Abstract
In the present study, we investigated the 3′ untranslated region (UTR) of the mouse core clock gene cryptochrome 1 (Cry1) at the post-transcriptional level, particularly its translational regulation. Interestingly, the 3′UTR of Cry1 mRNA decreased its mRNA levels but increased protein amounts. The 3′UTR is widely known to function as a cis-acting element of mRNA degradation. The 3′UTR also provides a binding site for microRNA and mainly suppresses translation of target mRNAs. We found that AU-rich element RNA binding protein 1 (AUF1) directly binds to the Cry1 3′UTR and regulates translation of Cry1 mRNA. AUF1 interacted with eukaryotic translation initiation factor 3 subunit B and also directly associated with ribosomal protein S3 or ribosomal protein S14, resulting in translation of Cry1 mRNA in a 3′UTR-dependent manner. Expression of cytoplasmic AUF1 and binding of AUF1 to the Cry1 3′UTR were parallel to the circadian CRY1 protein profile. Our results suggest that the 3′UTR of Cry1 is important for its rhythmic translation, and AUF1 bound to the 3′UTR facilitates interaction with the 5′ end of mRNA by interacting with translation initiation factors and recruiting the 40S ribosomal subunit to initiate translation of Cry1 mRNA.
INTRODUCTION
The expression of most genes is regulated temporally and spatially. Although most of the regulation of gene expression occurs at the transcription step, the regulation of mRNA stability, localization and modulation of translation are crucial steps, particularly in developmental processes (1) and biological clock systems (2–5). Expression profiles of mRNA or protein are not matched in many cases, implying that translational control is a dynamically regulated mechanism, which is not a silent step (6–8). Recent work had shown that the majority of regulation that dictates protein levels is at the level of translation (9). In most cases of transcript-specific translational regulation, mRNA-binding proteins bind to cis-elements in the 3′ untranslated region (UTR) of the target transcript, resulting in translational activation or repression (10). The 3′UTR is widely known to function as a cis-acting element that regulates the mRNA half-life (11–13), leading to greater or lesser protein levels. Along with the translational control by the 5′UTR, the 3′UTR is also important for translational initiation by making a ‘closed-loop’ or ‘circular’ structure of mRNA through association of poly(A)-binding protein (PABP) with eukaryotic translation initiation factor 4G (EIF4G) (14,15). In addition, the 3′UTR provides a target region for specific microRNAs, leading to translational repression (16)—this is an active area of investigation.
Here, we suggest a novel regulatory process involving the expression of CRY1 (which is encoded by one of the core circadian clock genes), Cry1 3′UTR-mediated translation. The physiological processes of all living organisms from bacteria to humans are governed by the daily cycle (17,18). Transcriptional and post-translational regulations are mainly focused on elucidating the underlying mechanisms of sustained oscillation (19–21). At the molecular level, a circadian rhythm is generated and regulated by various components—so-called ‘clock genes’—and the robustness of the periodicity is based on the interlocked transcriptional feedback loops of such participants. CRY is one of the most important clock components in the mammalian circadian circuit, functioning as a negative limb of the transcriptional feedback loop (22). CRY is necessary for dimerization of the PER (PERIOD)/CRY (CRYPTOCHROME) heterodimer, and its nuclear localization represses the transcriptional activity of the BMAL1 (aryl hydrocarbon receptor nuclear translocator-like)/CLOCK (circadian locomotor output cycles kaput) heterodimer. Additionally, CRY1 and CRY2 are indispensable for the maintenance of behavioral rhythmicity (23,24). In contrast to transcription and post-translational modification, few studies have focused on the post-transcriptional regulation of Cry1 (4). Cry1 is an important core clock gene and shows rhythmic expression that is not fully explained by transcription or protein modification. Nevertheless, translational control of Cry1 has not yet been studied. Here, we suggest another regulatory process of Cry1 expression, Cry1 3′UTR-mediated translation. The 3′UTR of Cry1 increased translation efficiency. Specifically, AU-rich element RNA-binding protein 1 (AUF1), also known as heterogeneous nuclear ribonucleoprotein D (HNRNPD), interacted with the 3′UTR of Cry1, and its knock-down decreased CRY1 levels. AUF1 showed circadian time-dependent cytoplasmic expression, and binding to the Cry1 3′UTR was rhythmic. Indeed, AUF1 associated with translation initiation factors and also directly interacted with the 40S ribosomal protein RPS3 or RPS14. In the present report, we concluded that the rhythmical RNA-binding protein AUF1 on the 3′UTR recruits the 40S ribosomal subunit to the 5′ end of mRNA by associating with EIF3B, leading to time-dependent expression of CRY1. Our study may expand the roles of the 3′UTR and RNA-binding proteins for the translation system.
MATERIALS AND METHODS
Plasmid construction
Mouse Cry1 3′UTR (Cry1-3′UTR) was amplified from the pcNAT-wt610 plasmid, which was previously reported (4), using Pfu polymerase (Solgent), and the sequence was confirmed by sequencing. The resulting products were cloned into the XhoI/NotI site of the control vector pRL, which expresses Renilla luciferase lacking the 3′UTR (psiCHECK™-2 vector; Promega).
For the in vitro binding assay/ultraviolet (UV)-crosslinking, fragments of mouse Cry1-3′UTR were amplified, and the polymerase chain reaction (PCR) products were digested and sub-cloned into the EcoRI/XbaI site of the pSK′ vector. For mRNA transfection, poly(A)30 was inserted following Renilla luciferase of pRL or following Cry1-3′UTR of the pRL-Cry1-3U vectors using the NotI site.
Plasmids for recombinant AUF1—pGEX-4T3-P37, pGEX-4T3-P40 and pGEX-4T3-P42—and the pGEX-4T3-P45 plasmid for glutathione-S-transferase (GST)-fused recombinant AUF1 isoforms were kindly provided by Dr Sung Key Jang at Pohang University of Science and Technology. To generate pProEX-Hta-RPS3, pProEX-Hta-RPS11, pProEX-Hta-RPS14 and pProEX-Hta-RPS18 for recombinant histidine (His) tag-fused 40S ribosomal proteins, full-length RPS3, RPS11, RPS14 and RPS18 were amplified by PCR from NIH 3T3 cells; the resulting DNA fragments were cloned into the EcoRI/XhoI-treated pProEX-Hta vector (Invitrogen).
To knock down Auf1 expression, we selected sequences targeting 3′UTRs of Auf1 from the Public TRC Portal (http://www.broadinstitute.org/rnai/public/). Knock-down–verified sense strand of Auf1 shRNA is 5′-tCCTGAATGGAAGTATGACGttcaagagaCGTCATA CTTCCATTCAGGttttttc-3′ (based on positions 1451 to 1470 of mouse Auf1). The 19-nucleotide (nt) Auf1 target sequences are indicated in uppercase letters. Sense strand and antisense strand were annealed and cloned into the HpaI-XhoI sites of pLL3.7 lentiviral vector.
Cell culture and drug treatment
HEK 293A and NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum (HyClone) and 1% antibiotics (WelGENE) and maintained in a humidified incubator with 95% air and 5% CO2.
The circadian oscillation of NIH 3T3 cells was synchronized by treatment with 100 nM dexamethasone. After 2 h, the medium was replaced with complete medium (3). To block the translation system, NIH 3T3 cells were treated with 100 μg/ml cycloheximide (3). For blocking transcription, NIH 3T3 cells were treated with actinomycin D (5 μg/ml) (4,25).
Transient transfection and RNA interference
For transient transfection of reporter plasmids, specific small interfering RNAs (siRNAs) and AUF1-overexpressing plasmids in NIH 3T3 cells, the Neon® Transfection System (Invitrogen) was used as recommended by the manufacturer.
siRNAs for endogenous Auf1 knock-down were purchased from Dharmacon (siGENOME SMART-pool HNRNPD M-042940-00). siRNAs for exon 2 or exon 7 of Auf1 (e2_si or e7_si) were purchased from Bioneer using previously reported sequences (26). The reporter mRNA transfection was performed as follows: NIH 3T3 cells were transiently transfected with 2 μg of the in vitro transcribed reporter mRNAs containing the cap structure at the 5′ end using Lipofectamine 2000 (Invitrogen), and incubated for 6 h prior to harvesting.
In vitro RNA synthesis, in vitro binding, UV-crosslinking, RNA affinity purification
For in vitro binding assays, [32P]UTP-labelled RNA was transcribed from XbaI-linearized recombinant pSK′ vectors using T7 RNA polymerase (Promega). In vitro binding and UV-crosslinking were performed as previously described (3,27). Briefly, equal amounts of labelled RNAs were incubated with 15 μg nuclear extracts or 30 μg cytoplasmic extracts of NIH 3T3 cells for 20 min. After incubation, the samples were UV irradiated on ice for 10 min using a CL-1000 UV crosslinker (UVP). Unbound RNA was digested with 5 μl of an RNase cocktail containing RNase A and RNase T1. The reaction mixtures were analysed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Streptavidin-biotin RNA affinity purification of Cry1-3′UTR-binding proteins was performed as previously reported (28). Briefly, cytoplasmic extracts prepared from NIH 3T3 cells were incubated with biotinylated Cry1-3′UTR, and were subjected to streptavidin resin adsorption. Resin-bound proteins were analysed by SDS-PAGE.
RNA quantification
Total RNA was extracted from NIH 3T3 cells using the TRI Reagent (Molecular Research Center). RNA was reverse-transcribed using ImProm-II™ (Promega) according to the manufacturer’s instructions. mRNA levels of endogenous or reporter plasmids were detected by quantitative real-time PCR using the StepOnePlus real-time PCR system (Applied Biosystems) with the FastStart Universal SYBR Green Master (Roche), as described previously (3). Specific primer pairs for Rluc, mouse Tbp, mouse Cry1, mouse Rpl32, mouse Per2, mouse Nr1d1 and mouse Dbp were used for real-time PCR (the primer sequences are shown in Supplementary Table S1).
Protein expression and purification
Escherichia coli BL21 (DE3) pLysS cells (Novagen) transformed with plasmids coding for His-RPS3, His-RPS11, His-RPS14 and RPS18 were grown to an absorbance at 600 nm (A600) of 0.4–0.6, and proteins were induced for 2 h at 37°C with 0.5 mM isopropyl β-d-1-thiogalactopyranoside. For purification of His-RPS3, RPS11, RPS14 and RPS18, cells were harvested and resuspended in a lysis buffer [50 mM Tris-Cl (pH 8.0), 300 mM NaCl, EDTA-free protease inhibitor cocktail from Roche]. The resuspended cells were further lysed by sonication, clarified by centrifugation and purified on Ni-NTA agarose (Invitrogen) with washing [50 mM Tris-Cl (pH 8.0), 300 mM NaCl, 10 mM imidazole], followed by elution [50 mM Tris-Cl (pH 8.0), 300 mM NaCl, 500 mM imidazole].
For preparation of GST-fused AUF1 isoforms, E. coli BL21 cells were cultured at 37°C to A600 = 0.6, and induced with isopropyl β-d-1-thiogalactopyranoside for 20 h at 18°C. Next, cells were resuspended in 25 mM Tris–HCl (pH 7.4), 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40 and 5 mM dithiothreitol, and lysed by sonication. GST-tagged AUF1 proteins were purified by Glutathione Sepharose 4B beads and eluted in 50 mM Tris-HCl (pH 8.0) and 20 mM l-Glutathione reduced (GE Healthcare).
Pulldown assay
Immunoprecipitation was performed with 2 μg antibodies and 30 μl protein A-Sepharose slurry (GE Healthcare) in cell lysis buffer [20 mM Tris (pH 8.0), 137 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% glycerol, protease inhibitor cocktail from Roche]. The GST pulldown assay was performed with 5 μg His- or GST-fused proteins using glutathione-Sepharose 4B (GE Healthcare).
Immunoblot analysis
Immunoblot analyses were performed using polyclonal anti-CRY1, monoclonal anti-AUF1 (Millipore), polyclonal anti-14-3-3ζ (Santa Cruz Biotechnology), monoclonal anti-RPS3 (Cell Signaling), polyclonal anti-RPS14 (Santa Cruz Biotechnology), polyclonal anti-EIF3B (Santa Cruz Biotechnology) and monoclonal anti-GAPDH (Millipore) as primary antibodies. Horseradish peroxidase-conjugated species-specific secondary antibodies (goat, Santa Cruz Biotechnology; guinea pig, Santa Cruz Biotechnology; mouse, Thermo Scientific; rabbit, Jackson ImmunoResearch Laboratories) were visualized using a SUPEX ECL solution kit (Neuronex) and a LAS-4000 chemiluminescence detection system (FUJIFILM), and the acquired images were analysed using Image Gauge (FUJIFILM) according to the manufacturer’s instructions.
Acquiring super-resolution structured illumination images
We used super-resolution structured illumination microscopy (SIM; Nikon N-SIM). The raw images were reconstructed to three-dimensional-SIM images using NIS-E software (Nikon). Images were acquired using an Eclipse Ti-E research inverted microscope with Nikon’s legendary CFI Apo TIRF 100× oil objective lens (NA 1.49) and 512 × 512 pixel resolution equipped with an iXon DU-897 EMCCD camera (Andor Technology). Multicolor fluorescence was acquired using a diode laser (488 nm, 561 nm).
Immunoprecipitation and reverse-transcription assays
We used a slightly modified method from that previously reported (29). The cytoplasmic extract was obtained as described previously (3). Immunoprecipitation was performed under RNase-free conditions and carried out in immunoprecipitation buffer containing 125 mM KCl, 20 mM HEPES (pH 7.4), 0.5 mM EDTA, 0.05% NP-40, 0.5 mM dithiothreitol, RNasin (Promega) and protease inhibitor cocktail (Calbiochem). Immunoprecipitation was performed using polyclonal anti-AUF1. RNA was extracted from the washed protein G-agarose bead pellet with an RNA isolation solution (Molecular Research Center). Reverse transcription and quantitative real-time PCR were performed as described above.
Polysome profiling
Dexamethasone treated or siRNA-transfected NIH 3T3 cells were treated with cycloheximide (100 μg/ml) for 5 min on the ice and then harvested. Cell extracts were subjected to sucrose gradient analysis, as previously described (30). Total RNA of each fraction was purified using TRI reagent (Molecular Research Center) and subjected to real-time PCR analysis for quantification.
Statistical analysis
All quantitative data are presented as the mean ± standard error of the mean (SEM). Comparisons between two groups were analysed by two-tailed unpaired Student’s t tests. For comparisons between more than two groups, a one-way analysis of variance (ANOVA) was used with a post hoc Tukey’s test. A P value less than 0.05 was considered statistically significant. A two-way ANOVA with a Bonferroni post-test was used to analyse the effects of con_si and Auf1_si transfection on circadian time points. CircWave v1.4 software (courtesy of Dr Roelof Hut, http://www.euclock.org/) was used to analyse the rhythmicity of expression patterns.
RESULTS
Cry1-3′UTR is involved in translation
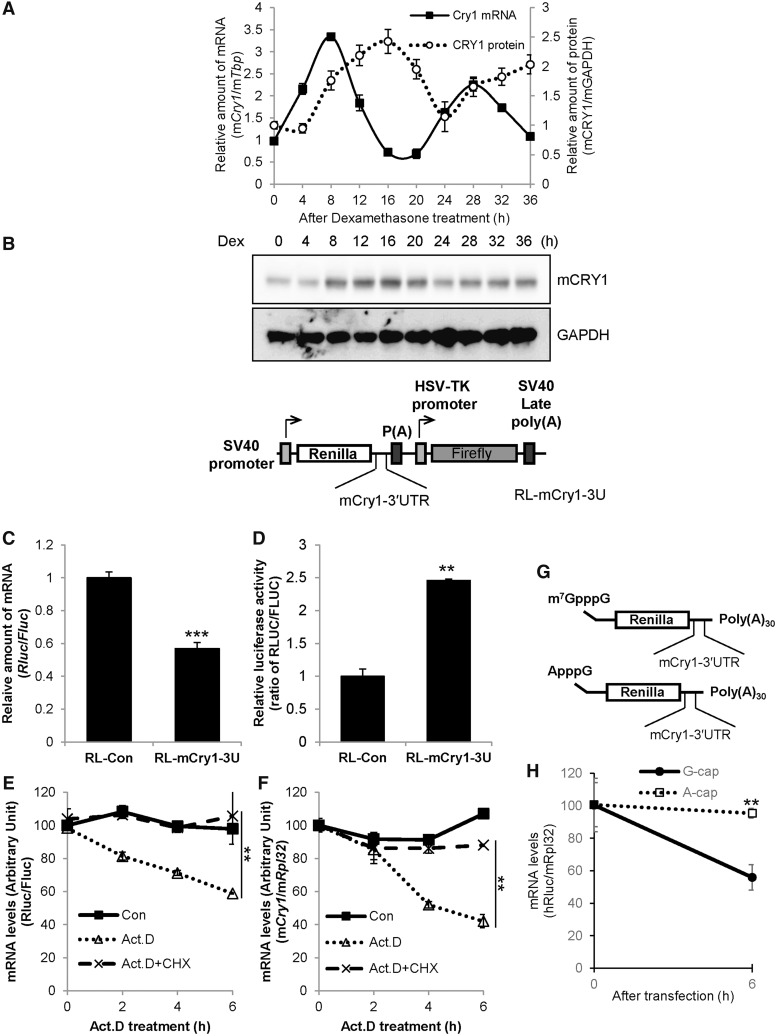
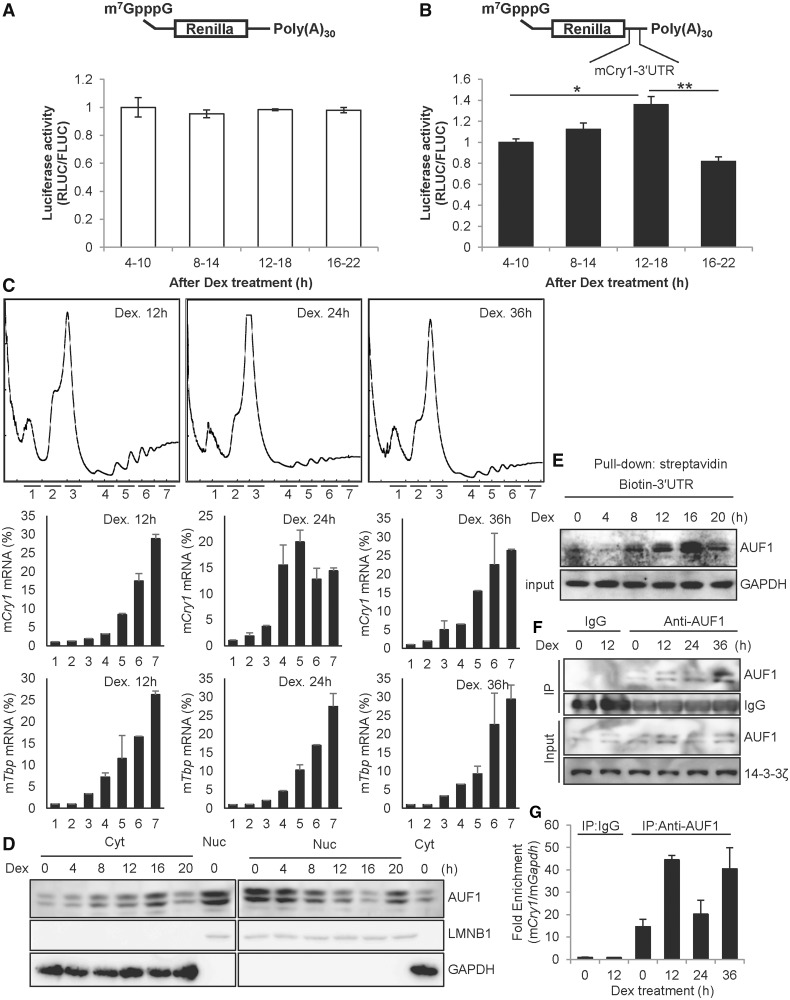
We observed and confirmed a distinctive relationship between mRNA and protein profiles of Cry1. The circadian rhythmicity of CRY1 protein showed a delayed peak or trough time of approximately 4 h compared with that of mRNA oscillation (Figure 1A and Supplementary Figure S1). The discrepant oscillation pattern between mRNA and protein of Cry1 indicates that an important regulation step may exist, particularly at the post-transcriptional step. Post-transcriptional regulation is mainly controlled by the UTR of mRNA; thus, we focused on the UTR of Cry1. First, Renilla luciferase (Rluc) was fused to Cry1-3′UTR (RL-Cry1-3U) to confirm the function of mouse Cry1-3′UTR in mRNA stability and translation compared with the Rluc control vector (RL) (Figure 1B). Generally, the 3′UTR is related to mRNA degradation, and we observed decreased luciferase mRNA levels of reporters that harbour Cry1-3′UTR (Figure 1C). We also determined protein levels of the reporters to identify the relationship between mRNA and protein levels of Cry1. Interestingly, reporters carrying Cry1-3′UTR showed increased luciferase activity compared with control reporters lacking 3′UTR sequences (Figure 1D). We thought that protein levels of the reporters mirrored mRNA levels; specifically, we assumed that Cry1-3′UTR-conjugated reporters may show reduced protein levels because Cry1-3′UTR decreases Cry1 mRNA levels. However, the results overturned a common sense that Cry1 mRNA is a prerequisite for CRY1 protein with a correlated expression pattern. We wanted to know whether mRNA stability of Cry1 could be affected by its translation or not. Transcription produces new mRNAs, and the newly produced mRNAs should be excluded when we measure the stability of pre-existing mRNAs. To check mRNA stability except transcriptional effect, we treated cells with Actinomycin D (Act.D), which blocks transcription elongation by binding to DNA at the transcription initiation complex (31). In the presence of Act.D endogenous Cry1 mRNA level was reduced together with Cry1-3′UTR reporter mRNA (Figure 1E and F). This means that 3′UTR is a destabilizing element in Cry1 mRNA. If the translation is linked to mRNA stability as we’ve hypothesized, mRNA level should be affected by translation status. Thus, we treated cells with CHX to block translation process and then checked the mRNA stability. In this study, we treated cells with Act.D and CHX together to investigate the exact mRNA level change related to translation without transcriptional effect. Co-treatment with Act.D and CHX retarded degradation of Cry1 mRNA and Cry1-3′UTR reporter mRNA (Figure 1E and F). However, CHX could block synthesis of a labile mRNA degradation factors. To complement this experiment, we performed another experiment. Transfection of Cry1 3′UTR fused reporter mRNAs containing a m7GpppG-cap, which activates translation, showed reduced reporter mRNA levels. Related to ApppG-capped Cry1-3′UTR reporters, which cannot bind to the translation initiation factor eIF4E, stability of reporter mRNA was enhanced (Figure 1G and H). From these results, we like to suggest that translation of Cry1 may be activated by the 3′UTR, and Cry1 mRNA levels can be regulated by 3′UTR-mediated translation.
Figure 1.
The 3′UTR of Cry1 is involved in translation. (A) NIH 3T3 cells were treated with dexamethasone (Dex), and cells were subjected to mRNA quantification or immunoblotting at the indicated time points. The relative Cry1 mRNA levels were expressed as the mean ± SEM (closed squares/solid line). The relative mCRY1 protein level (open circles/dotted line) were normalized to GAPDH and plotted. mCry1 mRNA (CircWave, P = 1 × 10−7) and mCRY1 protein (CircWave, P = 7 × 10−7) levels between 8–36h are significantly rhythmic. (B) Cry1-3′UTR was fused to Renilla luciferase (RL-Cry1-3U). Firefly luciferase was used as an internal control. (C) RL-con, which lacks the 3′UTR sequence, or RL-Cry1-3U plasmids were transfected into NIH 3T3 cells. After a 24-h incubation, total RNA was prepared, and mRNA levels were quantified by real-time PCR with Rluc- or Fluc-specific primers. mRNA levels were normalized to Fluc mRNA levels. The relative mRNA level of RL-con was set to 1 (n = 4, ***P < 0.0001). (D) From the same extracts of panel C, the luciferase assay was performed. Renilla luciferase activity (RLUC) was normalized to Firefly luciferase activity (FLUC), and RL-con activity (ratio of RLUC/FLUC) was set to 1 (n = 3, **P = 0.0059). (E) The Cry1-3′UTR reporter (RL-Cry1-3U) was transfected into NIH 3T3 cells. After a 24-h incubation, cells were treated with dimethyl sulfoxide (DMSO) vehicle (Con), actinomycin D (Act. D) or Act. D plus cycloheximide (CHX), and harvested at indicated time points. mRNA levels of Rluc or Fluc were measured by real-time PCR with Rluc- or Fluc-specific primers. (n = 3, **P < 0.05) (F) Untransfected NIH 3T3 cells were treated with DMSO, Act.D or Act.D plus CHX as shown in panel E. Endogenous Cry1 mRNA levels were determined by real-time PCR and normalized to Rpl32 mRNA levels at the indicated time points. The initial relative level of Cry1 mRNA was arbitrarily set to 100 (n = 3, **P < 0.05). (G) Schematic diagrams of the mRNA reporter of Cry1-3′UTR shows m7GpppG, ApppG and the 30-nt-long poly(A) tail [poly(A)30]. (H) m7GpppG and ApppG reporter mRNAs were transiently transfected and incubated 6 h, then mRNA levels were quantified. The initial relative mRNA levels of ApppG were set to 100 (n = 3, **P = 0.0021).
AUF1 regulates the translation of Cry1
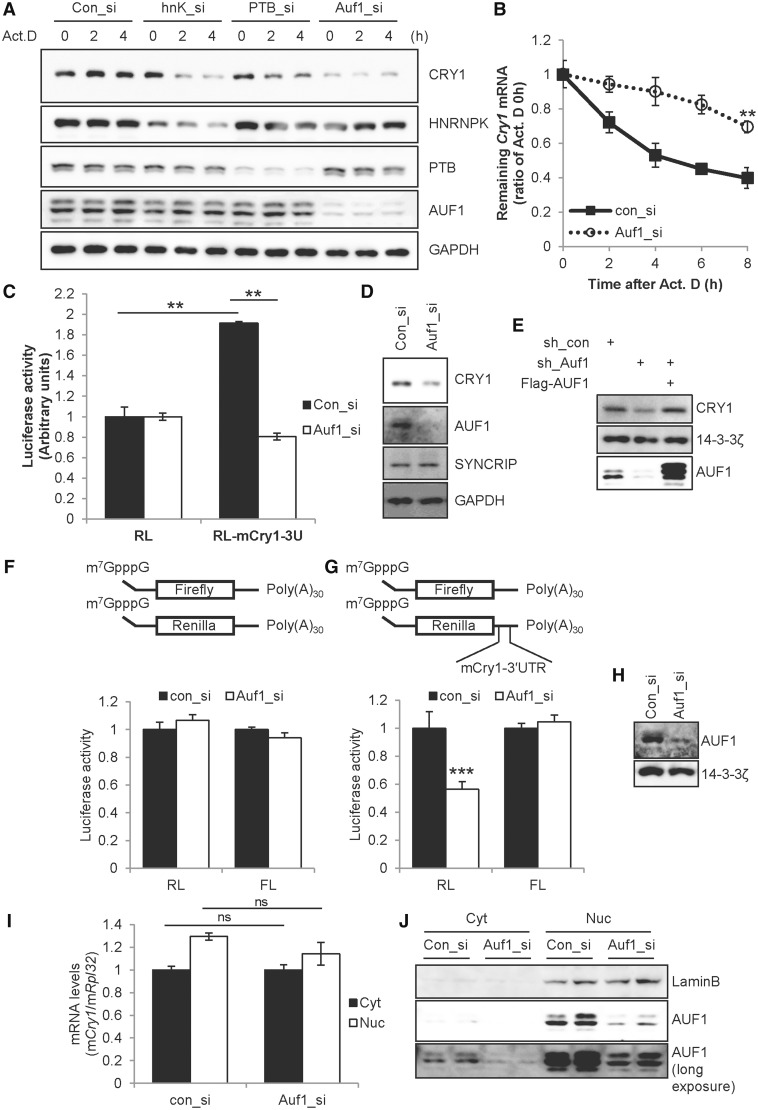
Post-transcriptional regulation such as mRNA transport, mRNA degradation and translation initiation is mainly regulated by RNA-binding proteins such as heterogeneous nuclear ribonucleoproteins (HNRNPs). To identify important RNA-binding proteins for Cry1 regulation, many HNRNPs were knocked down, and CRY1 protein levels were checked with Act.D treatment to exclude the transcriptional step. Among many RNA-binding proteins, knock-down of HnrnpK, HnrnpR, polypyrimidine tract binding protein 1 (Ptbp1) and Auf1 was effective against CRY1 expression, but not in the case of HnrnpA1 knock-down, comparing with the control siRNA (con_si)-transfected condition (Figure 2A and Supplementary Figure S2A). Knock-down of Syncrip (HnrnpQ) using siRNA (hnQ_si) also reduced CRY1 protein levels (Supplementary Figure S2B). Although all siRNAs for HnrnpK (hnK_si), Ptbp1 (PTB_si), Auf1 (Auf1_si) and Syncrip reduced CRY1 protein compared with the con_si-transfected condition, the knock-down of Auf1 was most effective. Indeed, we confirmed the function of AUF1 in Cry1 mRNA degradation: knock-down of Auf1 stabilized Cry1 mRNA (Figure 2B). Similar to the results shown in Figure 1C and D, Auf1 knock-down stabilized Cry1 mRNA but dramatically reduced CRY1 protein levels. This contradictory effect on mRNA and protein levels by AUF1 was confirmed using the reporter system. Knock-down of Auf1 decreased reporter activity that was elevated by Cry1-3′UTR, whereas Auf1 knock-down could not elicit a significant effect on the control reporter RL lacking the 3′UTR (Figure 2C). We confirmed the knock-down of Auf1 by immunoblotting (Figure 2D). Knock-down of Auf1 decreased not only the reporter activity of Cry1-3′UTR but also endogenous CRY1 protein levels. We also confirmed the function of AUF1 by knock-down and rescue experiment. Transfection of short hairpin RNA (shRNA) that targets 3′UTR of Auf1 (sh_Auf1) reduced CRY1 protein levels, but overexpression of Flag-AUF1 rescued CRY1 protein levels in sh_Auf1 transfected condition (Figure 2E). To exclude any transcriptional effect of Cry1-3′UTR, we generated reporter mRNA constructs containing a cap structure and poly(A)30 with or without the 3′UTR. The Firefly luciferase reporter mRNAs were used for transfection control. Regarding the control reporter mRNA construct, luciferase activity was not changed with knock-down of Auf1 (Figure 2F). By contrast, the reporter mRNA harbouring Cry1-3′UTR showed decreased translation under Auf1-reduced conditions (Figure 2G). We also confirmed knock-down of Auf1 as shown in Figure 2F–H). To exclude the possibility that the reduced translation under the Auf1 knocked-down condition originated from changes in mRNA transport or shuttling, we checked cytoplasmic and nuclear Cry1 mRNA levels under con_si- or Auf1_si-transfected conditions. We confirmed Auf1 knock-down and the obtained mRNA fraction (Figure 2J). We observed no alteration of the Cry1 mRNA fraction whether or not AUF1 was reduced (Figure 2I). Likewise, knock-down of Auf1 did not change the protein stability of CRY1 (data not shown). The results suggest that AUF1 accelerates CRY1 translation by acting on the 3′UTR.
Figure 2.
AUF1 regulates translation of Cry1. (A) Control siRNA (Con_si) or gene-specific siRNAs for HnrnpK (hnK_si), Ptbp1 (PTB_si) or Auf1 (Auf1_si) were transfected into NIH 3T3 cells. After a 12-h incubation, cells were treated with Act.D, and were harvested at the indicated time points and subjected to immunoblotting. (B) By using the same samples as those used in panel A, mRNA levels were quantified by real-time PCR with Cry1- or Rpl32-specific primers. Cry1 levels were normalized to levels of Rpl32, and the zero time point was set to 1 (n = 3, **P = 0.0033). (C) RL or RL-Cry1-3U reporter plasmids were co-transfected with control siRNA or Auf1_si into NIH 3T3 cells, and were subjected to the luciferase assay. The value of RL with con_si was set to 1 (n = 3, **P < 0.001). (D) Knock-down of Auf1 shown in panel C was confirmed by immunoblotting with indicated antibodies. (E) Plasmids that express control (sh_con) or Auf1 targeting (sh_Auf1) shRNA were transfected with Flag-tagged all AUF1 isoforms into NIH 3T3 cells. After 24-h incubation, cells were subjected to immunoblotting. (F and G) Schematic diagram of the mRNA reporter of Cry1-3′UTR shows 7-methyl-guanosine (m7G) and the 30-nt-long poly(A) tail [poly(A)30]. Firefly mRNA reporters for normalization were also used. Con_si or Auf1_si was transfected into NIH 3T3 cells and incubated for 12-h. Subsequently, cells were transfected with mRNA reporters harbouring Cry1-3′UTR or no UTR. After a 6-h incubation, cells were subjected to the luciferase assay (n = 3, P = 0.0001). (H) Immunoblotting was performed with specific antibodies using cell extracts used in panel F or G. (I) NIH 3T3 cells transfected with Con_si or Auf1_si was incubated for 12 h, and were fractionated into cytoplasmic and nuclear parts. mRNA levels were quantified by real-time PCR using Cry1- or Rpl32-specific primers. (J) Immunoblotting was performed with samples used in panel I. Lamin B expression was checked to verify fractionation.
AUF1 specifically binds to Cry1-3′UTR
Although the 3′UTR is an important element for translational modulation, the 5′UTR is generally considered to be the region for translational regulation with its binding proteins. Internal ribosome entry site (IRES) elements and IRES trans-acting factors (ITAFs) are important translational regulatory factors in 5′UTR-mediated translational modulation (3,32–35). Indeed, it was reported that AUF1 functions as an ITAF and activates IRES-mediated translation in a 5′UTR-dependent manner (30). We confirmed the IRES activity of Cry1-5′UTR to investigate whether the dynamic post-transcriptional regulation of Cry1 is modulated by the 5′UTR. To investigate the existence of an IRES in Cry1 mRNA, we inserted the 5′UTR of mouse Cry1 into a bicistronic reporter vector (3) (Supplementary Figure S3A). The vector produces a bicistronic RNA encoding Rluc in the first cistron and firefly luciferase (Fluc) in the second cistron. The translation of Rluc is served by cap-dependent translation, whereas the translation of the second cistron Fluc reflects the IRES activity of the inserted 5′UTR sequences. As previously reported, the 5′UTR of mouse Period homolog 1 (Per1) showed IRES activity (3); however, Cry1-5′UTR showed no IRES activity compared with the pRF control (Supplementary Figure S3B). From these results, we could exclude the 5′UTR for IRES-mediated translation of Cry1.
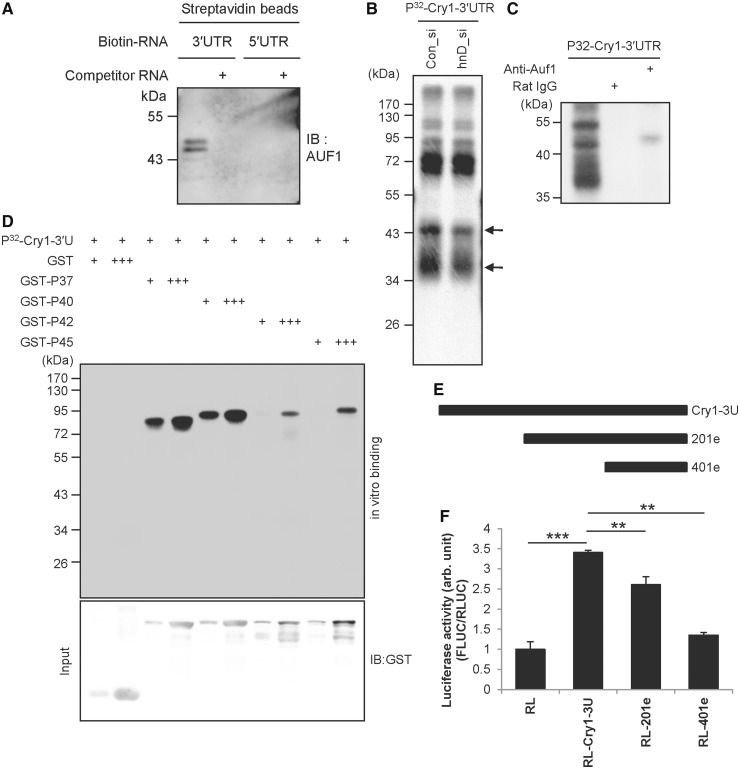
If AUF1 regulates translation of Cry1, 5′UTR-mediated translational regulation via AUF1 cannot be excluded. We checked the binding between the 3′ or 5′ UTR of Cry1 and AUF1 via biotin-RNA affinity precipitation. AUF1 was co-precipitated with biotin-labelled Cry1-3′UTR (Figure 3A). AUF1 bound to Cry1-3′UTR was dramatically decreased by non-labelled cold Cry1-3′UTR competitor RNA. Regarding Cry1-5′UTR, we observed no AUF1 binding. The results indicate that AUF1 specifically binds to Cry1-3′UTR but not to Cry1-5′UTR. We also checked other RNA-binding proteins to confirm the findings presented in Figure 3A. The binding of PTBP1, a known regulator of translation and splicing, was specific for Cry1-5′UTR but not for Cry1-3′UTR (Supplementary Figure S4A). Additionally, SYNCRIP (synaptotagmin binding, cytoplasmic RNA interacting protein), which can also reduce CRY1 protein levels, more specifically bound to Cry1-3′UTR than to Cry1-5′UTR (Supplementary Figure S4B). We also confirmed the binding of AUF1 to Cry1-3′UTR using a UV-crosslinking assay because UV-crosslinking detects only direct binding between RNA and protein: AUF1 was specifically bound to Cry1-3′UTR (Figure 3B). We confirmed the AUF1 band using an AUF1 knock-down approach. In UV-crosslinking, very small amount of protein can be detected. It means that knock-down effect is hardly visible in UV-crosslinking assay. As there are more AUF1 in the nucleus than cytoplasm, we could observe more dramatic difference of AUF1 intensity, so we used nuclear protein extract. When we used Auf1 knocked-down extract, the AUF1 band was decreased (Figure 3C, arrow). To confirm the binding of AUF1 to Cry1 3′UTR, immunoprecipitation of UV cross-linked proteins with radiolabelled in vitro-transcribed Cry1-3′UTR RNA was performed using anti-AUF1 antibody. AUF1 was immunoprecipitated from the UV cross-linked proteins by its specific antibody (Figure 3C, lane 3). No bands were detected when control IgG was used as a negative control (Figure 3C, lane 2). AUF1 is one of the ubiquitously expressed proteins that specifically binds to AU-rich RNA destabilizing elements (AREs) (36). AUF1 has four isoforms of different molecular weights (P37, P40, P42 and P45), all of which are generated by alternative precursor mRNA splicing (37,38) and shuttle between the nucleus and cytoplasm (39,40). Additionally, to confirm the direct binding of each AUF1 isoform to Cry1-3′UTR, purified recombinant GST-AUF1 was used in UV-crosslinking. GST-P37, GST-P40, GST-P42 and GST-P45 specifically bound to Cry1-3′UTR in a concentration-dependent manner, but GST did not (Figure 3D). These results suggest that AUF1 directly binds to Cry1-3′UTR but not to Cry1-5′UTR. This means that knock-down of Auf1 reduces CRY1 protein in a 3′UTR-dependent manner, and we could exclude the possibility of AUF1 binding to Cry1-5′UTR.
Figure 3.
AUF1 specifically binds to Cry1-3′UTR. (A) The in vitro transcribed Cry1 3′UTR or 5′UTR constructs were labelled with biotin-UTP and incubated with NIH 3T3 cell cytoplasmic extract. Streptavidin-affinity purified samples were separated by SDS-PAGE and subjected to immunoblotting with anti-AUF1. Abundant AUF1 was detected in the reaction with biotin-labelled mRNA. AUF1 binding decreased in the presence of 5-fold excess of non-labelled 3′UTR mRNA. (B) Radiolabelled 3′UTR of Cry1 was transcribed in vitro and subjected to in vitro binding and UV-crosslinking with nuclear extracts of Con_si- or Auf1_si-transfected NIH 3T3 cells. Samples were separated by SDS-PAGE for autoradiography. (C) Cytoplasmic extracts labelled by UV cross-linking with radiolabelled 3′UTR of mCry1 were subjected to immunoprecipitation with AUF1-specific antibody or normal Rat IgG as a control and then separated by SDS-PAGE for autoradiography. (D) In vitro transcribed 3′UTR was subjected to in vitro binding and UV-crosslinking assay with purified GST-tagged AUF1 isoforms (GST-P37, GST-P40, GST-P42 and GST-P45), and autoradiographic intensities were checked. In the lower panel, input levels were checked by immunoblotting with anti-GST. (E) Schematic diagram of the serially deleted mutation strategy. (F) Each deletion construct derived from full-length Cry1-3′UTR was transfected into NIH 3T3 cells, and luciferase assays were performed. The graph shows the relative luciferase activity derived from the RLUC/FLUC ratio (n = 3, ***P < 0.0001, **P < 0.001).
To determine the cis-acting element of the Cry1-3′UTR that is responsible for 3′UTR-mediated translation, we generated reporter constructs containing a truncated 3′UTR of Cry1 mRNA (Figure 3E and Supplementary Figure S5A). Full-length Cry1-3′UTR showed elevated reporter activity; however, the RL-201e reporter harbouring a deletion of the first 200 nts of Cry1-3′UTR showed diminished activity compared with full-length Cry1-3′UTR (Figure 3F). RL-401e, the shortest form of Cry1-3′UTR, showed dramatically reduced translation activity similar to the control RL reporter that does not harbour 3′UTR sequences. We assumed that AUF1 might bind to the cis-acting element for 3′UTR-mediated translation, and the binding pattern of AUF1 to Cry1-3′UTR might reflect reporter activity shown in Figure 3F. To verify our hypothesis, in vitro binding between [32P]UTP-labelled Cry1-3′UTRs and GST-fused recombinant AUF1 proteins was performed. Full-length Cry1-3′UTR showed a strong affinity to GST-AUF1 proteins, whereas RL-201e showed decreased binding affinity compared with the full-length construct (Supplementary Figure S5B). The binding between GST-AUF1s and the shortest Cry1-3′UTR (RL-401e) was dramatically decreased. We also made the 3′-truncated 3′UTR of Cry1, 1-250 construct (Supplementary Figure S5A). With 1-250 construct, AUF1 binding was increased relative to the 401e construct (Supplementary Figure S5C and D). The reporter activity of 1-250 was also increased relative to 401e construct (Supplementary Figure S5E). These results suggest that AUF1 directly binds to multiple AREs in the Cry1 3′UTR, and the number of AUF1 molecules bound to Cry1 mRNA determines the strength of translational activation.
AUF1 interacts with translation initiation factors
Because AUF1 has four isoforms of different molecular weights, we checked the function of each AUF1 isoform in Cry1-3′UTR-mediated translation. Overexpression of each AUF1 isoform elevated endogenous CRY1 protein levels (Supplementary Figure S6A). To clarify the function of each AUF1 isoform, Auf1 was knocked down by shRNA targeting 3′UTR of Auf1 and individual AUF1 isoforms were overexpressed. All isoform of AUF1 slightly increased CRY1 expression (Supplementary Figure S6B). From these results, we suggest that all AUF1 isoforms have functional roles in 3′UTR-mediated Cry1 translation.
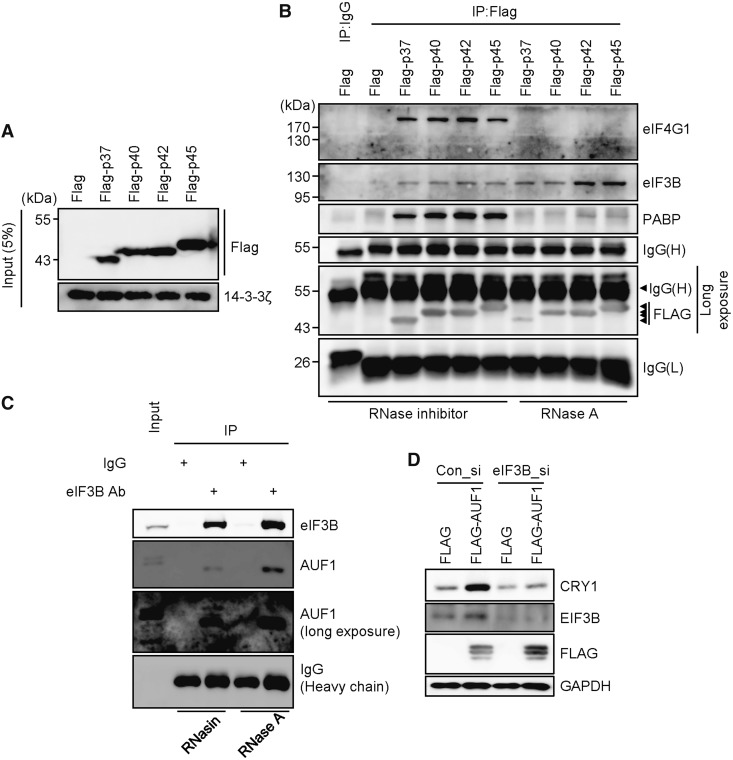
We want to know how AUF1 regulates translation of Cry1 in a 3′UTR-dependent manner because general translation is mainly regulated by the 5′UTR, not by the 3′UTR. We hypothesized that the 3′UTR-bound AUF1 recruits translation-related factors, and accelerates translation efficiency. To test this, we checked protein–protein interactions between AUF1 and translation-related factors. Flag-tagged AUF1 (Flag-AUF1) isoforms were overexpressed (Figure 4A). Flag-AUF1s were immunoprecipitated, and co-immunoprecipitated proteins, particularly translation initiation-related proteins, were evaluated. Interestingly, PABP and translation initiation-related factors EIF4G1 and EIF3B were co-immunoprecipitated with AUF1 (Figure 4B). Regarding eIF4G1 and PABP, interactions with AUF1 were strong under RNase-inhibited conditions. However, RNase A treatment decreased their binding affinity. Conversely, EIF3B still showed a strong affinity to the AUF1 even in the absence of RNA. To verify the interaction between AUF1 and EIF3B under endogenous conditions, EIF3B was immunoprecipitated using a specific antibody. We observed co-immunoprecipitated AUF1 in NIH 3T3 cells regardless of the presence of RNA (Figure 4C). We also observed the interaction between AUF1 and EIF3B in HEK 293A cells (Supplementary Figure S7). These data suggest that the interaction between AUF1 and EIF3B is a protein–protein interaction but not an RNA-mediated indirect interaction. We also overexpressed AUF1, and it increased CRY1 protein levels. But this increase of CRY1 expression was prevented by silencing EIF3B (Figure 4D). From these results, we suggest that AUF1 is involved in the translation machinery.
Figure 4.
AUF1 interacts with translation initiation factors. (A) NIH 3T3 cells were transiently transfected with Flag-tagged AUF1 isoforms Flag-P37, Flag-P40, Flag-P42 and Flag-P45. After a 24-h incubation, cells were subjected to immunoblotting. (B) With the same samples used in panel A, immunoprecipitation was performed with a Flag-specific antibody under RNA-containing or RNA-free conditions, and samples were subjected to immunoblotting. (C) Untransfected NIH 3T3 cells were subjected to immunoprecipitation with anti-EIF3B after RNase inhibitor or RNase A treatment. Immunoblotting was performed with specific antibodies. (D) Con_si or eIF3B targeting siRNAs (eIF3B_si) were transfected with Flag or all Flag-AUF1 isoforms into NIH 3T3 cells. After 24-h incubation, cells were subjected to immunoblotting with annotated antibodies.
AUF1 directly interacts with ribosomal proteins
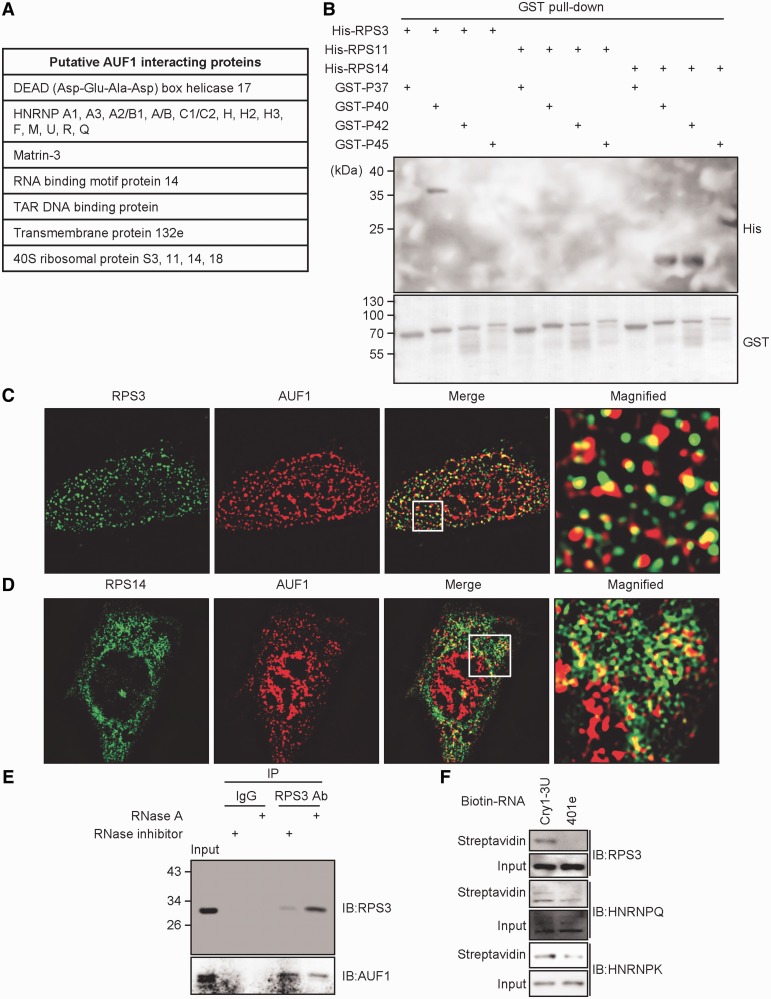
To identify AUF1-binding partners related to the translational process more objectively and comprehensively, Flag-tagged AUF1 isoforms were overexpressed and immunoprecipitated with the Flag antibody, and samples were analysed using the LTQ-orbitrap. Orbitrap analysis, the advanced mass spectrometry that uses orbital trapping of ionized protein fragments, can overcome resolution limit and provide us an accurate binding candidates compared with conventional MALDI-TOF mass spectrometry. From the data of the LTQ-orbitrap, we identified other HNRNPs as AUF1-binding proteins in an RNA-independent manner (Figure 5A). Interestingly, several ribosomal proteins, particularly 40S ribosomal proteins, were also identified as AUF1-binding proteins regardless of the presence of RNA. To verify the interaction between AUF1 and 40S ribosomal proteins identified by the LTQ-orbitrap, His-tagged recombinant ribosomal protein S3 (RPS3), ribosomal protein S11 (RPS11), ribosomal protein S14 (RPS14) and ribosomal protein S18 (RPS18) were prepared. RPS3, RPS11, RPS14 and RPS18 proteins are located on the solvent side of the 40S subunit, which have more accessible surface area to other proteins (41,42). After incubation with His-fused 40S ribosomal proteins and GST-fused AUF1, protein complexes were pulled-down using GST pull-down assays. When all GST-fused AUF1 isoforms were precipitated, His-fused RPS3 and RPS14 proteins were co-precipitated with AUF1 protein (Figure 5B; Supplementary Figure S8A–C). Regarding RPS11, His-RPS11-purified protein was not co-precipitated with AUF1. His-RPS18 recombinant protein also did not interact with AUF1 (data not shown). Taken together, these data indicate that AUF1 directly associates with the 40S ribosomal subunit through protein–protein interactions. However, AUF1 did not interact with all 40S ribosomal proteins; it interacted selectively.
Figure 5.
AUF1 directly interacts with ribosomal proteins. (A) The table for putative AUF1 interacting proteins was made base on the LTQ-orbitrap data. The full LTQ-orbitrap data file is in the Supplemental material. (B) Recombinant proteins of GST-AUF1 isoforms, His-RPS3, His-RPS11 or His-RPS14 were incubated and pulled down using GST-binding resins. Two micrograms of Histidine tag-fused RPS proteins or GST-fused AUF1 isoforms were applied. Each protein separated by SDS-PAGE was detected by the indicated antibodies. (C and D) Immunostaining was performed with AUF1-, RPS3-, or RPS14-specific antibodies. RPS3 or RPS14 was visualized using Alexa 488-conjugated secondary antibody. For AUF1, Alexa 594-conjugated secondary antibody was used for visualization with a super-resolution illumination microscopy (SIM). (E) NIH 3T3 cells were subjected to immunoprecipitation under RNA-free or RNA-containing conditions with RPS3-specific antibody, and samples were subjected to immunoblotting with indicated antibodies. (F) The in vitro-transcribed full-length or 401e with a truncated Cry1-3′UTR were labelled with biotin-UTP and incubated with NIH 3T3 cell cytoplasmic extract. Streptavidin-affinity purified samples were subjected to immunoblotting with indicated antibodies.
We also visualized the cellular interaction and co-localization of AUF1 with RPS3 or RPS14 by super-resolution microscopy using high-frequency SIM. AUF1 was detected to be co-localized with RPS3 or RPS14 (Figure 5C and D). In SIM-super-resolution images, not all AUF1 co-localized with RPS3 or RPS14, but some of AUF1 co-localized with RPS3 or RPS14. These data and previously shown data suggest that some portion of AUF1 that bound to specific target mRNA may interact with 40S ribosomal proteins. To verify the protein–protein interaction between AUF1 and 40S ribosomal proteins in cells, immunoprecipitation was performed using the RPS3 antibody with untransfected cell extracts. AUF1 proteins were co-precipitated with RPS3 under RNA-containing or RNA-free conditions (Figure 5E). Because the 401e construct, a deletion construct of Cry1-3′UTR, showed a dramatic decrease in AUF1 binding, we also checked the binding of RPS3 to the 401e-3′UTR construct. RPS3 appeared to bind to full-length Cry1-3′UTR but not to 401e-3′UTR Cry1 mRNA (Figure 5F). In the case of HNRNPK or SYNCRIP, the binding affinity to 401e-3′UTR was slightly reduced compared with full-length Cry1-3′UTR. These results suggest that AUF1 forms a complex with RPS3 under endogenous conditions, and RPS3 binding to Cry1-3′UTR may be mediated by AUF1. Because eukaryotic RPS3 is a highly conserved protein located on the solvent side of the 40S subunit on the beak of the head region (42), the protein–protein interaction between AUF1 and RPS3 may be reasonable. Taken together, we suggest that 3′UTR-bound AUF1 recruits 40S ribosomal proteins, and these interactions may activate 3′UTR-mediated translation.
Rhythmic cytoplasmic AUF1 regulates time-dependent CRY1 translation
We showed that AUF1 regulates translation of Cry1 in 3′UTR-dependent manner. Reporter mRNA transfection was also performed to exclude transcriptional regulation, and reporter mRNA harbouring Cry1-3′UTR only showed reduced reporter activity under AUF1-decreased conditions (Figure 2G). To verify the function of Cry1-3′UTR-mediated translational activation under physiological condition of circadian rhythm, dexamethasone was applied to achieve synchronization of circadian time in NIH 3T3 cells. When we transiently transfected reporter mRNAs to dexamethasone-treated cells at certain time intervals, the translational activity of reporter mRNAs harbouring Cry1-3′UTR seemed to be regulated rhythmically but not control reporter mRNAs lacking the 3′UTR (Figure 6A and B). In addition, the time of translation activity of Cry1-3′UTR was correlated with endogenous the CRY1 protein expression time (Figures 1A and 7B and C). The results suggest that Cry1-3′UTR activates translation of Cry1 rhythmically. To clarify the rhythmic translation of Cry1 mRNA, we studied polysome fractionation analysis. The overall profiles of ribosomes in sucrose gradient analyses were not altered by circadian time (Figure 6C, top panel). This was reflected in the levels of a control Tbp mRNA; the distribution pattern of Tbp mRNA was not changed by dexamethasone treatment (Figure 6C, third from top panel). However, a shift of Cry1 mRNA between heavy polysome and light polysome, reflecting a time-dependent translation of Cry1 mRNA, was observed in cells treated with dexamethasone (Figure 6C, second from top panel). To explain the rhythmic translation of Cry1, which occurred in a 3′UTR-dependent manner, we assumed the AUF1 level may follow the CRY1 protein profile; however, the total level of AUF1 was relatively constant (Figure 7B and Supplementary Figure S9A). We speculated whether the cytoplasmic AUF1 levels could be rhythmic, as translation occurs in the cytoplasm. To test our hypothesis, we harvested synchronized NIH 3T3 cells at indicated time points, and cytoplasmic and nuclear fractionated extracts were obtained. From these extracts, we checked the AUF1 expression pattern. As correlated with our hypothesis, cytoplasmic AUF1 showed rhythmicity in a time-dependent manner (Figure 6D). Indeed, the cytoplasmic AUF1 profile was matched to the CRY1 protein level. To further investigate this finding, we examined the interaction between cytoplasmic AUF1 and Cry1-3′UTR. We performed a biotin-labelled RNA pull-down assay with dexamethasone-treated cytoplasmic cell extracts. The binding affinity was correlated with the CRY1 protein phase (Figure 6E). We also checked if the interaction between AUF1 and endogenous Cry1 mRNA is rhythmic by using RNP immunoprecipitation and reverse-transcription assays. As cytosolic AUF1 showed rhythmic expression (Figure 6D), input lane of cytosolic AUF1 also showed circadian time-dependent expression (Figure 6F). The immunoprecipitated AUF1 levels were rhythmic during their respective circadian time frames, and co-immunoprecipitated mCry1 mRNA with AUF1 changed with dexamethasone treatment time (Figure 6G). Therefore, the results suggest that rhythmic cytoplasmic AUF1 expression leads to its rhythmic binding to the 3′UTR of Cry1 mRNA and subsequently triggers time-dependent 3′UTR-mediated translation.
Figure 6.
Rhythmic cytoplasmic AUF1 regulates time-dependent Cry1 translation. (A) NIH 3T3 cells were treated with dexamethasone (Dex), and the mRNA reporters lacking 3′UTR sequences were transiently transfected for 6 h at the indicated times, followed by measurement of luciferase activity. The relative values at 4–10 h were set to 1. (B) The mRNA reporters harbouring Cry1-3′UTR were transfected into Dex-treated NIH 3T3 cells (n = 4, P = 0.0097). (C) NIH 3T3 cells were treated with dexamethasone. After 12, 24, or 36 h of incubation, the cells were treated with cycloheximide. Then, the ribosomal distributions in sucrose density gradients were analysed in cell extracts (upper row). RNA samples were purified from fractions in the sucrose gradient. The amounts of mCry1 mRNA (middle row) and Tbp mRNA (bottom row) across the gradient were analysed by real-time PCR, and the relative amounts of RNA in each fraction are depicted by corresponding bars in the graphs. (D) NIH 3T3 cells were treated with 100 nM Dex and harvested at the indicated times, and cytoplasmic or nuclear extract was prepared. Next, immunoblotting was performed with specific antibodies. (E) NIH 3T3 cells were treated with Dex and harvested at the indicated times, and cytoplasmic extracts were prepared. Dex-treated cytoplasmic extracts were incubated with biotin-labelled Cry1-3′UTR, and samples were subjected to immunoblotting. (F) NIH 3T3 cells were treated with dexamethasone, and cytosolic extracts were prepared. Immunoprecipitation was performed using anti-AUF1 antibody and normal Rat IgG as a control. (G) The co-immunoprecipitated mRNAs with AUF1 shown in panel F were analysed by real-time PCR. AUF1-bound Cry1 mRNA levels were normalized to the levels of background Gapdh mRNA. The relative Cry1 mRNA level that immunoprecipitated with IgG at 0-h was set to 1.
Figure 7.
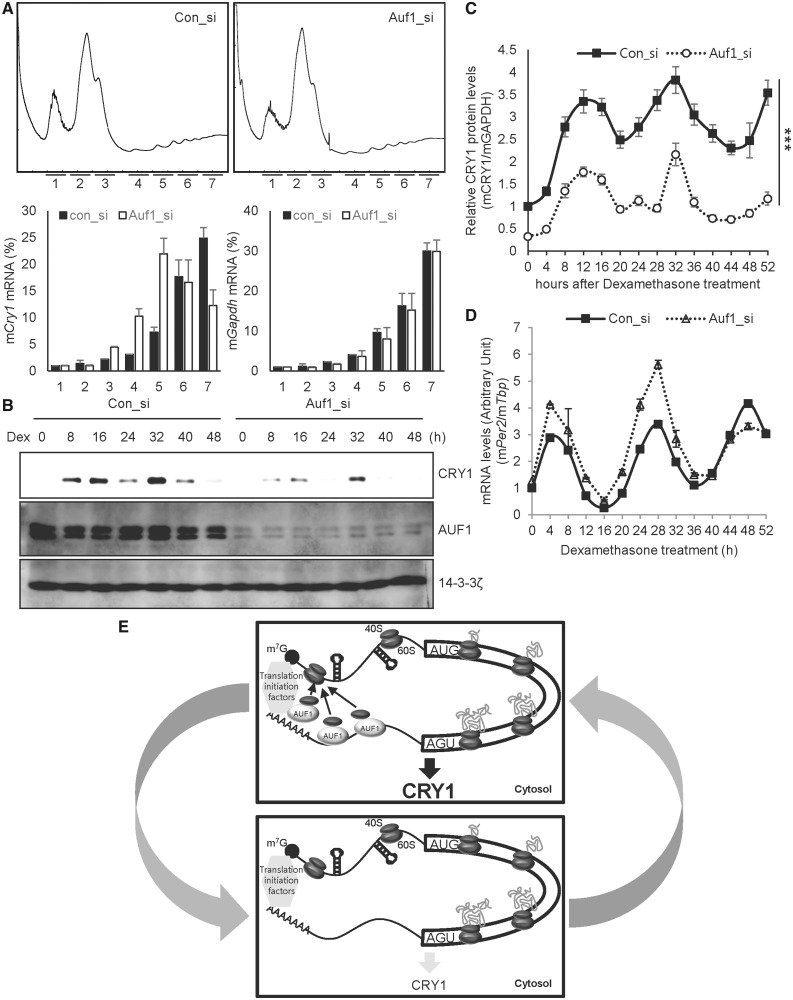
AUF1 regulates circadian expression of CRY1. (A) NIH cells were transfected with control (Con_si) or Auf1-specific siRNA (Auf1_si). After 24 h of incubation, the cells were treated with cycloheximide. Then, the ribosomal distribution in sucrose gradients were analysed in cell extracts (first row). Distribution of mRNA in sucrose gradients were analysed by real-time PCR, and the amounts of RNA in each fraction are depicted by corresponding bars in the graphs. (B) NIH 3T3 cells transfected with siRNAs targeting Auf1 (Auf1_si) or control siRNA (con_si) by microporation were incubated for 12 h and harvested at the indicated time points after treatment with Dex. Immunoblotting was performed with the indicated antibodies. (C) The relative mCRY1 levels in con_si transfected (closed squares/solid line) and Auf1_si transfected (open circles/dotted line) NIH 3T3 cells. mCRY1 protein levels were normalized to GAPDH and plotted. The P-value of variance between con_si and Auf1_si group was 0.0001. mCRY1 protein levels were significantly different between con_si and Auf1_si transfected groups at all dexamethasone treatment time points (P < 0.001). (D) Total RNA was prepared from the harvested cells as shown in panel C, and reverse transcription and real-time PCR were performed using specific primers (con_si vs. Auf1_si, P < 0.0001). mPer2 mRNA levels were significantly different between the con_si and Auf1_si-transfected groups (4, 20, 24, 28, 32, 48 h, P < 0.001; 12 h, P < 0.01; n = 3). (E) The upper box shows that increased cytoplasmic AUF1 binds to the 3′UTR of Cry1 mRNA. Ribosomal proteins, particularly the 40S ribosomal subunit that is released from termination codon UGA, would be recruited by AUF1, which can also associate with translation initiation factors. AUF1 may accelerate the reuse of ribosomal proteins or recruitment of the 40S ribosomal subunit to the 5′ end of Cry1 mRNA, which increases translation efficiency and CRY1 expression. The bottom box shows that the interaction between AUF1 and Cry1-3′UTR decreases because of reduced cytoplasmic AUF1 levels. Consequently, AUF1 would not recruit 40S ribosomal proteins to the 5′ end of mRNA, thus decreasing the translation efficiency and CRY1 expression.
AUF1 regulates circadian expression of CRY1
Auf1 knock-down decreased the activity of the reporter harbouring Cry1-3′UTR along with CRY1 protein levels; however, overexpression of AUF1 up-regulated Cry1-3′UTR reporter activity and CRY1 protein amounts (Figure 2A, C and D). We also studied the distribution change of Cry1 mRNA on polysome fraction, depending on the levels of AUF1. The overall profiles of ribosomes were not changed by decreased AUF1 as another previous paper reported (30) (Figure 7A, upper panel). On the other hand, a shift of Cry1 mRNA from heavy to light polysome fraction, reflecting a reduction in mCry1 mRNA translation, was observed in cells transfected with siRNA against Auf1 (Figure 7A, lower panel). To analyse the physiological role of 3′UTR-mediated translation of Cry1 by AUF1 in circadian expression, we used the knock-down approach in NIH 3T3 cells. Reduction of AUF1 resulted in lower CRY1 protein levels, suggesting that AUF1 is critical for Cry1 mRNA translation (Figure 7B and C). We also checked whether the decreased level of AUF1 changes the CRY1 expression phase. Knock-down of Auf1 did not alter the circadian expression phase of CRY1 (Supplementary Figure S9A). Regarding the Cry1 mRNA profile, in contrast to CRY1 protein oscillation, knock-down of Auf1 slightly increased Cry1 mRNA levels (Supplementary Figure S9C). From these results, we suggest that AUF1 regulates translation of Cry1, and it alters the circadian amplitude of CRY1. Furthermore, these data imply that decreased expression of CRY1 protein owing to Auf1 knock-down is mediated by reduced translational activity rather than transcriptional or other post-transcriptional modulation. Cry1 is a well-established rhythmically expressed gene, and thus we examined circadian oscillation of other clock genes under the condition of Auf1 knock-down to examine the effect of AUF1 in further detail. A decrease in the AUF1 level increased mouse Period2 (mPer2) mRNA levels, particularly at the peak time (Figure 7D). Knock-down of Auf1 gradually decreased mouse nuclear receptor subfamily 1 group D member 1 (Nr1d1), also known as Rev-erb alpha, mRNA levels (Supplementary Figure S9B). Reduction of AUF1 also changed oscillation patterns of mouse D site albumin binding protein (Dbp) mRNAs (Supplementary Figure S9D). Taken together, these results imply that the 3′UTR-dependent translational activation by AUF1 allows robust expression of CRY1 in relation to the circadian period, and it may have fine tuning effect on the circadian system.
DISCUSSION
Unexpectedly, we found that a reporter harbouring Cry1-3′UTR showed increased reporter translation activity. Knock-down of Auf1 decreased CRY1 protein levels. Although the main function of AUF1 is suggested to be acceleration of the degradation of diverse mRNAs by binding to AREs in the 3′UTR, some reports have describe the function of AUF1 in translational regulation such as in increasing IRES-mediated translation or translation-coupled mRNA degradation (30,43). However, little mechanistic understanding exists regarding about AUF1-dependent translational activation via 3′UTR (30,44–46). To elucidate the role of AUF1 in translation related to the circadian system, we checked AUF1-interacting proteins and found that AUF1 directly interacts with RPS3 or RPS14. AUF1 also interacted with translation initiation factors, particularly EIF3B. Because AUF1 was also reported to be an ITAF (30), our finding regarding a direct interaction between AUF1 and ribosomal proteins or translation initiation factors may provide clues for the IRES-mediated translational mechanism that is regulated by AUF1 and may elucidate the role of AUF1 in the elaborate translational system. Indeed, along with the ternary complex of PABP, cap-binding protein EIF4E, and EIF4G, the interaction between AUF1 and translation initiation factor EIF3B may circularize mRNAs and subsequently accelerate translation efficiency.
In this study, all isoforms of AUF1 were detected in immunoblotting or immunocytochemistry. These isoforms are usually in the same protein complex. We found that p37, p40, p42 and p45 directly interacted with Cry1 3′UTR as shown in Figure 3B–D. Indeed, AUF1 interacted with endogenous Cry1 mRNA (Figure 6F and G). From these results, all isoforms of AUF1 can interact with Cry1-3′UTR. Because AUF1 has four isoforms, we wanted to determine which isoform was important for 3′UTR-mediated translational activation. RPS3 strongly interacted with P40 isoforms compared with other AUF1 isoforms. Conversely, RPS14 showed strong affinity to P40 or P42 compared with other isoforms (Figure 5B and Supplementary Figure S8C). Indeed, RPS3 or RPS14 showed co-localization with AUF1, but only some of the AUF1 co-localized with RPS3 or RPS14. AUF1 isoforms may have differential interacting protein partners, which elicit different a translational function. Further study of interacting proteins of each AUF1 isoform will help to elucidate the role of AUF1 in translational activation. But the functions of each AUF1-interacting partners are related to translation. Indeed, AUF1 isoforms are mainly in the same protein complex; the function of AUF1 is needed to consider in the view of complex of AUF1 isoforms. So we think that AUF1 directly binds to Cry1-3′UTR and may recruit translational systems.
We believe that not all mRNAs are regulated by AUF1-mediated translation, but some portions of mRNAs are regulated by 3′UTR-dependent and AUF1-dependent translational activation. Knock-down of Auf1 slightly changed mRNA expression of clock genes. Nevertheless, knock-down of Auf1 did not lead to increasing Per2 mRNA after 40 h of Dex treatment (Figure 7D). Indeed, the first cycle and second cycle of Dbp mRNA are different (Supplementary Figure S9D). Circadian system possesses diverse adjustable regulation mechanisms, such as transcriptional, post-transcriptional, translational, and post-translational regulation. Moreover, there exist several factors such as clock gene homologs and another feedback loops, D-box loop and RORE loop, for circadian robustness. We think that the difference of other clock genes’ amplitude among the cycles may be originated from the tendency of compensation. But in this study we don’t know whether AUF1 directly regulates other clock genes. To clarify the function of AUF1 in global circadian system, further study is needed.
As shown in Supplementary Figures S2B and S4B, SYNCRIP also bound to only Cry1-3′UTR, and knock-down of Syncrip reduced CRY1 protein levels like AUF1. Indeed, SYNCRIP also directly interacted with 40S ribosomal proteins (47). Further study regarding SYNCRIP, AUF1 and other RNA-binding proteins may elucidate the concerted actions of 3′UTR-binding proteins and clarify the sophisticated mechanisms of 3′UTR-dependent translational activation.
RNA-binding proteins function as multiplayers. SYNCRIP was shown to rhythmically accelerate mRNA degradation of AANAT mRNA (28). In other cases, SYNCRIP functioned as an ITAF and increased IRES-mediated translation of target genes (3,27). PTBP1 also activates mRNA degradation (25) but increases translation of specific mRNA (48). These RNA-binding proteins generally bind to the UTR region, and many RNA-binding proteins can bind to UTR simultaneously. We think that multiple functions of RNA-binding proteins originate within the context of proteins bound to the target RNA. To clearly demonstrate the function of RNA-binding proteins, the context of interacting proteins and subsequent recruitment of functional machinery, such as the exosome or translation processing factors, must be considered. Further investigation to characterize functional roles of mRNA-binding proteins may clarify the RNA-binding protein-mediated translational regulation.
Although cytoplasmic AUF1 shows rhythmic expression, the effect of AUF1 on mCry1 mRNA binding appears stronger. Several signalling systems may modulate post-translational modifications of AUF1. Phosphorylation of AUF1 is altered concomitant with changes in RNA-binding activity and stability of ARE-containing mRNAs (49,50). Indeed, methylation of AUF1 may be essential for protein function (51). We think that post-translational modification of AUF1 may also have an effect on the RNA binding affinity of AUF1, and this may lead to discrepancy between expression of AUF1 and mRNA binding affinity of AUF1.
AUF1 accelerates translation but promotes rapid mRNA degradation mediated by AREs in their 3′UTRs. However, AUF1 alone does not exhibit ribonucleolytic activity. Binding of AUF1 to the ARE recruits exosomes for exonucleolytic mRNA degradation (52). This contradictory function of AUF1 on rhythmic Cry1 mRNA and protein expression can be explained as illustrated in our model (Figure 7E). AUF1 binds to preexisting Cry1 mRNAs. Ribosomes scan mRNA for translation or have a higher probability to meet the initiation codon immediately after their recruitment to the 5′ end of mRNA by interacting with AUF1. As EIF3B and other translation initiation proteins form circularized mRNA by interacting with PABP, AUF1 may help provide 40s ribosomal proteins to the 5′ end of mRNA, and translation efficiency may be increased. Thus, CRY1 protein gradually increases and finally reaches peak levels. AUF1 may also interact with the exosome and be displaced from the 3′UTR in a complex with PABP. This may allow the mRNA to be exposed to the exosome machinery. Modifications in AUF1 or a change in the context of interacting partners may reduce the cytoplasmic AUF1 level and concurrent binding to the 3′UTR of Cry1 mRNA. This may produce inefficient recruitment of the 40S ribosomal subunit, causing decreased translation activity of Cry1 and consequent decreased CRY1 protein levels.
The present study is the first report to reveal the functional role of AUF1 in 3′UTR-mediated translation and the circadian system. The present study elucidates a new mechanism of rhythmic 3′UTR-mediated Cry1 translation that may be an important step in the regulation of the circadian clock. From these results, we suggest that posttranscriptional regulation plays an important role in the circadian rhythm. Indeed, the 3′UTR-mediated translational activation mechanism will elucidate a new regulatory step in the circadian rhythmic system. Translation of other genes could be also regulated by the 3′UTR and in an HNRNP-dependent manner. The current study may help explain the time gap between mRNA and protein production with the regulation of amplitude in the circadian rhythm. We hope that our findings may reveal hidden crucial aspects of the complex molecular system for achieving the tightly regulated 24-h cycle in mammals with a distinct translation activation mechanism. Moreover, our study may expand the roles of 3′UTRs and RNA-binding proteins for the translation system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) [20110027957, 20120005830, 2013R1A1A2058135]; Next-Generation BioGreen 21 Program [PJ00950301]; Rural Development Administration, and the Inter-ER cooperation project from the Ministry of Trade, Industry and Energy (MOTIE); Korea Institute for Advancement of Technology (KIAT); Brain Korea 21 plus program [10Z20130012243] of the Korean Ministry of Education. Funding for open access charge: Brain Korea 21 plus program [10Z20130012243] of the Korean Ministry of Education.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Choogon Lee for providing anti-CRY1 antibody, and Dr Sung Key Jang for providing pGEX-4T3-P37, pGEX-4T3-P40, pGEX-4T3-P42 and pGEX-4T3-P45 plasmids.
REFERENCES
- 1.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee KH, Woo KC, Kim DY, Kim TD, Shin J, Park SM, Jang SK, Kim KT. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol. 2012;32:717–728. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol. Cell. Biol. 2010;30:197–205. doi: 10.1128/MCB.01154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl Acad. Sci. USA. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deery MJ, Maywood ES, Chesham JE, Sladek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS, et al. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr. Biol. 2009;19:2031–2036. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr. Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 10.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 11.Misquitta CM, Iyer VR, Werstiuk ES, Grover AK. The role of 3′-untranslated region (3′-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol. Cell. Biochem. 2001;224:53–67. doi: 10.1023/a:1011982932645. [DOI] [PubMed] [Google Scholar]
- 12.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J. Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warner JR, Rich A, Hall CE. Electron microscope studies of ribosomal clusters synthesizing hemoglobin. Science. 1962;138:1399–1403. doi: 10.1126/science.138.3548.1399. [DOI] [PubMed] [Google Scholar]
- 15.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 18.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 19.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 20.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 21.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 22.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 23.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 24.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008;82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobell HM. Actinomycin and DNA transcription. Proc. Natl Acad. Sci. USA. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- 33.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 34.Lee KH, Kim SH, Kim DY, Kim S, Kim KT. Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PloS One. 2012;7:e37936. doi: 10.1371/journal.pone.0037936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banihashemi L, Wilson GM, Das N, Brewer G. Upf1/Upf2 regulation of 3′ untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay. Mol. Cell. Biol. 2006;26:8743–8754. doi: 10.1128/MCB.02251-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dempsey LA, Li MJ, DePace A, Bray-Ward P, Maizels N. The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics. 1998;49:378–384. doi: 10.1006/geno.1998.5237. [DOI] [PubMed] [Google Scholar]
- 38.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 43.Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aharon T, Schneider RJ. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol. Cell. Biol. 1993;13:1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savant-Bhonsale S, Cleveland DW. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a > 20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 46.Curatola AM, Nadal MS, Schneider RJ. Rapid degradation of AU-rich element (ARE) mRNAs is activated by ribosome transit and blocked by secondary structure at any position 5′ to the ARE. Mol. Cell. Biol. 1995;15:6331–6340. doi: 10.1128/mcb.15.11.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SM, Paek KY, Hong KY, Jang CJ, Cho S, Park JH, Kim JH, Jan E, Jang SK. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res. 2011;39:7791–7802. doi: 10.1093/nar/gkr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galban S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blum JL, Samarel AM, Mestril R. Phosphorylation and binding of AUF1 to the 3′-untranslated region of cardiomyocyte SERCA2a mRNA. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H2543–H2550. doi: 10.1152/ajpheart.00545.2005. [DOI] [PubMed] [Google Scholar]
- 50.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 51.Fellows A, Deng B, Mierke DF, Robey RB, Nichols RC. Peptides modeled on the RGG domain of AUF1/hnRNP-D regulate 3′ UTR-dependent gene expression. Int. Immunopharmacol. 2013;17:132–141. doi: 10.1016/j.intimp.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.