Abstract
Single-stranded (ss) DNA binding (SSB) proteins play central roles in DNA replication, recombination and repair in all organisms. We previously showed that Escherichia coli (Eco) SSB, a homotetrameric bacterial SSB, undergoes not only rapid ssDNA-binding mode transitions but also one-dimensional diffusion (or migration) while remaining bound to ssDNA. Whereas the majority of bacterial SSB family members function as homotetramers, dimeric SSB proteins were recently discovered in a distinct bacterial lineage of extremophiles, the Thermus–Deinococcus group. Here we show, using single-molecule fluorescence resonance energy transfer (FRET), that homodimeric bacterial SSB from Thermus thermophilus (Tth) is able to diffuse spontaneously along ssDNA over a wide range of salt concentrations (20–500 mM NaCl), and that TthSSB diffusion can help transiently melt the DNA hairpin structures. Furthermore, we show that two TthSSB molecules undergo transitions among different DNA-binding modes while remaining bound to ssDNA. Our results extend our previous observations on homotetrameric SSBs to homodimeric SSBs, indicating that the dynamic features may be shared among different types of SSB proteins. These dynamic features of SSBs may facilitate SSB redistribution and removal on/from ssDNA, and help recruit other SSB-interacting proteins onto ssDNA for subsequent DNA processing in DNA replication, recombination and repair.
INTRODUCTION
Single-stranded (ss) DNA binding (SSB) proteins are ubiquitous and found in bacterial, archaeal and eukaryotic cells, mitochondria, phages and viruses (1–4). SSB proteins bind specifically to ssDNA with high affinity in a sequence-independent manner, protect ssDNA from nucleolytic digestion and prevent intrastrand pairing (i.e. hairpin formation), in order to keep ssDNA in a suitable conformation for the action of enzymes involved in DNA replication, repair and recombination (5). In addition, SSB proteins control the accessibility of ssDNA and can physically interact with a variety of cellular genome maintenance proteins through the highly conserved C-terminal tails of SSB, including nucleases, helicases, polymerases, DNA damage signaling and strand-exchange proteins (3–5), to stimulate their activities. For example, Escherichia coli SSB is able to interact directly with at least 14 other proteins, including DNA Polymerase II, III and V, primase, RecQ, RecO, RecJ, RecG, PriA, PriB, Exonuclease I and IX, Uracil DNA Glycosylase and phage N4 RNA polymerase (5), bringing them to their sites of function.
SSB proteins are characterized by the presence of structurally conserved oligosaccharide/oligonucleotide binding (OB)-fold domains (6–8) for ssDNA binding. The majority of bacterial SSB proteins have a single OB fold per polypeptide and function as homotetramers. EcoSSB is the best studied example of homotetrameric SSB proteins and has served as the prototypical SSB protein for decades of study (1,9,10). Eukaryotic human mitochondrial SSB, like EcoSSB, also encodes a single OB fold per monomer and functions as stable homotetramers (11). In contrast, replication protein A (RPA), the major SSB proteins in eukaryotes, is a heterotrimeric protein complex composed of three distinct subunits and contains six OB folds per heterotrimer (2,12). Interestingly, SSB proteins from the Thermus–Deinococcus genera of bacteria were recently identified as stable homodimers in solution with two OB folds per monomer, contrast to the homotetrameric form so far found in bacteria. The two OB folds of homodimeric SSBs within each monomer have distinct amino acid sequences and the functional SSB homodimer possesses only two highly conserved C-terminal tails whereas homotetrameric SSBs have four per tetramer (13–18).
Thermus thermophilus (Tth) SSB is a representative homodimeric SSB in the Thermus–Deinococcus group. The amino acid sequence of TthSSB shares 43%, 44%, 44%, 44%, 43%, 48% and 82% identity with Deinococcus radiodurans (Dra), Deinococcus radiopugnans, Deinococcus geothermalis, Deinococcus grandis, Deinococcus proteolyticus, Deinococcus murrayi and Thermus aquaticus SSBs, respectively (15,16,19–21). Structural analysis of homodimeric SSBs indicated that although homodimeric SSBs share a similar tertiary arrangement within each OB fold as seen in homotetrameric SSBs, the quaternary arrangement of the four OB folds in the protein’s active form is considerably different from that in homotetrameric SSBs (13,17,18). Escherichia coli maintains 200–3000 EcoSSB tetramers per cell and does not increase SSB levels significantly in response to DNA-damaging conditions, whereas in Deinococcus radiodurans the 20 000 DraSSB dimers per cell increase to 56 000 dimers in response to ionizing radiation (13). The large number of DraSSB per cell and its rapid change in expression level in response to ionizing radiation are responsible for the high tolerance of Dra to the DNA-damaging condition (22). These results indicate that the homodimeric SSBs found in extremeophiles may have an expanded role in DNA metabolism. A recently resolved DraSSB/ssDNA binding complex structure implied that although the ssDNA wraps around a DraSSB homodimer in a similar way as seen in the EcoSSB/ssDNA complex, there are considerably more unstructured ssDNA regions in the DraSSB-bound ssDNA, presumably due to an overall weaker protein–DNA interaction in the DraSSB/ssDNA complex (23). Biochemical studies indicated that the binding affinities of homodimeric SSBs to ssDNA are considerably lower than that of EcoSSB under similar conditions (Kd < 10−7 M for DraSSB, in 1 M NaCl; Kd < 10−9 M for EcoSSB, in 1 M NaBr); the affinity decreases as salt concentration increases, similar to EcoSSB (24,25).
Using EcoSSB as the prototypical SSB protein, we previously showed that EcoSSB can diffuse (or slide) on ssDNA spontaneously with an estimated diffusion coefficient of 300 (nucleotide)2/s at 37℃C, and that the diffusional migration of EcoSSB transiently melts DNA-hairpin structures and stimulates RecA filament elongation (26). We also showed that EcoSSB-interacting proteins can moderately slow EcoSSB diffusion on ssDNA by physically interacting with the last 8–10 amino acids within the conserved SSB C-terminal tail, raising the possibility that SSB acts as a mobile platform on ssDNA for the replication, repair and recombination machinery (27). However, it is not clear whether the diffusion activity observed for homotetrameric SSBs is general for other types of SSBs.
SSB binds ssDNA in different binding modes characterized by the length of ssDNA that it contacts and the number of ssDNA-binding domains involved (1,12,24,28). For EcoSSB, the relative stabilities of these binding modes largely depend on salt concentration and protein binding density: (i) at low monovalent salt concentrations (<20 mM NaCl) and high protein to DNA ratios, an EcoSSB tetramer occludes ∼35 nt with high inter-tetramer cooperativity using only two out of four OB folds [termed the (SSB)35 mode]. (ii) At high salt concentrations (≥200 mM NaCl), an EcoSSB tetramer occlude ∼65 nt with low cooperativity using all four OB folds [termed the (SSB)65 mode] (1,24). Furthermore, we previously observed direct transitions between the (SSB)35 and (SSB)65 binding modes in the presence of 10–100 nM EcoSSB and at low or intermediate salt concentrations (≤200 mM NaCl), and showed that 70-nt ssDNA with two EcoSSB tetramers bound can undergo dynamic structural rearrangement between two conformations [termed (SSB)35 and (SSB)35b modes] (29). The occluded site size of a homodimeric SSB from the Thermus–Deinococcus group shows much more reduced dependence on salt concentration than that of an EcoSSB tetramer (15,19–21,24,30–32). For example, the occluded site size of a DraSSB dimer is 45–47 nt at low salt concentrations (<20 mM NaCl) and increases to 50–55 nt at high salt concentrations (≥200 mM NaCl) (24,31,32). Therefore, it is unclear whether homodimeric SSBs can undergo similar binding mode transitions as seen in EcoSSB.
In this study, we use single molecule (sm) fluorescence resonance energy transfer (FRET) (33,34) to demonstrate that TthSSB, a representative homodimeric SSB, is capable of rapid diffusion along ssDNA, implying that the diffusion activity may be shared among different types of SSB proteins. We also show that the small-scale (tens of nucleotides) TthSSB diffusion along ssDNA is important in the fast redistribution of TthSSB on ssDNA after its initial binding to a random ssDNA location, and that TthSSB can transiently destabilize short DNA hairpins by diffusing in from ssDNA to an adjacent DNA hairpin. Furthermore, we present direct evidence that TthSSB undergoes rapid binding mode transitions in the presence of free SSB proteins, extending our previous observations on homotetrameric EcoSSB to homodimeric SSBs.
MATERIALS AND METHODS
DNA sequences and annealing procedures
DNA oligonucleotides used for the sm experiments are listed below. ‘/iAmMC6T/’ represents amine-modified thymine that is used to label the DNA with Cy3. Biotin modification was used for the surface immobilization.
5′-/Cy5/ GCC TCG CTG CCG TCG CCA-/biotin/-3′.
5′-TGG CGA CGG CAG CGA GGC (T)60 -/Cy3/-3′.
5′-TGG CGA CGG CAG CGA GGC (T)59 /iAmMC6T/ (T)n -3′, where n = 4, 8, 12 or 16.
5′-TGG CGA CGG CAG CGA GGC-/Cy3/-(T)58 -3′.
5′-/biotin/TGG CGA CGG CAG CGA GGC-/Cy5/-3’.
5′-GGG CGG CGA CCT /iAmMC6T/ (T)59 GCC TCG CTG CCG TCG CCA-3′.
5′-AGG TCG CCG CCC-3′.
5′-TGG CGA CGG CAG CGA GGC (T)65 /iAmMC6T/ TGT GAC TGA GAC AGT CAC TT-/Cy5/-T-3′.
‘/Cy3/’ and ‘/Cy5/’ represent the Cy3 and Cy5 fluorophores, respectively, which were attached directly to the DNA backbone using phosphoramidite chemistry. The partial duplex DNA substrates carrying Cy3 and Cy5 were annealed by mixing ∼5 μM of biotinylated strand and ∼7 μM of non-biotinylated strand in 10 mM Tris–HCl (pH 8.0) and 50 mM NaCl, followed by slow cooling from 90℃C to room temperature for ∼2 h.
Protein expression and purification
Thermus thermophilus (Tth) HB8 SSB proteins were expressed and purified as previously described (35,36). All protein concentrations cited in the text refer to TthSSB dimers.
Sample assembly and data acquisition
smFRET experiments were performed at (23 ± 1)℃C unless specified otherwise. Of partial duplex DNA substrates, 50–100 pM were immobilized on a quartz slide surface which is coated with polyethylene glycol (PEG) (mPEG-SC, Laysan Bio) in order to eliminate nonspecific surface adsorption of proteins (34). The immobilization was mediated by biotin–Neutravidin binding between biotinylated DNA, Neutravidin (Pierce) and biotinylated PEG (Bio-PEG-SC, Laysan Bio). For the experiments shown in Figures 1–5, 1nM TthSSB were added into the sample chamber and incubated with the surface-tethered DNA substrates for 5 min in Buffer A containing 20 mM Tris–HCl (pH 8.0), 500 mM NaCl and 0.1 mg/ml BSA. A buffer wash step was then performed using Buffer A to flush out the excess unbound proteins. Finally, buffer B containing 20 mM Tris–HCl (pH 8.0), 0.1 mg/ml BSA, 2% (v/v) glycerol, 1 % (w/v) D-glucose, 165 U/ml glucose oxidase, 2170 U/ml catalase, 3 mM Trolox and indicated amount of NaCl were injected into the sample chamber before data acquisition. For the experiments shown in Figures 6 and 7, buffer B with desired TthSSB concentration was directly injected into the sample chamber for data acquisition after immobilizing the DNA substrates. The single molecule data were acquired using total internal reflection fluorescence (TIRF) microscope (37) with a time resolution of ∼30 ms. In some circumstances where the experiment needs to be performed at lower temperatures, a water-circulating bath circulator (NESLAB RTE-7 Digital One; Thermo Scientific) was used to cool down the sample stage, objective and prism-holder all together. The temperature of the outer surface of the sample chamber was measured by a handhold digital thermometer (Omega) as the best estimation of the experimental temperature inside the sample chamber.
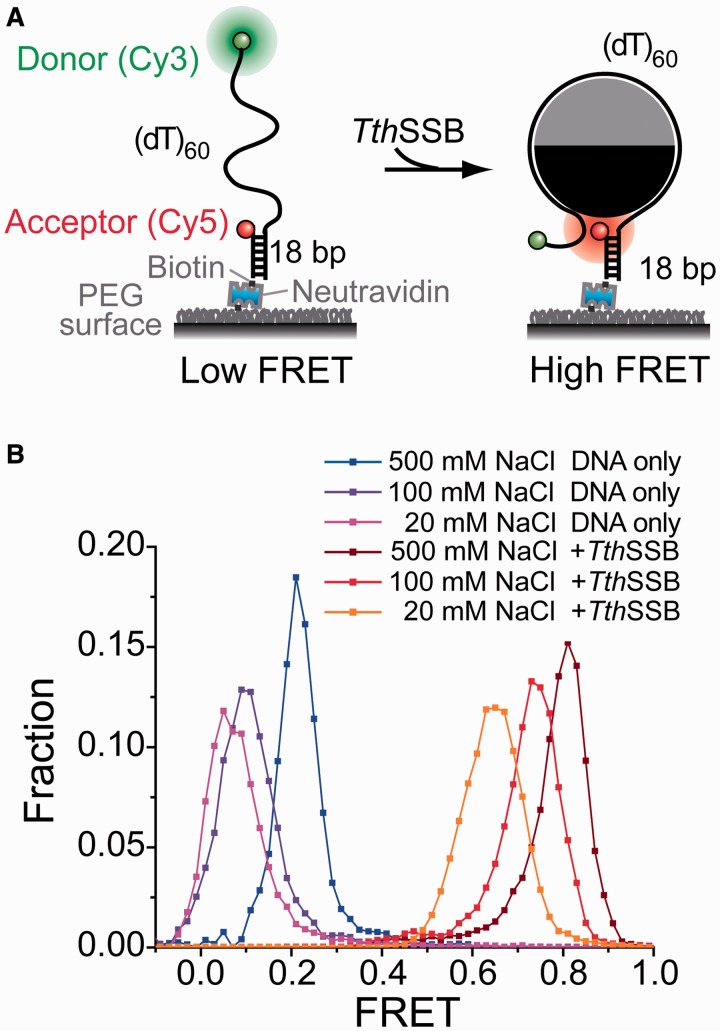
Figure 1.
smFRET assays that report the conformations of TthSSB-bound ssDNA. (A) A schematic illustration of our sm FRET experimental design for TthSSB. A partial duplex DNA substrate [(dT)60] was immobilized on a PEG-coated surface. Cy5 and Cy3 were attached to the ss–dsDNA junction and the end of the ssDNA overhang, respectively. (B) The smFRET histograms for (dT)60 DNA alone and TthSSB binding to (dT)60 at different salt concentrations. TthSSB proteins were loaded onto (dT)60 under high salt condition (500 mM NaCl) and unbound proteins were removed before data acquisition (see also Materials and methods section).
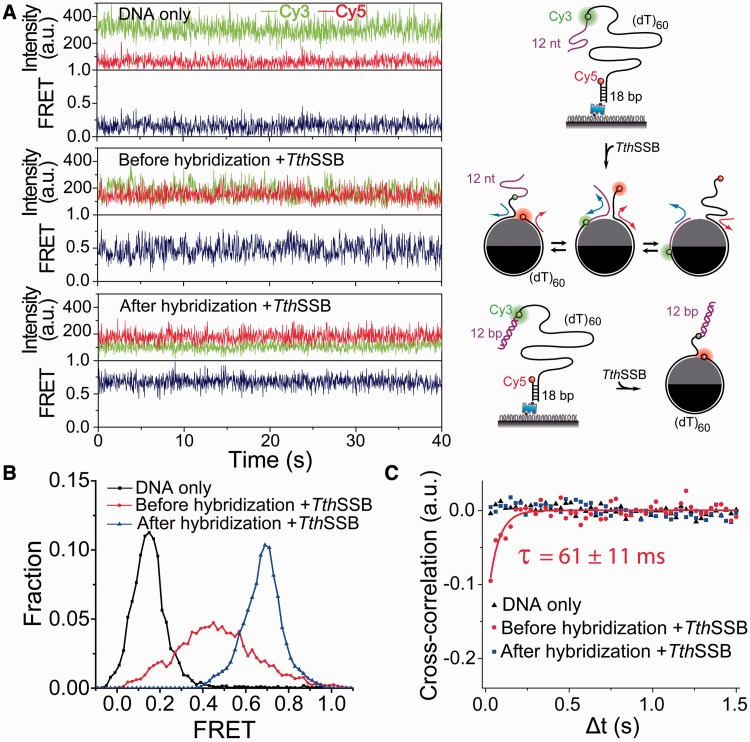
Figure 2.
Protein diffusion detection assays based on DNA hybridization. (A–C) Representative single-molecule time traces (A), smFRET histograms (B) and average cross-correlation curves (C), for (dT)60+12m DNA alone and TthSSB-bound (dT)60+12m with and without the hybridization to the 12-nt mixture sequence ssDNA region. FRET fluctuations beyond measurement noise were detected only when the 12-nt extension is available for TthSSB binding. Unbound proteins were removed before data acquisition such that the FRET flucatuations reflect only the repositioning of the bound TthSSB along (dT)60+12m. For simplification, the 18-bp duplex DNA region is not shown for TthSSB-bound (dT)60+12m in (A). The solid line in (C) is a fit to a single exponential function.
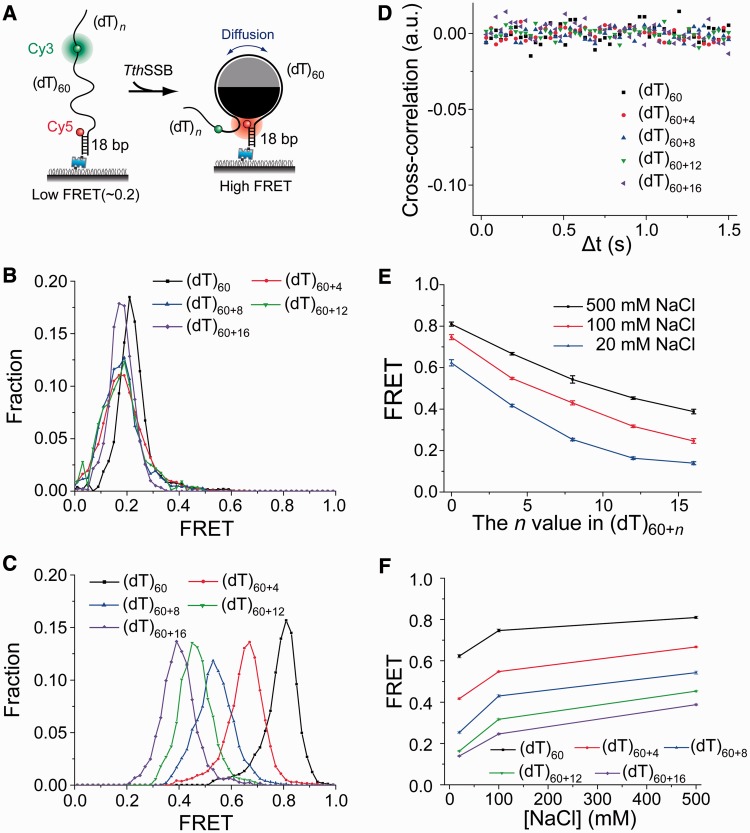
Figure 3.
TthSSB appears to be positioned near the center of the (dT)60+n ssDNA overhangs though rapid diffusion. (A) A schematic illustration of our experimental design for (dT)60+n (n = 0, 4, 8, 12 or 16). Cy3 and Cy5 were attached to the ss–dsDNA junction and the middle of the ssDNA overhang, respectively, separated by (dT)60. (B) The smFRET histograms of (dT)60+n DNA in the absence of proteins, obtained at 500 mM NaCl. (C) smFRET histograms for TthSSB-bound (dT)60+n, obtained at 500 mM NaCl, showing a single narrow FRET peak. (D) Average cross-correlation curves for TthSSB-bound (dT)60+n, obtained at 500 mM NaCl, indicating no significant FRET fluctuations. (E) The FRET value at the FRET peak position versus the n value in (dT)60+n, obtained at 20, 100 or 500 mM NaCl. (F) The FRET value at the FRET peak position versus the salt concentration for different (dT)60+n substrates. TthSSB proteins were loaded onto (dT)60 under high salt condition (500 mM NaCl) and unbound proteins were removed before the buffer containing a lower NaCl concentration was added to the sample chamber.
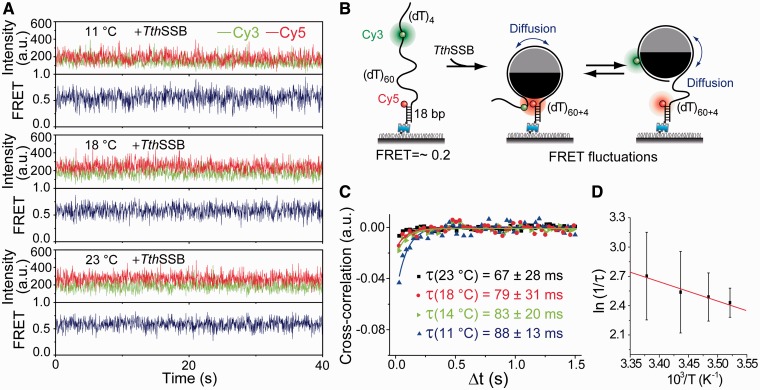
Figure 4.
Lower temperatures slow TthSSB diffusion on (dT)60+4. (A) Representative single-molecule time traces for TthSSB-bound (dT)60+4 obtained at different temperatures (23, 18, 14 and 11℃C) and at 100 mM NaCl. (B) A schematic illustration of our experimental design, showing how TthSSB diffusion may result in FRET fluctuations. (C) Average cross-correlation curves for TthSSB-bound (dT)60+4 obtained at different temperatures and at 100 mM NaCl. More significant FRET fluctuations beyond measurement noise were detected at lower temperatures. The time scales of the FRET fluctuations were determined from fits to a single exponential function (solid lines). Unbound proteins were removed before data acquisition. (D) Arrhenius plot of apparent rates as a function of 1/T.
Figure 5.
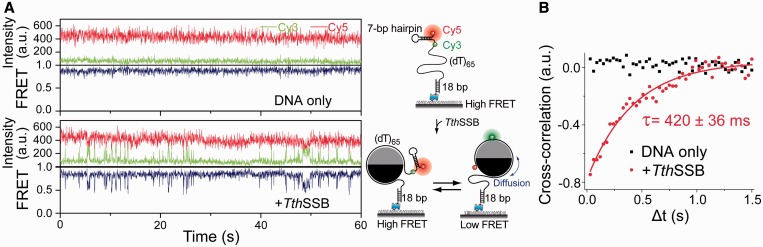
TthSSB diffusion can transiently destabilize short DNA hairpin structures. (A and B) Representative single-molecule time traces (A) and average cross-correlation curves (B), for (dT)65+hp+3 alone and TthSSB-bound (dT)65+hp+3, obtained at 500 mM NaCl. Unbound proteins were removed before data acquisition. The solid line in (B) is a fit to a single exponential function.
Figure 6.
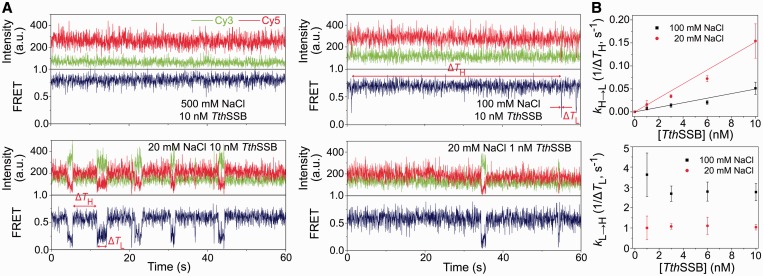
Transitions between different binding modes for TthSSB binding to (dT)60 in the presence of proteins in solution. (A) Representative sm FRET-time traces for TthSSB-bound (dT)60, obtained at 500 and 100 mM NaCl when 10 nM TthSSB were present, and those obtained at 20 mM NaCl when 10 or 1 nM TthSSB were present in the sample camber. 1 nM TthSSB were first incubated with the surface-tethered (dT)60 in 500 mM NaCl, followed by a buffer wash to remove unbound TthSSB proteins. The buffer containing the indicated TthSSB and NaCl concentration was then injected into the chamber for data acquisition. ΔTH and ΔTL represent the durations for the high and low FRET state, respectively. (B) Transtion rates of the interconversions between high and low FRET state (1/ΔTH and 1/ΔTL represent the high to low FRET-state transiton and the low to high FRET-state transition, respectively), as a function of the TthSSB concentration. Solid lines are the linear fits for high to low FRET-state transition rate as a function of TthSSB concentration.
Figure 7.
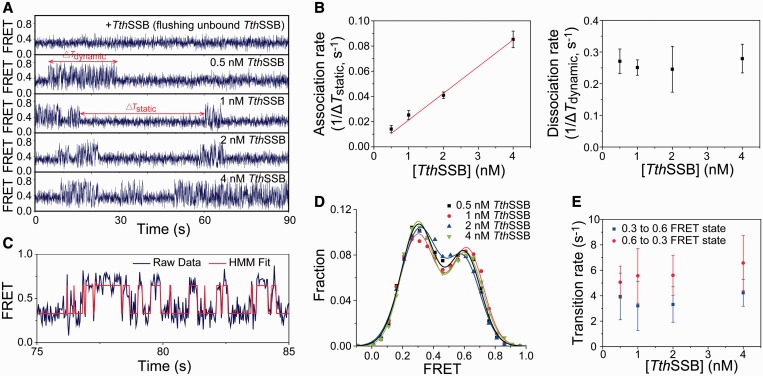
Transitions between different binding modes for TthSSB binding to (dT)60+16 in the presence of proteins in solution. (A) Representative sm FRET-time traces for TthSSB-bound (dT)60+16, obtained at 100 mM NaCl and when 0, 0.5, 1, 2 or 4 nM TthSSB were present in the sample camber. ΔTdynamic and ΔTstatic represent the durations for the ‘dynamic’ and ‘static’ states, respectively. (B) Association and dissociation rates of the second TthSSB to/from (dT)60+16 (1/ΔTstatic and 1/ΔTdynamic, respectively), as a function of the TthSSB concentration. (C) Hidden Markov model (HMM)-derived idealized FRET trajectory (red) superimposed on the FRET-time trace (blue), for a selected time period during which the molecule is in the ‘dynamic’ state. Two FRET states were determined from the HMM fit (∼0.3 and ∼0.6 FRET). (D) FRET-efficiency distributions within the ‘dynamic’ state at different TthSSB concentrations (averaged from >50 molecules). Solid lines are the fits to a double-Gaussian function. (E) Transition rates between the 0.3 and 0.6 FRET state, determined from the HMM fit at different TthSSB concentrations.
FRET efficiency calculation
Apparent FRET efficiency was calculated from the fluorescence intensities of the donor (ID) and acceptor (IA) using the formula EFRET = IA/(IA + ID). The background and the cross-talk between the donor and acceptor were corrected for as previously described (34). The smFRET histograms were built from >5000 surface-tethered DNA molecules.
Cross-correlation analysis
The cross-correlation analysis was performed as previously described (38,39). The cross-correlation curves were calculated between donor and acceptor fluorescence intensity-time traces obtained from each DNA molecule carrying a donor and an acceptor, and were then averaged over >100 DNA molecules. All cross-correlation curves presented are average curves. We determined the characteristic time, τ, by fitting the average cross-correlation curve to a single exponential function.
RESULTS
TthSSB binding brings the two ends of bound ssDNA in close proximity
First, we employed smFRET, a single-molecule method to sensitively monitor the redistribution and changes of distance between a donor and an acceptor fluorophore in the range of 3–8 nm (34), to examine the binding of a single TthSSB dimer to a short stretch of ssDNA. A partial duplex DNA substrate containing a 3′ 60-nt Poly(T) overhang [referred as (dT)60] was immobilized on a surface passivated with PEG. A donor (Cy3) and an acceptor (Cy5) were attached near the two ends of the 60-nt ssDNA overhang, respectively, so that the FRET efficiency between them reports on the conformations of the 60-nt ssDNA (Figure 1A). The occluded site size of a TthSSB dimer is expected to be ∼45–55 nt with a slight dependence on salt concentration, similar to other homodimeric SSBs in the Thermus–Deinococcus group (24,30,31). The 60-nt ssDNA overhang can hence accommodate only one TthSSB dimer in its fully wrapped binding mode. Indeed, previous studies using gel mobility shift assays have shown that (dT)60 can efficiently accommodate only one TthSSB dimer (14).
Before adding the proteins, a low FRET peak was observed for (dT)60 in the smFRET histograms (Figure 1B) due to the large end-to-end distance of the 60-nt ssDNA region. As we increased salt concentration from 20 to 500 mM NaCl, the FRET peak position for (dT)60 alone moved from ∼0.05 to ∼0.2 due to the increased ssDNA flexibility (or compaction) at higher salt concentrations (40). 1 nM TthSSB dimers were then added into the sample chamber and incubated with the surface-tethered DNA substrates for 5 min at 500 mM NaCl, followed by a buffer wash to flush out excess unbound TthSSB proteins. Upon TthSSB binding, a much higher FRET peak centered at ∼0.8 was observed at 500 mM NaCl even after the buffer wash (Figure 1B). Because TthSSB remains bound to (dT)60 for more than 2 h in the absence of unbound proteins (data not shown), we subsequently flowed in buffers containing different salt concentrations (but with no free SSB proteins) for data acquisition. The observed FRET peak values were ∼0.74 and ∼0.63 for 100 and 20 mM NaCl, respectively (Figure 1B). The significantly increased FRET efficiencies observed upon TthSSB binding in a broad range of salt concentrations (20–500 mM NaCl) indicate that the binding of a TthSSB dimer brings the two ends of the ssDNA region into close proximity, similar as seen in homotetrameric SSBs (10,29,41), and it is in good agreement with the recently reported structure of the DraSSB/ssDNA complex (23). The slight decrease in FRET at decreased salt concentrations may result from either a difference in the TthSSB occluded site size (24,30,31), a difference in the flexibility of unbound ssDNA regions (40), or both.
TthSSB is capable of diffusion along ssDNA
To test whether TthSSB can diffuse along ssDNA, we used the single-molecule diffusion detection assay previously developed for EcoSSB (26,37). In this assay, we used a partial duplex DNA with a 5′ 72-nt ssDNA overhang which consists of a (dT)60 region and a 12-nt extension of mixture sequence [referred as (dT)60+12m]. Cy3 and Cy5 were attached near the two ends of the (dT)60 region (Figure 2A). Before adding the proteins, a low FRET peak centered at ∼0.15 was observed for (dT)60+12m in the smFRET histogram (Figure 2B). Upon TthSSB binding to (dT)60+12m, a broad FRET peak centered at ∼0.43 was observed (Figure 2B) and FRET-time traces obtained from single TthSSB–DNA complexes displayed fast FRET fluctuations beyond measurement noise (Figure 2A), similar to those previously observed for EcoSSB diffusion on (dT)69+12m (26). These FRET fluctuations were markedly suppressed and a steady FRET value (∼0.7) was observed when the 12-nt mixture sequence in the 72-nt ssDNA overhang was hybridized to its complementary DNA strand (Figures 2A and 2B). To exclude binding and dissociation of additional TthSSB molecules as the cause of fluctuations, excess unbound TthSSB was removed by a buffer wash step as described above for (dT)60. We also ruled out local melting of the duplex portion as a source of FRET fluctuations (Supplementary Figure S1). Therefore, the FRET fluctuations are likely caused by transient excursions of TthSSB from the (dT)60 region to the 12-nt extension, i.e. one-dimensional TthSSB diffusion.
To further quantify the time scale of FRET fluctuations, we calculated the average cross-correlation of the donor and acceptor fluorescence intensities from >100 fluorescence intensity-time traces of single (dT)60+12m molecules in each condition (27,39). Anti-correlation between donor and acceptor intensities was only observed for TthSSB-bound (dT)60+12m without hybridization of the 12-nt extension, and the single exponential fit to the average cross-correlation curve yielded a time scale of FRET fluctuations, τ= (61 ± 11) ms (Figure 2C). Cross-correlation analysis showed no significant anti-correlation for either unbound (dT)60+12m or TthSSB-bound hybridized (dT)60+12m (Figure 2C).
The time-averaged position of a diffusing TthSSB is near the center of ssDNA
To further test TthSSB diffusion on ssDNA, we designed a series of partial duplex DNA substrates containing 3′ Poly(T) overhangs of different lengths [referred as (dT)60+n]. The ssDNA overhang is slightly longer than the occluded site size of a DraSSB dimer, and consists of a (dT)60 region with Cy3 and Cy5 attached near its ends (Figure 3A) and a (dT)n extension (n = 0, 4, 8, 12 or 16). When n = 0, (dT)60+n is the substrate we used in Figure 1. Before adding the protein, all of the (dT)60+n substrates showed a single low FRET peak centered at ∼0.2 in the smFRET histograms at 500 mM NaCl (Figure 3B). After incubation with 1 nM TthSSB dimer in 500 mM NaCl and the buffer wash step, a single peak centered at a higher FRET value was observed in the smFRET histograms for all five DNA substrates (Figure 3C). Interestingly, the average cross-correlation curves determined from >100 fluorescence intensity-time traces of single TthSSB-bound (dT)60+n molecules showed no significant anti-correlation between Cy3 and Cy5 fluorescence intensities (Figure 3D), indicating that FRET fluctuations, which are expected from TthSSB diffusion, could not be well detected with our experimental time resolution (∼30 ms). Given that TthSSB has high affinity to Poly(T) and would generally not slide off the ssDNA tail, we considered two possible explanations for this observation: (i) after its initial binding to a random position along the ssDNA overhang, TthSSB is unable to move along (dT)60+n, resulting in a time-independent single FRET state for each TthSSB-bound (dT)60+n molecule. (ii) After its initial binding to a random position along ssDNA, TthSSB is able to move along (dT)60+n, but the movement is too fast to be detected within our 30-ms time resolution, resulting in a time-averaged FRET value for each TthSSB-bound (dT)60+n molecule. For the first explanation, one would expect that the sm FRET-efficiency distribution of (dT)60+n with a larger n value should be broader with TthSSB bound and should include all the FRET efficiency values observed for TthSSB-bound (dT)60+m (here m < n), because a longer overhang should provide more possible binding positions (i.e. more diverse FRET efficiency values) for TthSSB. However, the single peak of the smFRET histogram, instead of becoming broader, is shifted towards a lower FRET value as n increases (Figure 3C), disfavoring the first explanation. Therefore, we favor the second explanation in which TthSSB rapidly diffuses along the entire length of the ssDNA overhang and appears to be positioned near the center of the overhang within our 30-ms time resolution. This time-averaging effect has also been observed in other smFRET studies (42,43).
All of the measurements above for (dT)60+n were conducted at room temperature (23℃C) and in 500 mM NaCl. As a further test for the second explanation, we repeated the experiments at room temperature (23℃C) and lower temperatures (18, 14 and 11℃C) and in 100 mM NaCl for (dT)60+4 (Figure 4). Previously, we demonstrated that lowering the temperature slows EcoSSB diffusion on ssDNA (26). Indeed, we were able to observe FRET fluctuations beyond measurement noise in the FRET-time traces of TthSSB-bound (dT)60+4 at 18, 14 and 11℃C (Figure 4A). We excluded the possibility of local wrapping–unwrapping of ssDNA on TthSSB by conducting the experiment at the same condition (100 mM NaCl, 11℃C) on the (dT)60 substrate with Cy3 and Cy5 attached near the two ends of the ssDNA overhang (Supplementary Figure S3). FRET fluctuations were due to anti-correlated fluctuations of the donor and acceptor intensities, as confirmed by the cross-correlation analysis (Figure 4C), which yielded the fluctuation time scales: τ(23℃C) = (67 ± 28) ms, τ(18℃C) = (79 ± 31) ms, τ(14℃C) = (83 ± 20) ms, and τ(11℃C) = (88 ± 13) ms. The Arrhenius fit of ln(1/τ) versus 1/T (Figure 4D) gave an apparent activation energy of Ea = (16 ± 3) kJ/mol for TthSSB diffusion, which is smaller than the Ea determined for EcoSSB diffusion on ssDNA [∼(81 ± 7) kJ/mol] (26).
Having demonstrated that TthSSB is capable of diffusion on (dT)60+n under high and intermediate salt condition (500 and 100 mM NaCl), we next tested TthSSB diffusion at low salt concentrations. After loading a TthSSB dimer onto surface-tethered (dT)60+n in 500 mM NaCl, we flushed out unbound proteins with a buffer also containing 500 mM NaCl. A buffer containing either 100 or 20 mM NaCl was subsequently flowed into the sample chamber before data acquisition. At both 20 and 100 mM NaCl, we observed similar results as at 500 mM NaCl: a single narrow peak was observed in the smFRET histogram (data not shown) and is shifted towards a lower FRET value as n increases (Figure 3E), suggesting that TthSSB diffuses on (dT)60+n in a wide range of salt concentrations (20–500 mM NaCl). As discussed above for (dT)60, the slight decrease in FRET efficiency at decreased salt concentrations for each (dT)60+n substrate (Figure 3F) may result from either a difference in the TthSSB occluded site size (24,30,31), a difference in the flexibility of unbound ssDNA regions (40), or both.
TthSSB diffusion transiently destabilizes short DNA-hairpin structures
The single molecule studies have shown homotetrameric SSBs are able to transiently disrupt short DNA-hairpin structures (26,44). To further demonstrate the functional role of TthSSB diffusion, we examined whether TthSSB can use its diffusion activity to transiently destabilize DNA-hairpin structures. We designed a partial duplex DNA containing a 3′ (dT)65 ssDNA overhang with a 7-bp internal hairpin located at the 3′-end of the overhang [referred as (dT)65+hp+3]. Cy3 and Cy5 were attached to the ends of the hairpin sequence such that FRET reports the formation or melting of the hairpin (Figure 5A). Before adding the proteins, (dT)65+hp+3 exhibited steady high FRET (∼0.9) in the smFRET-time traces obtained at 500 mM NaCl, suggesting that the hairpin itself is stably formed at room temperature (Figure 5A). After incubation with 1 nM TthSSB dimer in 500 mM NaCl and the buffer wash step, a number of brief excursions to a low FRET state (∼0.5) were observed in FRET-time traces, indicating transient unzipping and reformation of the hairpin. Cross-correlation analysis yielded the time scale of FRET fluctuations τ = (420 ± 36) ms (Figure 5B). As the (dT)65 region is slightly larger than the occluded site size of a TthSSB dimer, one TthSSB dimer is expected to remain bound to the (dT)65 region after the buffer wash. Since the unbound proteins were flushed out before the data acquisition, the transient melting of the hairpin must be due to the destabilization of the hairpin segment by TthSSB.
Direct observation of binding mode transition and cooperative binding of TthSSB
Previous ensemble studies have suggested that homodimeric SSBs, like homotetrameric SSBs, may undergo a transition between different binding modes, although the occluded site size of a homodimeric SSB showed a much more reduced salt concentration dependence than that of a homotetrameric SSB (24,30). Using smFRET, we previously showed that a partial duplex DNA substrate containing a (dT)70 overhang may accommodate either one EcoSSB tetramer in the (SSB)65 mode or two EcoSSB tetramers in the (SSB)35 mode, and that a (dT)70 molecule undergoes transitions between these two scenarios in the presence of EcoSSB proteins in solution (29). Given that TthSSB dimers have a less salt-dependent occluded site size change (45–55 nt) compared to EcoSSB, we asked whether similar results can be obtained for TthSSB on (dT)60. Instead of flushing out the unbound proteins, we kept 1, 3, 6 or 10 nM TthSSB dimers in solution and acquired the data at 20, 100 or 500 mM NaCl accordingly. Figure 6A shows representative FRET-time traces obtained from single TthSSB-bound (dT)60 molecules. Clear fluctuations between two discrete FRET states (referred as the high/low FRET state) were observed at 20 and 100 mM NaCl, but not at 500 mM NaCl. We determined the average transition rates between the two FRET states (Figure 6B).
Our observations agree with previous biophysical studies indicating that homodimeric SSBs show an enhanced intermolecular binding cooperativity at lower salt concentrations (23,30,31). Additionally, previous biochemical studies have shown that a homodimeric SSB binds up to two ssDNA molecules of (dT)25 with a salt-dependent negative cooperativity: at either 20 and 200 mM NaCl, the second molecule of (dT)25 has a weaker affinity to DraSSB than the first (dT)25 molecule, but such negative cooperativity (i.e. the difference in the affinities of the first and the second bound (dT)25) is much larger at 20 mM NaCl than at 200 mM NaCl (24). The salt-dependent negative cooperativity observed for homodimeric SSBs predicts that the decrease in salt concentration would favor the transition from single bound protein to two bound proteins per (dT)60 because larger negative cooperativity would favor a single TthSSB dimer binding to about half of (dT)60. Indeed, the transition rate from high FRET to low FRET states (kH→L) is larger at higher TthSSB concentrations and at lower salt concentrations, whereas the transition rate from low to high FRET states (kL→H) does not depend on TthSSB concentration but is smaller at lower salt concentrations (Figure 6B). We hence assigned the high FRET state to (dT)60 with one TthSSB dimer bound and the low FRET state to (dT)60 with two TthSSB dimers bound. In the low FRET state, two TthSSB dimers bind to the same (dT)60 molecule, with each TthSSB dimer occupying 30-nt ssDNA region on average.
Next, we tested another DNA substrate with a longer ssDNA overhang (dT)60+16 in the presence of 0.5–4 nM TthSSB dimers in solution. Two-state transitions were also observed in the single molecule FRET-time traces at 20 and 100 mM NaCl, but not at 500 mM NaCl (Figures 7A and Supplementary Figure S2). One of the two states in the FRET-time traces displayed fast FRET fluctuations (referred as ‘dynamic’ state), whereas the other state displayed a steady FRET value (referred as ‘static’ state). We determined the average transition rates between the ‘dynamic’ and ‘static’ states, 1/ΔTdynamic and 1/ΔTstatic (ΔTdynamic and ΔTstatic represent average durations in the ‘dynamic’ and ‘static’ states, respectively). We found 1/ΔTstatic, similar to kH→L, displays a linear dependence on TthSSB concentration, whereas 1/ΔTdynamic, similar to kL→H, is independent of TthSSB concentration (Figure 7B). Therefore, we assigned the ‘static’ state to (dT)60+16 with one TthSSB dimer bound and the ‘dynamic’ state to (dT)60+16 with two TthSSB dimers bound. 1/ΔTstatic and 1/ΔTdynamic hence gave an association rate constant of (2.1 ± 0.1) × 107M-1·S-1 and a dissociation rate constant of (0.26 ± 0.07) S-1] for the second TthSSB dimer binding and dissociation, respectively. Furthermore, we quantified the FRET fluctuations within the ‘dynamic’ state using an HMM-based statistical approach that determines the most likely time sequence of FRET states (Figure 7C) (45). FRET efficiency distributions within the ‘dynamic’ state can be fit well to a double-Gaussian function (Figure 7D), indicating the existence of two distinct FRET substates (referred as 0.3 and 0.6 FRET states, respectively). The transition rate from the 0.3 FRET to 0.6 FRET state was determined to be (3.7 ± 1.6) s-1, and the transition rate from 0.6 FRET to 0.3 FRET state was determined to be (5.7 ± 1.8) s-1, both of which are independent of TthSSB concentration (Figure 7E). We therefore assigned the two FRET substates to two distinct structural arrangements of (dT)60+16 with two TthSSB dimers bound. The association of the second TthSSB dimers results in a fast ‘dynamic’ FRET state for (dT)60+16 but not for (dT)60, probably because the dynamic structural rearrangement from the DNA molecule with two TthSSB dimers bound requires the (dT)16 extension.
DISCUSSION
In this study, we employed smFRET to study the dynamics of TthSSB, a representative homodimeric SSB, binding to short ssDNA molecules ranging from 60 to 76 nt. Although their quaternary arrangements of the OB folds are different (13,17,18), both homotetrameric and homodimeric SSBs bind ssDNA in a way that brings the two ends of bound DNA in close proximity under high salt conditions, consistent with the recently resolved crystal structure of DraSSB–ssDNA complex (23). Both types of SSBs are capable of rapid diffusion along ssDNA. Previously, we showed that even when the SSB–ssDNA structure is not fully wrapped and a small force (1–5 pN) is used to unwrap some ssDNA from the protein surface, EcoSSB diffusion on ssDNA still persists (27). Our results imply that the diffusion activity may be a shared property for different types of ssDNA-binding proteins. Besides SSB proteins, some other proteins have also been reported to show similar one-dimensional random walk along ssDNA, including POT1-TTP diffusion on telomeric DNA (46) and RAD52 diffusion during homology search (47,48).
Similar to homotetrameric SSBs, TthSSB can diffuse continually as long as there is an available extension of ssDNA beyond its occluded site size. The activation energy for TthSSB diffusion is ∼(16 ± 3) kJ/mol, smaller than that of EcoSSB diffusion [∼(81 ± 7) kJ/mol] under a similar condition (26). The reduced activation energy of TthSSB diffusion might result from the different residues of TthSSB that interact with ssDNA compared to those of EcoSSB, given that the amino acid sequence of the two OB folds in each TthSSB monomer (i.e. the N- and C-terminal OB folds) only share 32% and 40% identity, respectively, with EcoSSB’s OB fold (14,15). SSB OB folds bind to ssDNA through a combination of base-stacking, electrostatic and polar interactions between protein residues and ssDNA, all of which are dependent on the amino acid sequence in the OB fold (23). For example, researchers have identified in EcoSSB four residues (W40, W54, F60, W88) engaged in the base-stacking interaction between its OB folds and ssDNA (13–15); in TthSSB, the corresponding residues are L, Y, L, W for N-teminal OB fold and F, W, W for C-terminal OB fold (the residue corresponding to W40 could not be identified via sequence alignment), respectively (14,15). The different amino acid sequence present in Thermus–Deinococcus SSBs should be accounted for the lower DNA-binding affinity as well as the reduced activation energy for SSB diffusion on ssDNA. Besides, in the resolved crystal structure of the DraSSB–ssDNA complex, the electron density of a considerable portion of the ssDNA that wraps around SSB protein was lost, indicating a relatively dynamic characteristics of the ssDNA at these positions, consistent with the low activation energy for the diffusion (23). Additionally, TthSSB diffusion appears to be faster than that of EcoSSB, and is too fast to be detected for some DNA substrates [e.g. (dT)60+n] with the 30-ms time resolution. This is consistent with the previous observation that the binding affinity of ssDNA for homodimeric SSBs is weaker than for EcoSSB (24) because SSB diffusion requires breakage and reformation of SSB–DNA interactions (27).
SSB diffusion may have multiple functional roles. First, rapid SSB diffusion along DNA should be important in redistributing SSB on ssDNA after its initial binding to a random DNA location, because for proteins with such high affinities, redistribution would be difficult if it required complete dissociation and reassociation. Second, SSB diffusion can transiently destabilize short DNA hairpin structures, probably due to trapping of spontaneously opened ends of the duplex region. We have shown that the hairpin removal by EcoSSB diffusion is responsible for the facilitated RecA filament growth (26). For TthSSB, it has been shown that TthSSB enhances the synthesis rate of DNA polymerases from Tth and Pyrococcus (49), and many different types of SSBs have been used to increase the amplification efficiency for the polymerase chain reaction (PCR) (50). Because specific interactions between polymerases and SSBs are not required for the stimulated activity, the observation is presumably due to SSB diffusion that removes DNA-hairpin structures. Third, SSB diffusion may provide a mechanism for how SSBs recruit other SSB-interacting enzymes onto ssDNA for subsequent DNA processing. We previously showed that specific interactions between EcoSSB and EcoRecO did not abolish but moderately slowed EcoSSB diffusion on ssDNA (27).
Furthermore, we presented direct evidence that TthSSB undergoes rapid binding mode transitions in the presence of free SSB proteins in solution. Although the occluded site size of a homodimeric SSB displays less dependence on the salt concentration than that of an EcoSSB tetramer and ranges from 45 to 55 nt with increase in salt concentration (15,19–21,24,30–32), we observed transitions between a single bound protein and two bound proteins per ssDNA on (dT)60 and (dT)60+16 at low and intermediate salt concentrations (20–100 mM NaCl). When two TthSSB dimers bind to the same 60- or 76-nt ssDNA molecule, the second TthSSB has a weaker affinity to DNA than the first bound TthSSB (i.e. negatively cooperative binding). Additionally, 76-nt ssDNA with two TthSSB dimers bound may undergo dynamic structural rearrangement between two conformations. Similar observations were made previously for EcoSSB binding to (dT)70 (29).
SSB-binding mode transition, depending on the local ionic strength and SSB concentrations, might play a role in controlling the accessibility of ssDNA in various biological processes by tuning the binding site size per SSB and intermolecular cooperativity of SSB proteins, exposing or shielding ssDNA region for other proteins to bind. In addition, SSB-binding mode transition can be regulated by SSB–protein interactions via highly conserved SSB C-terminal tails. Previous studies showed that PriC, a key protein for E. coli DNA replication restart, alters the structure of SSB/ssDNA binding complex and shifts the binding equilibrium towards the highly cooperative (SSB)35 mode over the less cooperative (SSB)65 mode, by interacting with EcoSSB’s C-termial tails (51).
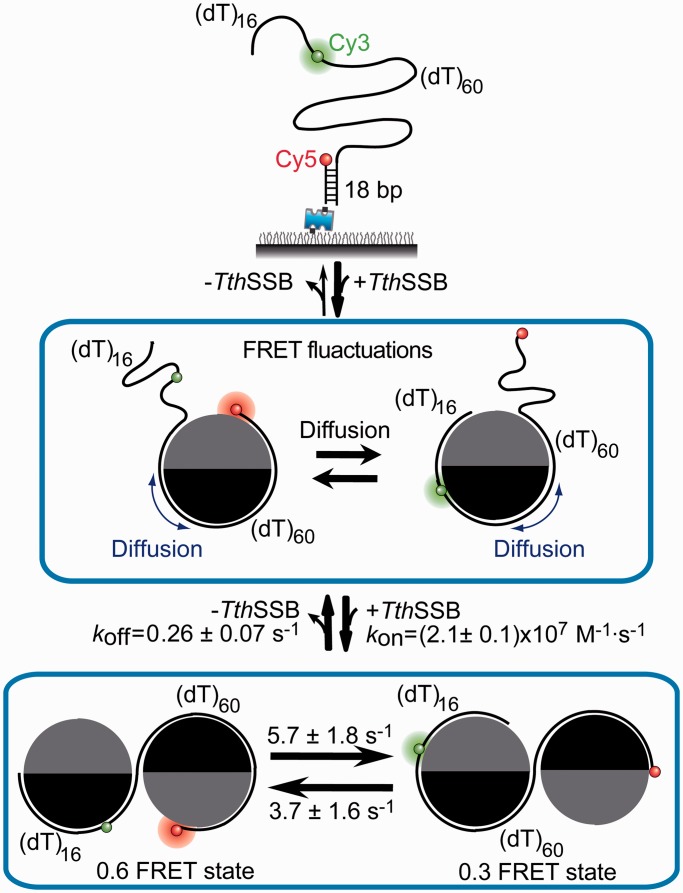
In summary, the data presented in this study extend our previous observations on homotetrameric EcoSSBs to homodimeric SSBs, and suggest that TthSSB/DNA complexes are highly dynamic (Figure 8). Importantly, the covalent linkage between multiple non-identical OB-fold domains in TthSSB dimers, which is also the signature of eukaryotic RPAs, does not preclude SSB diffusion or binding mode transitions. When only one TthSSB dimer binds to an ssDNA region that is slightly longer than its occluded site size, it diffuses rapidly while remaining stably bound, which should be useful to cover and protect even small ssDNA gaps between two bound TthSSBs and remove DNA hairpin structures. Additionally, TthSSB/DNA complexes undergo dynamic interconversions among different binding modes and/or structural rearrangements if there are unbound proteins nearby. These dynamic features of SSB–DNA complex should be important for the functional roles of SSBs in genome maintenance and are possibly shared among different types of SSBs.
Figure 8.
Model of TthSSB dynamics on ssDNA. (dT)60+16 is illustrated as the ssDNA template for TthSSB binding. For simplification, only the ssDNA region is shown in the cartoon for TthSSB-bound (dT)60+16.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank all the members of Ha laboratory for experimental help and discussions.
FUNDING
National Institutes of Health [RR025341 and GM065367 to T.H.]; National Science Foundation [0822613 and 0646550 to T.H.]; JSPS KAKENHI Grant [22570147]; Howard Hughes Medical Institute (to T.H.). Funding for open access charge: NIH [RR025341 and GM065367 to T.H.]; National Science Foundation [0822613 and 0646550 to T.H.]; JSPS KAKENHI [22570147].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 2.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 3.Nam EA, Cortez D. SOSS1/2: Sensors of Single-Stranded DNA at a Break. Mol. Cell. 2009;35:258–259. doi: 10.1016/j.molcel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit. Rev. Biochem. Mol. Biol. 2009;44:98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 5.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Crit. Rev. Biochem. Mol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl Acad. Sci. USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 11.Yang C, Curth U, Urbanke C, Kang CH. Crystal structure of human mitochondrial single stranded DNA binding protein at 2.4 angstrom resolution. Nat. Struct. Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 12.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein DA, Eggington JM, Killoran MP, Misic AM, Cox MM, Keck JL. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl Acad. Sci. USA. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabrowski S, Olszewski M, Piatek R, Brillowska-Dabrowska A, Konopa G, Kur J. Identification and characterization of single-stranded-DNA-binding proteins from Thermus thermophilus and Thermus aquaticus - new arrangement of binding domains. Microbiology. 2002;148:3307–3315. doi: 10.1099/00221287-148-10-3307. [DOI] [PubMed] [Google Scholar]
- 15.Dabrowski S, Olszewski M, Piatek R, Kur J. Novel thermostable ssDNA-binding proteins from Thermus thermophilus and T. aquaticus-expression and purification. Protein Expr. Purif. 2002;26:131–138. doi: 10.1016/s1046-5928(02)00504-1. [DOI] [PubMed] [Google Scholar]
- 16.Eggington JM, Haruta N, Wood EA, Cox MM. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 2004;4:2. doi: 10.1186/1471-2180-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedorov R, Witte G, Urbanke C, Manstein DJ, Curth U. 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 2006;34:6708–6717. doi: 10.1093/nar/gkl1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jedrzejczak R, Dauter M, Dauter Z, Olszewski M, Dlugolecka A, Kur J. Structure of the single-stranded DNA-binding protein SSB from Thermus aquaticus. Acta Crystallogr. D. 2006;62:1407–1412. doi: 10.1107/S0907444906036031. [DOI] [PubMed] [Google Scholar]
- 19.Filipkowski P, Koziatek M, Kur J. A highly thermostable, homodimeric single-stranded DNA-binding protein from Deinococcus radiopugnans. Extremophiles. 2006;10:607–614. doi: 10.1007/s00792-006-0011-8. [DOI] [PubMed] [Google Scholar]
- 20.Filipkowski P, Duraj-Thatte A, Kur J. Novel thermostable single-stranded DNA-binding protein (SSB) from Deinococcus geothermalis. Arch. Microbiol. 2006;186:129–137. doi: 10.1007/s00203-006-0128-2. [DOI] [PubMed] [Google Scholar]
- 21.Filipkowski P, Kur J. Identification and properties of the Deinococcus grandis and Deinococcus proteolyticus single-stranded DNA binding proteins (SSB) Acta. Biochim. Pol. 2007;54:79–87. [PubMed] [Google Scholar]
- 22.Lockhart JS, DeVeaux LC. The essential role of the Deinococcus radiodurans ssb gene in cell survival and radiation tolerance. PLoS One. 2013;8:e71651. doi: 10.1371/journal.pone.0071651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George NP, Ngo KV, Chitteni-Pattu S, Norais CA, Battista JR, Cox MM, Keck JL. Structure and cellular dynamics of Deinococcus radiodurans single-stranded DNA (ssDNA)-binding protein (SSB)-DNA complexes. J. Biol. Chem. 2012;287:22123–22132. doi: 10.1074/jbc.M112.367573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozlov AG, Eggington JM, Cox MM, Lohman TM. Binding of the dimeric Deinococcus radiodurans single-stranded DNA binding protein to single-stranded DNA. Biochemistry. 2010;49:8266–8275. doi: 10.1021/bi100920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002;41:6032–6044. doi: 10.1021/bi020122z. [DOI] [PubMed] [Google Scholar]
- 26.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witte G, Fedorov R, Curth U. Biophysical analysis of Thermus aquaticus single-stranded DNA binding protein. Biophys. J. 2008;94:2269–2279. doi: 10.1529/biophysj.107.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witte G, Urbanke C, Curth U. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 2005;33:1662–1670. doi: 10.1093/nar/gki310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggington JM, Kozlov AG, Cox MM, Lohman TM. Polar destabilization of DNA duplexes with single-stranded overhangs by the Deinococcus radiodurans SSB protein. Biochemistry. 2006;45:14490–14502. doi: 10.1021/bi061178m. [DOI] [PubMed] [Google Scholar]
- 33.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue J, Shigemori Y, Mikawa T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006;34:e69. doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue J, Honda M, Ikawa S, Shibata T, Mikawa T. The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Res. 2008;36:94–109. doi: 10.1093/nar/gkm1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Ha T. Single-molecule analysis of SSB dynamics on single-stranded DNA. Methods Mol. Biol. 2012;922:85–100. doi: 10.1007/978-1-62703-032-8_5. [DOI] [PubMed] [Google Scholar]
- 38.Zhou R, Kozlov AG, Roy R, Zhang JC, Korolev S, Lohman TM, Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:485–485. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl Acad. Sci. USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antony E, Weiland EA, Korolev S, Lohman TM. Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J. Mol. Biol. 2012;420:269–283. doi: 10.1016/j.jmb.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karymov M, Daniel D, Sankey OF, Lyubchenko YL. Holliday junction dynamics and branch migration: single-molecule analysis. Proc. Natl Acad. Sci. USA. 2005;102:8186–8191. doi: 10.1073/pnas.0407210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joo C, McKinney SA, Lilley DM, Ha T. Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J. Mol. Biol. 2004;341:739–751. doi: 10.1016/j.jmb.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 44.Antony E, Kozlov AG, Nguyen B, Lohman TM. Plasmodium falciparum SSB Tetramer Binds Single-Stranded DNA Only in a Fully Wrapped Mode. J. Mol. Biol. 2012;420:284–295. doi: 10.1016/j.jmb.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honda M, Okuno Y, Yoo J, Ha T, Spies M. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J. 2011;30:3368–3382. doi: 10.1038/emboj.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothenberg E, Grimme JM, Spies M, Ha T. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl Acad. Sci. USA. 2008;105:20274–20279. doi: 10.1073/pnas.0810317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perales C, Cava F, Meijer WJJ, Berenguer J. Enhancement of DNA, cDNA synthesis and fidelity at high temperatures by a dimeric single-stranded DNA-binding protein. Nucleic Acids Res. 2003;31:6473–6480. doi: 10.1093/nar/gkg865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kur J, Olszewski M, Dlugolecka A, Filipkowski P. Single-stranded DNA-binding proteins (SSBs) - sources and applications in molecular biology. Acta Biochim. Polonica. 2005;52:569–574. [PubMed] [Google Scholar]
- 51.Wessel SR, Marceau AH, Massoni SC, Zhou R, Ha T, Sandler SJ, Keck JL. PriC-mediated DNA replication restart requires PriC complex formation with the single-stranded DNA-binding protein. J. Biol. Chem. 2013;288:17569–17578. doi: 10.1074/jbc.M113.478156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.