross-linked by 1% formaldehyde (12 min) and quenched by 0.125 M glycine, washed in PBS, collected into buffer I (10 mM HEPES/KOH pH 7.9, 85 mM KCl, 1 mM EDTA, 1 × protease inhibitor cocktail (Roche) and lysed in buffer I adding 10% Nonidet P-40o/n; 10 min. Cell nuclei were pelleted (700 × g; 5 min) and lysed in buffer II (50 mM Tris/HCl pH 7.4, 1% SDS, 0.5% Empigen BB, 10 mM EDTA pH 8.0, 1× protease inhibitor cocktail (Roche)). Samples were sonicated using a Bioruptor sonicator (Diagenode) to 300–800 bp DNA fragments. Cell debris was spun (16 100 × g; 5 min) and cleared chromatin stored at −80°C. Approximately 2 × 106 cells were used for one Immunoprecipitation (IP) reaction. 20% of each sample was removed as Input fraction. Sheared chromatin was diluted 1:10 in dilution buffer (150 mM NaCl, 20 mM Tris–HCl pH 8.1, 1.2 mM EDTA, 1% Triton X-100, 0.01% SDS and EDTA-free complete protease inhibitor cocktail (Roche)) and precleared for 2 h with 60 μl protein A agarose/salmon sperm DNA slurry (Millipore) at 4°C. Precleared chromatin was incubated with the appropriate antibody (7.5 µg of α-RNA polymerase II C-terminal domain (CTD) repeat YSPTSPS (phospho S2) antibody (Abcam, ab5095), 7.5 µg α-Pol II (N-20) (Santa Cruz, sc-899 X) or 7.5 µg Polyclonal rabbit Anti-FLAG antibody (Sigma Aldrich, F7425) at 4°C o/n with gentle rotation. Antibody-chromatin complexes were precipitated with 60 μl protein A agarose/salmon sperm DNA slurry for 4 h at 4°C with gentle rotation, followed by centrifugation (5 min; 500 × g; 4°C). Antibody-chromatin complexes were washed with 1 ml low-salt buffer (20 mM Tris [pH 8.1], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), high-salt buffer (20 mM Tris [pH 8.1], 0.5 M NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), lithium chloride (LiCl) buffer (10 mM Tris [pH 8.1], 0.25 M LiCl, 1 mM EDTA, 1% Igepal-CA630, 1% deoxycholic acid) and twice with TE buffer. After final aspiration,washed samples were supplemented with 10% Chelex (Biorad) and boiled (15 min). Proteinase K (100 µg/ml) was added to the Chelex/protein A bead suspension and incubated for 1.5 h at 56°C while shaking, followed by another 15 min of boiling. The suspension was then applied onto Micro Bio-Spin Columns (Bio-Rad) and centrifuged at 500 × g for 5 min for purification of nucleic acids. The eluate was used directly as a template in qPCR. Primer sequences are available on request. Data were analysed using the 2−ΔΔCT method and reported as Output to Input fraction.
Generation of mmip-1α chimera
mmip-1α chimera were generated by fusion PCR using pmMIP-0 and pmMIP-42 as templates. Primer sequences are available on request. Chimera were cloned into pcDNA5 containing the cytomegalovirus (CMV) promoter via HindIII and BamHI.
Genome-wide analysis of CpG frequency
For genome-wide correlations, whole cell polyA+ CAGE data were downloaded from the ENCODE/RIKEN consortium for H1-hESC (GSM849357), HepG2 (GSM849335) and HeLa-S3 (GSM849342). Only transcripts scoring an irreproducible discovery rate (IDR) of <0.05 were considered significant. The datasets were filtered for annotated TSSs and sorted according to their RPKM values. By this, we identified the 5% highest and 5% lowest expressed genes in the three cell lines. In absolute numbers, each generated dataset consist of 1000–1500 genes. For every gene, we extracted three 500 bp regions: TSS +500 bp, +/–250 bp around the center of each gene and −500 bp to the transcription termination site (TTS). The CG dinucleotide frequency (CpG frequency) and the overall %GC (GC content) of every position was computed using the annotatePeaks script as part of the HOMER software package (34). Tracks for CpG islands, conserved TF binding sites (tfbsConsSite) and actual TF binding as determined by ChIP-Seq (tfbsClusteredV2) were downloaded from the ENCODE consortium. CpG islands classified by the CpG_MI algorithm (35) were obtained from http://bioinfo.hrbmu.edu.cn/cpgmi/. All datasets used correspond to the human genome built hg19. Line plots were generated with annotatePeaks. Dot plots and linear regressions were carried out with the statistics program R (36). The overlap of high and low expressed gene sets in the three different cell lines was determined and plotted with BioVenn (37).
RESULTS
Generation of cytokine genes with differing intragenic CpG amount
The human and murine genes MIP-1α (humip-1α and mmip-1α) and granulocyte macrophage-colony stimulating factor (hugm-csf and mgm-csf) were analysed with regard to variable intragenic CpG content and its consequence on transgene expression in mammalian cells. Gene variants with differing CpG content were generated based on the wt sequences. To avoid biased transgene expression of CpG-modified genes as a result of an altered codon usage, the nucleotide sequences of all reporter genes used were first adapted with regard to an optimized codon usage (opt) for mammalian cells (38). On the basis of these optimized (opt) gene variants, the nucleotide sequences were further modified to quantitatively deplete CpGs (–CpG) or maximize (+CpG) intragenic CpG content within the ORF (Table 1). For mmip-1α, four synthetic gene variants were generated, containing either 7 CpGs (wt), 13 CpGs (codon optimized), 0 CpGs (codon optimized, –CpG) and 42 CpGs (codon optimized, +CpG), respectively (Supplementary Figure S1). An increase of CpG content resulted in a decreased CAI, which represents the relative adaptiveness of the respective gene’s codon usage toward the codon usage of highly expressed genes (Table 1, Supplementary Figure S1B) (3).
Table 1.
Sequence characteristics of the different cytokine gene variants
|
mmip-1α |
humip-1α |
|||||||
|---|---|---|---|---|---|---|---|---|
| wt | opt | –CpG | +CpG | wt | opt | –CpG | +CpG | |
| CpG | 7 | 13 | 0 | 42 | 8 | 11 | 0 | 43 |
| CAI | 0.77 | 0.96 | 0.92 | 0.73 | 0.77 | 0.93 | 0.90 | 0.72 |
| GC (%) | 51 | 58 | 53 | 63 | 57 | 56 | 57 | 63 |
| TpA | 7 | 7 | 8 | 5 | 6 | 10 | 7 | 4 |
|
mgm-csf |
hugm-csf |
|||||||
|---|---|---|---|---|---|---|---|---|
| wt | opt | –CpG | +CpG | wt | opt | –CpG | +CpG | |
| CpG | 11 | 21 | 0 | 61 | 10 | 12 | 0 | 63 |
| CAI | 0.75 | 0.99 | 0.94 | 0.75 | 0.82 | 0.96 | 0.92 | 0.70 |
| GC (%) | 50 | 61 | 53 | 61 | 57 | 59 | 53 | 63 |
| TpA | 17 | 6 | 11 | 6 | 7 | 10 | 9 | 5 |
For each gene variant, the amount of CpG dinucleotides, the CAI, the GC content (%) and the number of TpA dinucleotides are specified.
Unmethylated CpG dinucleotides significantly increase gene expression in transiently and stably transfected mammalian cells
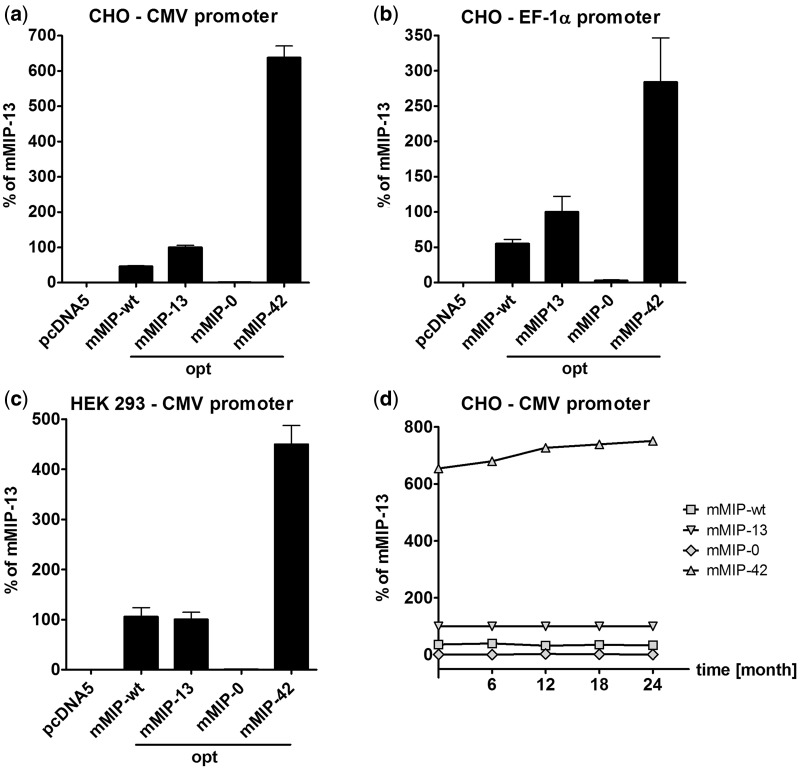
To evaluate the expression efficiency of the CpG variants, cytokine constructs were first transiently transfected into mammalian H1299 cells. Although CpG-depletion resulted in a decrease of expression efficiency in mMIP-1α and huMIP-1α, maximal CpG frequency led to clearly increased cytokine levels in all cases, compared to the respective optimized reference genes (Supplementary Figure S2). To assess the impact of differential intragenic CpG content on long-term expression and regulation mechanisms, mmip-1α gene variants were stably transfected into HEK 293 and CHO cell lines, which are widely employed for recombinant protein production (39). To exclude promoter specificity of the observed gene expression variations, the CMV promoter and the promoter controlling the expression of the human Elongation Factor-1α (EF-1α) were tested in parallel. The Flp-In recombination system was used to establish cells incorporating single copies of mmip-1α transgenes within an identical genetic background (40). Analogous to transient transfections, cytokine expression of CpG-modified mMIP-1α variants clearly correlated with intragenic CpG content. Although CpG-depletion resulted in a drastic expression decline, mMIP-42 exhibited a 6.4-fold (CHO, CMV promoter), 3-fold (CHO, EF-1α promoter) and 4.5-fold (HEK 293, CMV promoter) increase of transgene expression compared to the optimized reference mMIP-13 (Figure 1a–c). Cytokine expression levels in stable CHO Flp-In cells persisted over a period of at least 2 years under selection pressure, indicating that intragenic CpG sites and the promoter are not susceptible to DNA methylation leading to gene silencing (Figure 1d). Indeed, bisulphite conversion and genomic sequencing of the mip-1α expression cassettes in CHO Fpl-In cells after 2 years of cell passaging revealed a completely unmethylated state of mmip-1α and the CMV promoter (data not shown).
Figure 1.
Influence of intragenic CpG content on mMIP-1α expression by different promoters and in different mammalian cell lines. Quantification of mMIP-1α levels by ELISA in the cell culture supernatants of (a) CHO Flp-In cells—mMIP-1α expression controlled by the CMV promoter; (b) CHO Flp-In cells—mMIP-1α expression controlled by the EF-1α promoter; and (c) HEK 293 Flp-In cells—mMIP-1α expression controlled by the CMV promoter. Protein levels were expressed as percentage of the codon optimized mMIP-13. Standard deviations are indicated by error bars and result from duplicates each (a, b, c). pcDNA5 without insert served as negative control. (d) Expression of mMIP-1α variants controlled by the CMV promoter in stably transfected CHO Flp-In cells was monitored over the period of 2 years.
Intragenic CpG frequency has no influence on translational efficiency or post-transcriptional processing of the mRNA but increases de novo transcriptional activity
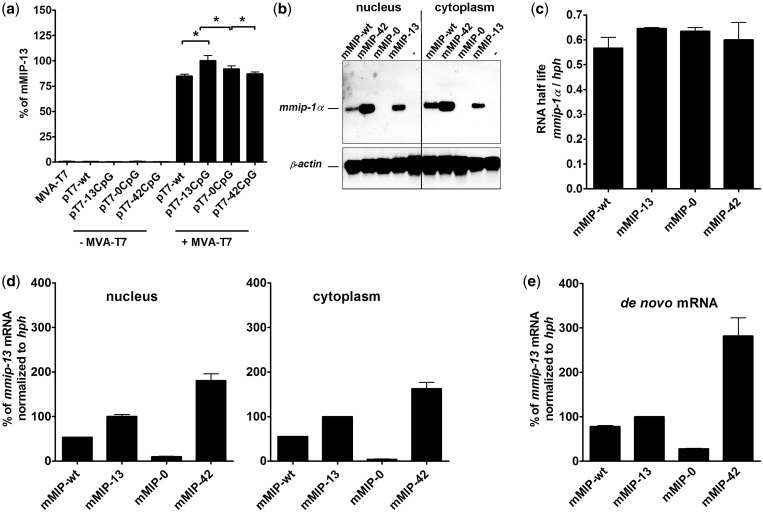
In order to identify potential effects of modified reporter gene sequences on translational events, we infected 293T cells with a modified vaccinia ankara strain known to replicate exclusively in the cytoplasm in order to limit T7 polymerase expression to the cytoplasmic compartment (MVA T7). Upon infection, this system allows cytoplasmic transcription of a transfected reporter gene controlled by the T7 promoter and thereby uncouples mmip-1α transcription from all nuclear processes. Thus, cytokine levels can directly be ascribed to translational processes. The expressed cytokine levels in HEK 293T cells infected with MVA-T7 and transfected with pT7-mMIP-wt/-0/-13 and −42, respectively, directly correlated with the CAI of the respective gene variant (Figure 2a and Table 1). Thus, a major impact of CpG content on translational efficiency can be widely excluded. To elucidate whether post-transcriptional processes were responsible for CpG-based differential gene expression, we further investigated mRNA levels and integrity of mmip-1α variants in stably transfected CHO Flp-In cells controlled by the CMV promoter (Figure 1a). Northern blot analysis revealed one distinct mmip-1α signal corresponding to the expected size of the full-length cytokine mRNA arguing against alternative splicing events (Figure 2b). Due to the need of tightly regulated cytokine expression, cytokine mRNAs tend to have a rather short half-live (41). To assess whether CpG modifications might have changed the degrading characteristics of RNA, transcript stability of all mmip-1α variants was determined by blocking de novo RNA synthesis of stably transfected cells using Actinomycin D followed by mRNA quantification via reverse transcription quantitative PCR (RT-qPCR). Using this assay, the relative RNA level decline of each gene variant over 24 h was evaluated (Figure 2c). Remarkably, the changes in the nucleotide sequence had no major effect on mRNA stability, which is in agreement with the fact that the absolute number of TpA dinucleotides known to contribute to RNA degradation is low for all constructs tested (Table 1). RNA steady state levels in the nucleus and the cytoplasm of the stably integrated mmip-1α variants were furthermore quantified separately via RT-qPCR (Figure 2d). mmip-0 transcripts were reduced by 25-fold in the cytoplasm and by 10-fold in the nucleus, compared to mmip-13. In contrast, a 1.6-fold (cytoplasm) and 1.8-fold (nucleus) increase, respectively, was observed for mmip-42 compared to mmip-13. This indicates that defects in RNA export can be excluded as cause for the differences observed in steady state mRNA levels. Our results imply a CpG effect on the level of gene transcription, which could be verified by altered de novo RNA synthesis rates of mmip-1α variants as quantified by a nuclear run-on assay (Figure 2e). De novo synthesized mmip-0 RNA transcripts were reduced 3.5–fold, whereas mmip-42 levels were increased 2.7-fold, compared to mmip-13. The observed discrepancies between absolute mRNA and protein levels might be due to a non-linear correlation between transcribed and translated mRNA (Table 2). To study the functional impact of CpG content on transcription, further analyses were focused on mmip-0 and mmip-42, which exhibit strong differences in expression levels. The wt gene mmip-wt was used as internal control, as it represents the natural situation.
Figure 2.
Influence of intragenic CpG content on post-transcriptional mechanisms and translational efficiency (a) mMIP-1α quantification by ELISA of the supernatant of 293T cells infected with MVA-T7 and transfected with pT7-mMIP-wt, −13, −0 and −42. Standard deviations indicate the mean of three independent experiments. Statistics were calculated by unpaired two-tailed t-test, P < 0.05. (b) Northern blot analysis of mmip-1α variants stably expressed in CHO Flp-In cells. ß-actin was used as loading control. One representative experiment is shown. (c) Influence of intragenic CpG content on RNA half-live. Stably transfected CHO Flp-In cell lines were incubated with Actinomycin D and total RNA was isolated and quantified by RT-qPCR. The mean and standard deviation of three measurements is shown. (d) Influence of CpG content on steady state RNA levels. Cytoplasmic and nuclear RNA fractions prepared from stably transfected CHO Flp-In cells were subjected to RT-qPCR. The amount of mmip-1α-specific transcripts was normalized to hph. The mean and standard deviation of triplicates is shown. (e) Influence of CpG content on de novo synthesis of mmip-1α transcripts as quantified by a nuclear run-on assay. The mean and standard deviation of two independent experiments is shown.
Table 2.
Summary of protein and RNA levels of mmip-1α variants expressed by the CMV promoter in stably transfected CHO Flp-In cells
| mmip-wt (%) | mmip-13 (%) | mmip-0 (%) | mmip-42 (%) | |
|---|---|---|---|---|
| Protein in cell supernatant (ELISA) | 46 | 100 | 1 | 638 |
| mRNA in the nucleus (northern blot) | 111 | 100 | 33 | 289 |
| mRNA in the cytoplasm (northern blot) | 100 | 100 | 20 | 400 |
| mRNA in the nucleus (rtPCR) | 53 | 100 | 10 | 179 |
| mRNA in the cytoplasm (rtPCR) | 56 | 100 | 6 | 167 |
| de novo RNA (nuclear run-on) | 77 | 100 | 23 | 270 |
Intragenic CpG levels affect chromatin structure and accessibility
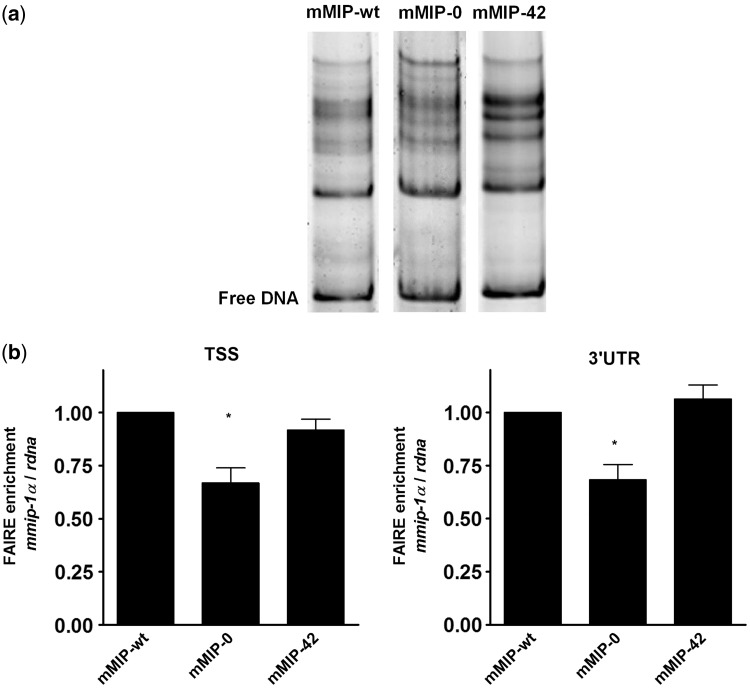
Sequence patterns can directly affect nucleosome positioning by determining biophysical properties of DNA like the bending flexibility around a histone octamere (6,42,43). Based on these facts, we tested whether changes in CpG content alter the chromatin structure in vitro. Competitive nucleosome reconstitutions were performed with CpG-modified mmip-1α variants by salt dialysis (32) and nucleosome positions were resolved by native PAGE. The position of a histone octamere within the DNA fragment affects its electrophoretic mobility: centrally located nucleosomes migrate slower than nucleosomes located at the end of a DNA fragment (44).
Although the CpG frequency within the ORF of mmip-1α did not influence the affinity for histone octamers in vitro (Supplementary Figure S3), a comparison of mononucleosomal electrophoretic migration patterns of mmip-wt, mmip-0 and mmip-42 revealed differences in the positioning of nucleosomes (Figure 3a). Since sequence differences can also direct nucleosome positioning in vivo (6), the chromatin structure of stably integrated CpG variants was also examined in vivo using the FAIRE assay. In this assay, chromatin is cross-linked with formaldehyde, sheared by sonication and phenol–chloroform extracted (33). This procedure results in preferential enrichment of accessible and rather nucleosome-depleted genomic regions that can be quantified by qPCR. Both at the transcription start site (TSS) and at the 3′ untranslated region (3′ UTR), mmip-wt and mmip-42 exhibited very similar levels of nucleosomal density, whereas mmip-0 showed a significantly lower degree of nucleosome depletion (ANOVA; P < 0.05; Figure 3b). Thus, intragenic CpG-depletion in mmip-0 directly correlates with a higher degree of chromatinization, which corresponds to the very low transcriptional activity. The variants mmip-wt and mmip-42 exhibited similar chromatin accessibility suggesting that once a gene is active, the overall accessibility of the higher order structures of chromatin is similar, irrespective of the transcription rate.
Figure 3.
Influence of intragenic CpG content on nucleosome positioning and chromatin accessibility. (a) Nucleosome positioning of mmip-wt, mmip-0 and mmip-42 in vitro. Fluorescently labeled PCR fragments of mmip-wt (DY550), mmip-0 (DY550) and mmip-42 (DY647) were reconstituted to mononucleosomes by salt dialysis, followed by PAGE and detection by fluorescence imaging. One representative reconstitution is shown. (b) Chromatin accessibility of mmip-1α variants stably expressed in HEK 293 Flp-In cells as analysed by FAIRE. Enrichment for nucleosome-depleted chromatin by FAIRE extraction was quantified by qPCR at the TSS and 3′ UTR of mmip-1α variants relative to rdna and normalized to mmip-wt. Data are presented as the ratio of DNA recovered from cross-linked cells divided by the amounts of the same DNA in the corresponding non-cross-linked samples. The mean and standard deviation of three independent FAIRE preparations each is shown. Statistics were calculated by one-way ANOVA Tukey’s multiple comparison test, P < 0.05).
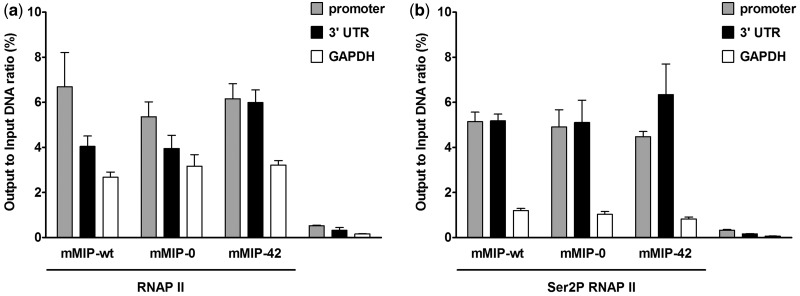
Intragenic CpG dinucleotides increase elongation rate
The CTD of the largest subunit of the eukaryotic RNA polymerase II (RNAP II) contains several YSPTSPS heptad repeats that are unphosphorylated in the preinitiation complex of RNAP II and become phosphorylated at multiple sites upon initiation (45). Phosphorylation of Serine 2 (Ser2P) is found in the elongating RNAP II and the 3′ end processing of the transcript, therefore serving as a marker for the fraction of elongating polymerases (46). To test whether the occupancy of elongating polymerases of mmip-1α variants is dependent on intragenic CpG content, HEK 293 Flp-In cell lines expressing the respective mmip-1α CpG variants were used for Chromatin Immunoprecipitation (ChIP) experiments with antibodies targeting the N-terminus of RNAP II (total RNAP II) and the specifically phosphorylated CTD of the elongating RNAP II (Ser2P RNAPII). Total RNAP II at the promoter was equally abundant between mmip-wt, mmip-0 and mmip-42 (Figure 4a). In contrast, the 3′ UTR of mmip-wt and mmip-0 revealed reduced occupancy of RNAP II compared to the 3′ UTR of mmip-42. Correlating with total RNAP II, similar amounts of elongating Ser2P RNAP II were detected at the promoter between mmip-wt, mmip-0 and mmip-42. However, the 3′ UTR exhibited increased Ser2P RNAP II occupancy in mmip-42 compared to mmip-0 and mmip-wt (Figure 4b). Since the mmip-1α gene is too short to be occupied by more than one polymerase at a time, we suggest that the increased amount of de novo transcripts shown by the nuclear run-on assay is due to an increased elongation rate with increased CpG content.
Figure 4.
RNAP II (a) and Ser2P RNAP II (b) binding at the promoter and 3′ UTR of mmip-1α variants analysed by ChIP. ChIP was performed by cross-linking of HEK 293 Flp-In cells stably expressing mmip-1α variants, sonication, incubation with the appropriate antibody and collection of bound DNA by sepharose A beads. Precipitated DNA was quantified at the promoter and 3′ UTR of mmip-wt, mmip-0 and mmip-42. The first exon–intron-junction of gapdh was used as internal control. Results were expressed as input to output ratio. Polyclonal rabbit Anti-FLAG antibody served as a negative control. The mean and standard deviation of three independent experiments each is shown.
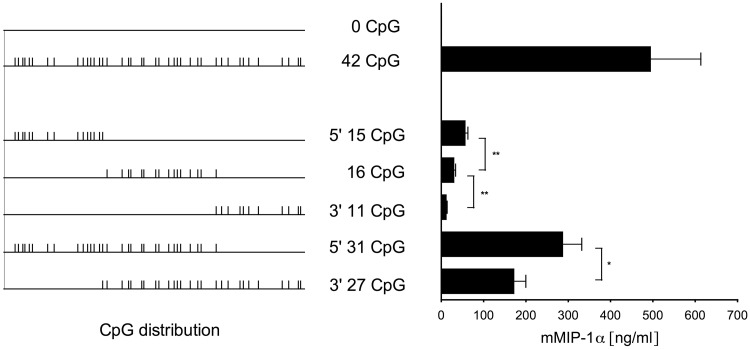
CpG dinucleotides situated in the 5′ region increase transgene expression to a greater extent compared to CpG dinucleotides in the 3′ region
To shed light on the positional and functional relevance of CpGs within the ORF, we generated chimera based on mmip-0 and mmip-42 to obtain genes with CpG clusters in either 5′, central or 3′ regions of the ORF, followed by stable CHO Flp-In integration. The protein levels expressed by stable CHO Flp-In transfectants revealed that not only the mere amount but also the proximity of CpG dinucleotides to the 5′ end of the gene is required for the increase in expression rates (Figure 5). This positional relevance of intragenic CpG dinucleotides could be confirmed with the previously applied GFP variants gfp-0 and gfp-60 in CHO Flp-In cells (Supplementary Figure S5).
Figure 5.
Expression levels of mmip-1α chimera. Expression of mmip-1α chimera in stably transfected CHO Flp-In cells was analysed by ELISA. The mean and standard deviation of triplicates is shown. Statistics were calculated by unpaired two-tailed t-test, *P < 0.05; **P < 0.005.
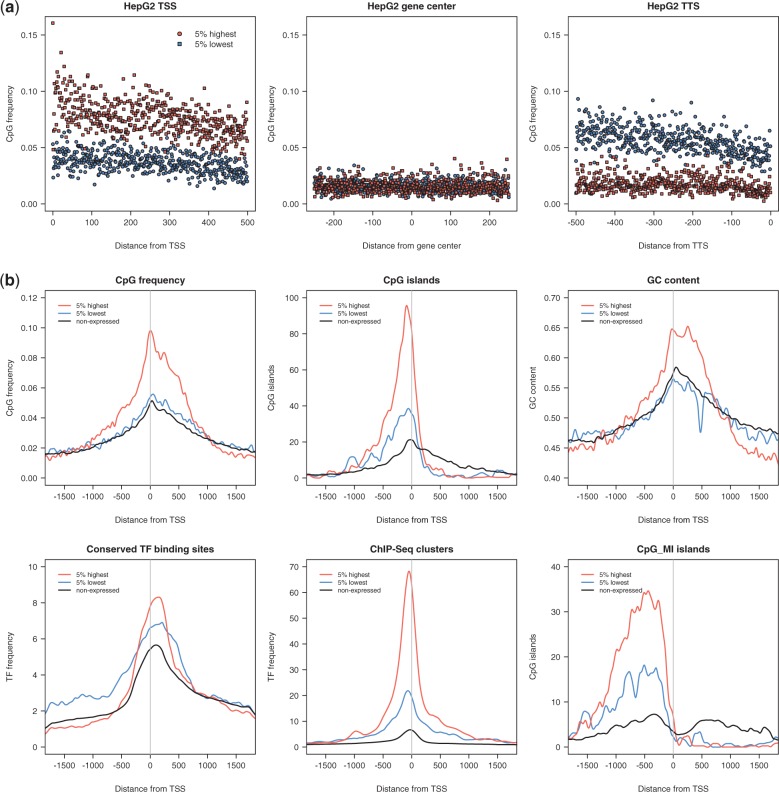
Highly expressed genes display a 2-fold increased CpG frequency at the TSS compared to lowly expressed genes
Based on the observation that intragenic CpG dinucleotides promote transgene transcription in correlation to their TSS-proximity, we analysed whether such an association can be found with actively transcribed genes in vivo. Three human cell lines of different origin, including the human embryonic stem cell line H1-hESC, the human liver carcinoma cell line HepG2 and the widely used HeLa cervical cancer cell line were selected for genome-wide correlations between expression levels and CpG frequency. Using the transcriptome quantifications from the ENCODE/RIKEN consortium, the 5% highest and 5% lowest expressed genes in the three cell lines were selected (47). In absolute numbers, each dataset consists of 1000–1500 genes (1029 genes for each high and low datasets were assayed for H1-hESC, 1451 for HepG2 and 977 for HeLa). For every gene, we extracted three 500 bp regions: TSS +500 bp, +/–250 bp around the center of each gene and −500 bp to the TTS (Figure 6a and Supplementary Figure S6a). Corresponding to our experimental data, we observed a 2-fold higher CpG frequency at the TSS of highly transcribed genes compared to the low expressing genes for all three cell lines (Figure 6a and Supplementary Figure S6a). The high levels of CpG dinucleotides of the strongly expressed genes decrease toward the gene center to reach the level of CpGs found in low expressing genes, perfectly mirroring the experimental data showing that only a high CpG content at the 5′ end is required for high level expression. To our surprise, the CpG levels of the low expressed genes increase toward the TTS, whereas the CpG level of the active genes remains low (Figure 6a and Supplementary Figure S6a). This observation may hint to the requirement of low CpG levels for efficient transcription termination. Even though the overlap of highly transcribed genes in the three cell lines is only 27.6% these genes display the same CpG characteristics, as well as the low expressing genes exhibiting a minor overlap (0.56%) but showing a comparable CpG pattern at the termination region.
Figure 6.
Genome-wide correlation of CpG frequency and expression levels. (a) CpG frequency of the 5% highest and 5% lowest expressed genes in HepG2 cells. Frequencies are exemplarily displayed within the first 500 bp, starting from the TSS, +/– 250 bp around the gene center and the last 500 bp of all genes, ending with the TTS. Every symbol indicates the CpG frequency at the corresponding position. At the TSS, the CpG frequency of high expressed genes is up to 2-fold higher compared to low expressed genes. The occurrence of CpG dinucleotides decreases toward the gene center and stays at a low level for highly expressed genes, whereas low expression correlates to increased CpG values around the TTS. (b) Frequency of CpG dinucleotides, CpG islands, GC content, conserved binding sites for TFs and actual binding of TFs as determined by ChIP-Seq (ChIP-Seq clusters) of the 5% highest, the 5% lowest and all non-expressed genes in HeLa cells. Displayed are regions of +/– 1700 bp around the TSS and frequencies are calculated in 10 bp intervals. Tracks for sequence elements were downloaded from the ENCODE consortium. CpG islands were additionally classified by the CpG_MI algorithm (35).
A closer inspection of the genomic data shows that within the 5% highest expressed genes CpG dinucleotides are strongly enriched at the promoter region up to 700 bp downstream of the TSS (Figure 6b, CpG frequency). Along with increased CpG frequency, the overall GC content of these genes is significantly elevated in this region (Figure 6b, GC content). Interestingly, elevated levels of CpG dinucleotides do not necessarily demarcate the presence of CpG islands as defined by the ENCODE consortium (G + C content ≥ 50%, CpG Observed/Expected ≥ 0.6, length ≥ 200 bp (48) which peak preceding the newly identified CpG-rich domain, covering gene promoters (and promoter-proximal regulatory elements) (Figure 6b, CpG islands). As an alternative approach we applied the CpG MI algorithm, which calculates the average mutual information of the physical distances between two neighboring CpGs and provides the highest prediction accuracy for functional CpG islands to date (35). Similar to the encode set, the CpG_MI program fails to classify the highly frequent CpG dinucleotides downstream of the TSSs as CpG islands (Figure 6b, CpG_MI islands). Strongly expressed genes are significantly enriched in CpG islands, however our experimental data show that expression levels correlate rather to intragenic CpG frequency. Therefore, the CpG-rich area from the TSS to about position +700 bp possibly represents a functional domain associated with high levels of transcription. In addition, the region downstream of the promoter shows only a slight enrichment for conserved TF binding sites in highly expressed genes (Figure 6b, conserved TF binding sites), although known TFs are concentrated in promoter regions (Figure 6b, ChIP-Seq clusters). In summary, this study shows that the downstream region of strongly expressed genes encompasses a 700 bp long CpG-rich domain region downstream of the gene promoter that is not classified as a CpG island and is not enriched for known TFs. Hence, we suggest a new mechanism of the CpG content influencing transcription levels.
DISCUSSION
Previous studies in our laboratory have shown that the depletion of CpGs from different transgenes—humanized GFP, the capsid protein of HIV and murine erythropoietin (mEPO)—resulted in a drastic loss of expression yields in mammalian cell lines and mice (21,49). Here, we demonstrate for the first time that vice versa the accumulation of intragenic CpG dinucleotides at the 5′ region of genes results in increased gene expression in mammalian cells. The increase of CpG levels correlates with altered chromatin structure and increased levels of elongating RNA Pol II. Confirming our experimental data, we show in genome-wide studies that this is a general feature of highly expressed genes. High levels of gene expression correlate with the presence of a domain of ∼700 bp with high CpG content downstream of the TSS that does not overlap with CpG islands and TF binding. We suggest that these 5′ CpG domains are required to distort the chromatin structure and to increase the level of elongating RNA Pol II.
The cytokine genes murine and human mip-1α (mmip-1α and humip-1α), as well as murine and human gm-csf (mgm-csf and hugm-csf) were subjected to state-of-the-art computer-assisted optimization strategies with focus on CpG-depletion or CpG-enrichment within the nucleotide sequence of the ORF. Intragenic CpG-depletion resulted in decreased protein expression in transiently transfected H1299 (mMIP-1α and huMIP-1α) as well as in stably transfected Flp-In HEK 293 and CHO cells. Inversely, we could also show that intragenic CpG-enrichment led to a clear increase in protein expression levels in all cell types tested and under control of different promoters (CMV, EF-1α promoter). The CpG-rich gene variant mmip-42 showed the highest expression levels despite low CAI, whereas the CpG-depleted gene variant mmip-0 gave the lowest protein yield though having a CAI notably higher than the wt and the CpG–rich gene. The expression levels do clearly correlate with the level of intragenic CpG content, suggesting that the CAI as well as the relative codon adaptiveness underlying the CAI is not sufficient as a gene design parameter.
Intrinsic DNA structure mediated by the DNA sequence affects nucleosome positioning in vitro and in vivo (5,42,50,51). Genome-wide analysis of nucleosome positions suggests that ∼50% of the nucleosomes are positioned with respect to the underlying DNA sequence (6). The nucleosomal configuration of the endogenous human mip-1α reveals a strong positioning of the +1 nucleosome at the 5′ end of the coding sequence that is shifted 40 nt upstream when activated upon inflammation (Längst,G., unpublished data). Nucleosome positioning and chromatin structure are major determinants of gene activity and we suggest that the positioning of nucleosomes, in particular the +1 nucleosome, plays an important role in transcriptional control of mip-1α. In vivo chromatin structure analyses at the TSS and the 3′ UTR of mmip-1α variants stably expressed by Flp-In HEK 293 cells revealed similar chromatin accessibilities between mmip-wt and mmip-42. However, mmip-0 exhibiting reduced CpG levels showed a significantly higher degree of chromatinization, both at the TSS and the 3′ UTR. In accordance with this finding, we identified different nucleosome positioning patterns among the CpG variants in vitro, suggesting that increasing the CpG content alters the chromatin structure at the transgene.
Ramirez-Carrozzi et al. found a high CpG content in the promoters of some primary response genes to be responsible for the assembly of very unstable nucleosomes. Accordingly, transcription of this group of genes can be induced without the requirement of chromatin-remodeling complexes (52). A similar destabilizing mechanism could apply for the CpG-rich transcribed region of highly transcribed genes. In our model system the CpG-enrichment in mmip-42 did inversely not result in enhanced chromatin de-compaction compared to mmip-wt. This is most likely due to the features of the Flp-In system which mediates transgene integration into an already very open chromatin structure (40). Our study suggests that altered positioning of the 5′ nucleosomes of the 5′ coding region does not correlate with an overall increased RNAP II density, however we detected increased levels of transcribing RNAP II at the 3′ UTR of mmip-42 compared to mmip-0 and mmip-wt. Genome-wide studies of transcription regulation in human cells have demonstrated that ∼20% of unexpressed genes are constantly occupied by RNAP II prior to transcription initiation (53). Potentially, the CpG richness allows efficient promoter escape of the Polymerase and more RNAP II switches to the elongating form, resulting in high levels of transcription. A genome-wide study by Choi et al. identified a group of genes in human cells with exceptionally high frequency of mainly unmethylated CpGs at the 5′ end of the gene (54). These genes exhibited higher expression rates than genes controlled by a promoter CpG island. The authors suggested that enhanced expression levels could result from facilitated RNAP II elongation. This hypothesis is in agreement with our findings, suggesting that the deposition of nucleosomes downstream of the TSS is influenced by CpGs and their modification, playing an important role in determining transcription levels. Expression analyses of mmip-1α chimera showed that not only the total amount of CpG dinucleotides, but rather the localization of the CpG dinucleotides close to the start codon is required for high levels of gene expression. Given the direct correlation between levels of protein expression and transcript amounts, we suggest that CpGs in the 5′ region increased gene transcription to a greater extent than in the 3′ region. Interestingly, genome-wide studies revealed that human protein coding genes display a significant excess of CpG dinucleotides in the 5′ ends of exons (16,54–56). Our detailed correlation analysis of transcription strength and CpG density of human genes in three different cell lines showed a strong overlap between highly expressed genes and CpG density at the region between the TSS and 700 bp downstream. This region does not classify as CpG island and is not preferentially bound by specific DNA-binding factors, as revealed by annotation of ChIP-Seq traces. In addition, we observed a similar strong correlation at the ends of the coding region for weakly expressed genes, revealing an increased CpG frequency within a region of 700 bp directly upstream to the TTS. Our experimental results give an explanation for the 5′ enrichment of CpG frequency with highly transcribed genes, but the role of high CpG content with weakly transcribed genes has to be addressed in future studies.
ACCESSION NUMBERS
The wt sequences of cytokine genes used in this study can be found in the NCBI gene bank under following accession numbers: mmip–1α (NM_011337), hugm–csf (M11220), humip–1α (NM_002983) and mgm–csf (X03020).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bayerische Forschungsstiftung (ForProtect) [AZ-862-09 to R.W.]; Bill and Melinda Gates Foundation [38599 to R.W.]; Bayerisches Genomforschungsnetzwerk (BayGene to G.L.). Funding for open access charge: Bayerische Forschungsstiftung (ForProtect).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gerd Sutter (Paul Ehrlich Institute, Langen, Germany) for kindly providing the MVA-T7 virus strain. We also thank Marcus Graf (GeneArt AG) for calculation of codon frequencies.
REFERENCES
- 1.Welch M, Villalobos A, Gustafsson C, Minshull J. Designing genes for successful protein expression. Methods Enzymol. 2011;498:43–66. doi: 10.1016/B978-0-12-385120-8.00003-6. [DOI] [PubMed] [Google Scholar]
- 2.Maertens B, Spriestersbach A, von Groll U, Roth U, Kubicek J, Gerrits M, Graf M, Liss M, Daubert D, Wagner R, et al. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci. Publ. Protein Soc. 2010;19:1312–1326. doi: 10.1002/pro.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp PM, Li WH. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fath S, Bauer AP, Liss M, Spriestersbach A, Maertens B, Hahn P, Ludwig C, Schäfer F, Graf M, Wagner R. Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression. PLoS ONE. 2011;6:e17596. doi: 10.1371/journal.pone.0017596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 6.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutler E, Gelbart T, Han JH, Koziol JA, Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc. Natl Acad. Sci. USA. 1989;86:192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 11.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 12.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 13.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 15.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ball MP, Li JB, Gao Y, Lee J-H, LeProust EM, Park I-H, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 19.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat. Rev. Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson RT. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog. Nucleic Acid Res. Mol. Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 21.Bauer AP, Leikam D, Krinner S, Notka F, Ludwig C, Langst G, Wagner R. The impact of intragenic CpG content on gene expression. Nucleic Acids Res. 2010;38:3891–3908. doi: 10.1093/nar/gkq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sportès C, Gress RE. Interleukin-7 immunotherapy. Adv. Exp. Med. Biol. 2007;601:321–333. doi: 10.1007/978-0-387-72005-0_35. [DOI] [PubMed] [Google Scholar]
- 23.Braund R, Hook S, Medlicott NJ. The role of topical growth factors in chronic wounds. Curr. Drug Deliv. 2007;4:195–204. doi: 10.2174/156720107781023857. [DOI] [PubMed] [Google Scholar]
- 24.Taylor-Robinson A. Schistosomiasis-induced IL-10 suppresses allergy prevalence. Trends Parasitol. 2001;17:62. doi: 10.1016/s1471-4922(00)01865-1. [DOI] [PubMed] [Google Scholar]
- 25.Karin N. Induction of protective therapy for autoimmune diseases by targeted DNA vaccines encoding pro-inflammatory cytokines and chemokines. Curr. Opin. Mol. Ther. 2004;6:27–33. [PubMed] [Google Scholar]
- 26.White L, Krishnan S, Strbo N, Liu H, Kolber MA, Lichtenheld MG, Pahwa RN, Pahwa S. Differential effects of IL-21 and IL-15 on perforin expression, lysosomal degranulation, and proliferation in CD8 T cells of patients with human immunodeficiency virus-1 (HIV) Blood. 2007;109:3873–3880. doi: 10.1182/blood-2006-09-045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards CJ, Feldman JL, Beech J, Shields KM, Stover JA, Trepicchio WL, Larsen G, Foxwell BM, Brennan FM, Feldmann M, et al. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Mol. Med. Camb. Mass. 2007;13:40–58. doi: 10.2119/2006-000056.Edwards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenborn ET, Goiffon V. Calcium phosphate transfection of mammalian cultured cells. Methods Mol. Biol. 2000;130:135–145. doi: 10.1385/1-59259-686-x:135. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Ravazzolo R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques. 2000;29:1012–1017. doi: 10.2144/00295st02. [DOI] [PubMed] [Google Scholar]
- 31.Leclerc GJ, Leclerc GM, Barredo JC. Real-time RT-PCR analysis of mRNA decay: half-life of Beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2002;2:1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes D, Laskey RA. Assembly of nucleosomes and chromatin in vitro. Methods Enzym. 1989;170:575–585. doi: 10.1016/0076-6879(89)70065-3. [DOI] [PubMed] [Google Scholar]
- 33.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su J, Zhang Y, Lv J, Liu H, Tang X, Wang F, Qi Y, Feng Y, Li X. CpG_MI: a novel approach for identifying functional CpG islands in mammalian genomes. Nucleic Acids Res. 2010;38:e6. doi: 10.1093/nar/gkp882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean CB, Nielsen JD. Generalized linear mixed models: a review and some extensions. Lifetime Data Anal. 2007;13:497–512. doi: 10.1007/s10985-007-9065-x. [DOI] [PubMed] [Google Scholar]
- 37.Hulsen T, de Vlieg J, Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raab D, Graf M, Notka F, Schödl T, Wagner R. The GeneOptimizer Algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst. Synth. Biol. 2010;4:215–225. doi: 10.1007/s11693-010-9062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldi L, Hacker DL, Adam M, Wurm FM. Recombinant protein production by large-scale transient gene expression in mammalian cells: state of the art and future perspectives. Biotechnol. Lett. 2007;29:677–684. doi: 10.1007/s10529-006-9297-y. [DOI] [PubMed] [Google Scholar]
- 40.Karimi M, Goldie LC, Ulgiati D, Abraham LJ. Integration site-specific transcriptional reporter gene analysis using Flp recombinase targeted cell lines. BioTechniques. 2007;42:217–224. doi: 10.2144/000112317. [DOI] [PubMed] [Google Scholar]
- 41.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan N, Hughes TR, Lieb JD, Widom J, Segal E. Contribution of histone sequence preferences to nucleosome organization: proposed definitions and methodology. Genome Biol. 2010;11:140. doi: 10.1186/gb-2010-11-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Langst G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl Acad. Sci. USA. 2007;104:15635–15640. doi: 10.1073/pnas.0702430104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komarnitsky P, Cho E-J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 47.The ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 49.Kosovac D, Wild J, Ludwig C, Meissner S, Bauer AP, Wagner R. Minimal doses of a sequence-optimized transgene mediate high level and longterm EPO expression in vivo: challenging CpG-free gene design. Gene Ther. 2010;18:189–198. doi: 10.1038/gt.2010.134. [DOI] [PubMed] [Google Scholar]
- 50.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 51.Gencheva M, Boa S, Fraser R, Simmen MW, Whitelaw CBA, Allan J. In vitro and in vivo nucleosome positioning on the ovine beta-lactoglobulin gene are related. J. Mol. Biol. 2006;361:216–230. doi: 10.1016/j.jmb.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 52.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol. Cell. Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi JK, Bae J-B, Lyu J, Kim T-Y, Kim Y-J. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol. 2009;10:R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 56.Medvedeva YA, Fridman MV, Oparina NJ, Malko DB, Ermakova EO, Kulakovskiy IV, Heinzel A, Makeev VJ. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC Genomics. 2010;11:48. doi: 10.1186/1471-2164-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.