Abstract
Splicing factor SRSF10 is known to function as a sequence-specific splicing activator. Here, we used RNA-seq coupled with bioinformatics analysis to identify the extensive splicing network regulated by SRSF10 in chicken cells. We found that SRSF10 promoted both exon inclusion and exclusion. Motif analysis revealed that SRSF10 binding to cassette exons was associated with exon inclusion, whereas the binding of SRSF10 within downstream constitutive exons was associated with exon exclusion. This positional effect was further demonstrated by the mutagenesis of potential SRSF10 binding motifs in two minigene constructs. Functionally, many of SRSF10-verified alternative exons are linked to pathways of stress and apoptosis. Consistent with this observation, cells depleted of SRSF10 expression were far more susceptible to endoplasmic reticulum stress-induced apoptosis than control cells. Importantly, reconstituted SRSF10 in knockout cells recovered wild-type splicing patterns and considerably rescued the stress-related defects. Together, our results provide mechanistic insight into SRSF10-regulated alternative splicing events in vivo and demonstrate that SRSF10 plays a crucial role in cell survival under stress conditions.
INTRODUCTION
Alternative splicing (AS) is a process by which different combinations of exons can be joined together to produce a diverse array of messenger RNA (mRNA) isoforms from a single primary transcript. This process is a common mechanism for controlling gene expression and generating protein diversity in higher eukaryotic cells. In humans, >95% of multiexonic protein-coding genes undergo AS (1); however, a precise understanding of how these events are regulated and coordinated in vivo is still lacking. Given that AS plays a key role in the regulation of gene expression and that aberrant splicing has been implicated in a wide range of human diseases (2,3), it is important to fully understand the regulatory mechanisms by which AS is regulated in vivo.
The inclusion of an alternative exon in mature mRNA is largely dependent on the recognition and usage of the flanking splice sites by the splicing machinery. This involves many cis-acting RNA elements, known as exonic or intronic splicing enhancers and silencers, which are bound by different regulatory proteins. Serine/arginine-rich (SR) and heterogeneous nuclear ribonucleoprotein (hnRNP) proteins are two well-characterized RNA-binding proteins that interact with the enhancers or silencers and subsequently stimulate or repress the inclusion of an alternative exon, respectively (4–6). However, recent genome-wide analyses revealed that these regulatory proteins can modulate both exon activation and repression in vivo, which is likely dependent on their binding location within the pre-mRNA (7–9). Therefore, it seems clear that RNA-binding proteins widely use this strategy to regulate splicing in the cell. However, splicing-sensitive arrays and CLIP-seq analysis of SRSF1- and SRSF2-regulated splicing revealed that specific splicing outcomes were not directly linked to the positional effect of SR protein binding in vivo. Rather, these outcomes were mainly determined by the balance of cooperation and competition between the SR proteins and other regulatory proteins in vivo (10).
SRSF10 (formally known as SRp38) is an atypical SR protein that functions differently from the standard SR proteins. Although its domain organization is similar to that of other SR proteins, SRSF10 was found to be unable to activate the splicing of a model substrate, β-globin pre-mRNA, in an in vitro assay (11). SRSF10 was initially identified as a general splicing repressor that is activated by dephosphorylation (11,12). Subsequent studies showed that phosphorylated SRSF10 acts as a sequence-specific activator (13). Consistent with this role, SRSF10 was further demonstrated to be a key regulator of AS, influencing the selection of mutually exclusive exons in the GluR-B pre-mRNA in a sequence-specific manner (13,14).
In this study, we took advantage of the previously generated SRSF10-deficient (KO) chicken DT40 cells (12) and globally analyzed the AS events regulated by SRSF10 using an RNA-seq approach. We found that SRSF10 activates both exon inclusion and exclusion, and it is involved in all of the common modes of AS in vivo. Motif analysis revealed that SRSF10-activated and -repressed exons could be distinguished by the arrangement of SRSF10-enriched sequence motifs. SRSF10 binding to the alternative exon resulted in exon inclusion, whereas SRSF10 binding in the region of the downstream constitutive exon led to exon exclusion. This positional effect was further demonstrated by mutating the SRSF10 binding motifs of two minigene reporters. Importantly, the insertion of SRSF10 consensus sequences within an SRSF10-repressed exon was sufficient to induce exon inclusion. Our results thus provide substantial insight into mechanisms of SRSF10-regulated AS events in vivo.
Functionally, a large body of literature revealed that many of SRSF10-regulated alternative exons are associated with pathways of stress and apoptosis. Consistent with this finding, SRSF10 KO cells displayed elevated levels of apoptosis in response to endoplasmic reticulum (ER) stress than wild-type (WT) cells. Strikingly, induction of SRSF10 in KO cells recovered the WT splicing patterns for the majority of SRSF10-verified splicing events and considerably rescued the ER stress-related defects. These findings thus demonstrate that SRSF10 is required for cell survival under stress conditions, probably reflecting abilities of SRSF10 in regulating a large subset of stress/apoptosis-related splicing events.
MATERIALS AND METHODS
RNA sequencing
The complementary DNA (cDNA) libraries for paired-end sequencing were prepared using mRNA-Seq Sample Prep Kit (Illumina) according to the manufacturer’s instructions. After selecting ∼200-bp fragments by agarose gel electrophoresis, Illumina paired-end sequencing adapters were ligated to the DNA fragments for polymerase chain reaction (PCR) amplification and sequencing with an Illumina HiSeq™ 2000 system. The raw sequence data and processed data have been submitted to Gene Expression Omnibus with accession number GSE53354.
Mapping of paired-end reads
Before read mapping, clean high-quality reads were obtained from the raw reads by removing the adaptor sequences, reads with >5% ambiguous bases (noted as N) and low-quality reads containing more than half of the bases with qualities of <20. The clean reads were then aligned to the reference chicken genome (release: galGal4) and the corresponding recently annotated genes (galGal4.0) using the TopHat program (v2.0.6). In the alignment, preliminary experiments were performed to optimize the alignment parameters (–read-mismatches 3 –read-edit-dist 4 –read-realign-edit-dist 0 –mate-inner-dist 20 –min-anchor 6 –coverage-search –microexon-search –solexa1.3-quals –b2-very-sensitive) to provide the most information on the AS events.
Transcriptome reconstruction
The recent transcript annotation of the chicken genome sequence (galGal4.0) was obtained from ftp://ftp.ncbi.nlm.nih.gov/genomes/Gallus_gallus/GFF/. KO and WT transcriptomes were reconstructed, respectively, by cufflinks using the parameters (–GTF-guide galGal4.0.gff –frag-bias-correct galGal4.fa –multi-read-correct –upper-quartile-norm). Then these two transcriptomes were merged with the original version by cuffmerge using the parameters (–ref-gtf galGal4.0.gff –ref-sequence galGal4.fa KO_WT_transcriptomes.txt) to yield comprehensive reannotated transcripts for subsequent AS analysis.
AS analysis
After mapping, an ‘accepted_hits.bam’ file was generated, which includes information regarding the chromosome position for exonic reads and exon–exon junction reads. All junction reads must meet two criteria: (i) at least 6 nt of a read must match perfectly to each of the two flanking regions of a potential junction site, and (ii) a junction site that has >3 non-redundant reads in both the WT and KO samples must be filtered. Afterwards, a series of Java programs implemented into the software named ASD (AS detector) were developed to fulfill the following tasks: (i) to reconstruct exon-clusters based on the aforementioned reannotated chicken transcriptome for identification of five common modes of AS events for each exon-cluster, (ii) to count the number of junction reads that align either to the inclusion or exclusion isoforms in both the WT and KO samples, respectively, and calculate a P-value using junction read-counts between KO and WT samples by Fisher exact test, (iii) to calculate read coverage for the alternative exon and its corresponding gene in both WT and KO samples, respectively, and calculate a second P-value by Fisher exact test based on the alternative exon read coverage relative to its gene read coverage between WT and KO samples, (iv) to combine the two P-values to get an adjusted P-value using a weighted arithmetic equation (6,15) (see Supplementary Method) for assessing the statistical difference of AS between the two samples. ASD is available at http://www.novelbio.com/asd/ASD.html.
Detection of GA-rich region and motif analysis of SRSF10-regulated exons
Based on SELEX-identified (11) and RNAcompete-predicted consensus sequences (16) for SRSF10, and also taking into account the degenerate nature of RNA-binding sequences, we arbitrarily used GA-rich 6-mers that include at least one G and two As with GA content >80% as potential SRSF10 binding sites. Significantly, enrichments of GA-rich regions were identified by counting the occurrences of each 6-mer (46 total occurrences = 4096 combinations) within five segments of each transcript with a SRSF10-regulated cassette exon (including the cassette exon and the two flanking constitutive exons and introns). The cassette exons, which were not affected by SRSF10, were treated as the control group. A hypergeometric test (Fisher's right-tailed exact test) was performed to detect the enrichment of each hexamer sequence within the sequences undergoing SRSF10-regulated inclusion (or exclusion) events against the control sequences. Among the 4096 combinations, a Bonferroni-adjusted P-value (or false discovery rate, FDR) was calculated by multiplying the actual total number of tests. Subsequently, the sums of the minus log2-transformed P-values were calculated for GA-rich 6-mers as a metric to determine the peaks of GA-rich 6-mers within the five regions around the regulated cassette exons. Among the identified GA-rich peaks, the overrepresented GA-rich 6-mers (P < 0.0001) along with flanking 5 nt on each side (a total of 16 nt) were further collected and submitted to Multiple EM for Motif Elicitation (MEME) suite (17) for possible GA-rich motif analysis. The parameter setting for MEME was a number of single motif per sequence of 0 or 1, a minimum motif width of 5, a maximum motif width of 8, a maximum number of different motifs of 3 and a search of only the given strand. Finally, MEME searching motif enrichments were manually edited to have a length of 6-mer.
RNA isolation and RT-PCR validation
RNA extraction, reverse-transcription and PCR amplification were all carried out as previously described (14). Briefly, PCR primers were designed to amplify two or multiple isoforms with different sizes. For complex splicing events, isoform-specific restriction digestion and/or DNA sequencing was used to validate PCR product identity. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for normalization. Sequences for primer pairs used in this study are listed in the Supplementary Table S3.
Plasmids construction, cell culture and transfection
Minigenes were constructed by amplifying genomic sequences spanning exon 12–14 of KDM6A and exon 8–10 of EIF4ENIF1, which were then inserted into CMV-Tag2b between the BamH1 and Xho1 sites, respectively. Creating SRSF10-binding motifs on the minigene constructs was made as previously described (7). Specifically, three copies of the consensus SRSF10 binding sequences or random sequences were inserted into the middle of exon 13 of KDM6A. Similar strategy was used to construct other plasmids used in the study. For deletion mutation constructs, sequences for primer pairs were available upon request. DT40 cells of different background including WT, SRSF-deficient (KO) and SRSF10-deficient containing exogenously expressed hemagglutinin-tagged SRSF10 (KO-re) were maintained essentially as described previously (12). Lipofectamine2000 (Invitrogen) was used for plasmid transient transfection according to manufacturer’s instructions.
Cells treated with ER stress inducer, apoptosis assay and western blotting
Cells were treated with the ER stress inducer (either thapsigargin, 0.5 uM, or tunicamycin (TM), 2 ug/ml) for 28 h. Cell apoptosis was determined by propidium iodide staining and flow cytometry. For recovery, after 16 h treatment with ER stress inducer, cells were collected and washed with phosphate-buffered saline once, then maintained in fresh medium. Numbers of surviving cells were determined by trypan blue exclusion. Actin monoclonal antibodies were purchased from Santa Cruz; human eIF2 [pSer52] antibody was from Biosource. Western blotting was performed as previously described (18).
Statistic analysis
The data represent mean ± SD format from at least three independent experiments except where indicated. Statistical analysis was performed by Student’s t-test at a significant level of P < 0.05.
RESULTS
RNA-seq and transcriptome analysis of WT and SRSF10-deficient DT40 cells
We have previously shown that SRSF10 functions as a sequence-specific splicing activator that is capable of regulating AS both in vitro and in vivo (13,14). To understand the global profile of the AS networks regulated by SRSF10, we used an RNA-seq approach to analyze the effects of SRSF10 depletion on splicing regulation in chicken DT40 cells (12). Briefly, WT cells and SRSF10-KO cells were cultured in standard medium to 90% confluence. Total RNA was isolated from these cells, and cDNA libraries were prepared individually by performing a series of procedures, including poly(A) enrichment, RNA fragmentation, random hexamer-primed cDNA synthesis, linker ligation, size selection and PCR amplification (Supplementary Figure S1A). The libraries were then sequenced in a 2 × 100-bp format using an Illumina HiSeq instrument.
After quality filtering, we generated a total of 53 001 728 and 53 138 030 clean reads for the WT and KO cells, respectively (Table 1). Then, we used TopHat (v2.0.6) to align these reads against the chicken genome sequence (galGal4) and its corresponding annotated genes (galGal4.0.gff3) (Supplementary Figure S1B). The paired-end reads had an approximate insert size of 192 ± 16 bp (Supplementary Figure S2), which is consistent with the size of the cDNA selected during the library preparation. Given that accurate mapping of reads that span splice junctions is a critical component for the identification of AS events, a series of experiments were performed to optimize the alignment parameters that would ensure the number of reads mapping to splice junctions (see the ‘Materials and Methods’ Section).
Table 1.
Summary of RNA-seq data mapping results
| Sample | WT | KO |
|---|---|---|
| Clean reads | 53 001 728 (100.0%) | 53 138 030 (100.0%) |
| Mapped reads | 46 152 593 (87.1%) | 45 354 730 (85.4%) |
| Unique mapped readsa | 45 512 948 (98.6%) | 44 766 437 (98.7%) |
| Multi-mapped readsb | 639 645 (1.4%) | 588 293 (1.3%) |
| Junction readsc | 15 314 481 (33.2%) | 15 505 045 (34.2%) |
| Unmapped reads | 6 847 282 (12.9%) | 7 782 321 (14.6%) |
aThat include paired-end reads with an unalignable mate.
bWith multiple alignments.
cAll junction reads have ≥3 offset alignments.
After proper optimization, a total of 46 152 593 (87%) and 45 354 730 (85%) mapped reads were generated for the WT and KO cells, respectively. Almost 99% of both the WT and KO reads aligned to the reference genome in a unique manner, whereas <1% filtered as multi-mapped reads. Among the mapped reads, ∼15 314 481 WT (33.2%) and 15 505 045 KO (34.2%) reads were aligned against splice junctions (Table 1). A comprehensive annotation of the potentially novel transcripts relative to the known transcripts is indispensable for dissecting potential AS events occurring in the chicken transcriptome. To this end, Cufflink packages were used to reconstruct the chicken transcriptome based on the read distribution of the genomic sequences and then merge it with the current reference database. The final merged chicken transcriptome had a significantly expanded number of annotated transcripts and was sufficient for the following AS analysis (Supplementary Figure S1B).
Identification of SRSF10-affected splicing events
There are five major modes of AS events described in metazoan organisms, including cassette exons, alternative 5′ splice sites, alternative 3′ splice sites, mutually exclusive exons and retained introns (Figure 1A). To identify the changes in these five AS events on the loss of SRSF10, we developed a Java program that can search and locate sets of alternative exons and junctions from the genomic coordinates of all annotated chicken transcripts. Exonic reads and junction reads that align either to the inclusion or exclusion isoforms in both the WT and KO samples were counted, respectively.
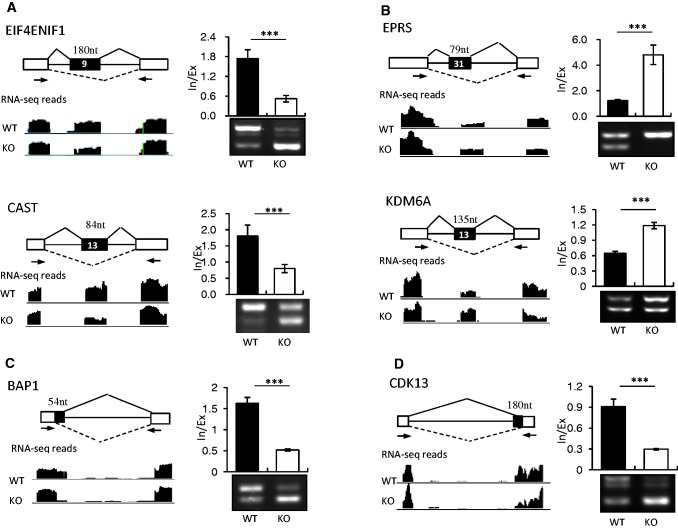
Figure 1.

Common AS events and the SRSF10-affected splicing events observed in DT40 cells. (A) Diagrams of the exclusion and inclusion isoforms for the five modes of AS events that were examined. White boxes, flanking constitutive exons; black boxes, alternative spliced exons/regions; solid lines, splice junctions supporting the inclusion isoform; dotted lines, splice junctions supporting the exclusion isoform. (B) Summary of the different AS events identified in DT40 cells and the splicing events affected by SRSF10 depletion, as revealed by analysis of RNA-seq data.
Comparison of the junction read counts between the WT and KO samples can reveal whether a change in the splicing pattern has occurred due to the depletion of SRSF10; this would be further adjusted by comparison of exon read coverage between the WT and KO cells (see ‘Materials and Methods’ Section). With an adjusted P-value cutoff of <0.05, we were able to identify 173 AS events from a total number of 10 652 splicing events that changed significantly in the KO samples. Specifically, the majority of the affected splicing events (130) belonged to the cassette exon category; 37 of the remaining events fell into other splicing modes, including 16 for alternative 5′ splice sites, 17 for alternative 3′ splice sites, 3 for mutually exclusive exons and 1 for intron retention. In addition, another six events (including coordinated two cassette exons and complicated splicing events) were placed into the ‘undefined’ category, as they could not be classified as any AS mode (Figure 1B, and Supplementary Table S1). In summary, SRSF10 is involved in all of the common modes of AS, and cassette exons are the most frequent targets for SRSF10 regulation.
Validation of SRSF10-affected splicing events
To verify whether these predicted AS events are real targets regulated by SRSF10, we designed primer pairs that were able to detect alternative exons in 45 potential target transcripts, which were chosen based on both P-values and read coverage signals. These primers were used to analyze the RNA isolated from the WT and KO cells by reverse transcriptase-polymerase chain reaction (RT-PCR). Although there were no or only minor changes in the splicing patterns in the 13 transcripts examined (t-test, P > 0.05, date not shown), 32 were significantly different between the WT and KO samples (t-test, P < 0.05, Supplementary Table S2), indicating a high consistency (73%) between the RNA-seq predictions and RT-PCR results in our study. Unexpectedly, 24 (72%) events contained unannotated splice junctions. The high percentage of unannotated transcripts indicated that the currently available chicken transcript annotations are incomplete. Table 2 shows the top 12 splicing events that were dramatically affected by SRSF10 (t-test, P < 0.001). Figure 2 shows representative examples of these 12 events, with RNA-seq read coverage, RT-PCR results and quantification of their RNA products measured as the inclusion/exclusion (In/Ex) ratio. Specifically, the WT cells predominantly expressed cassette exon-containing mRNA isoforms, such as for transcripts encoding EIF4ENIF1 and CAST; however, in the KO cells, SRSF10 depletion significantly increased exon skipping to nearly 90% (ETF4ENIF1) and 60% (CAST) (Figure 2A).
Table 2.
List of top 12 splicing events affected by SRSF10
| Gene symbol | Gene description | AS type | Annotation of two spliced variantsa |
|---|---|---|---|
| EIF4ENIF1 | Eukaryotic translation initiation factor 4E nuclear import factor 1 | Exon inclusion | A + U |
| CAST | Calpastatin | Exon inclusion | A + U |
| CASP1 | Caspase 1, apoptosis-related cysteine peptidase | Exon inclusion | A + U |
| PHF20L1 | PHD finger protein 20-like 1 | Exon inclusion | A + U |
| HSF2 | Heat shock transcription factor 2 | Exon inclusion | A + A |
| EPRS | Glutamyl-prolyl-tRNA synthetase | Exon exclusion | A + A |
| KDM6A | Lysine (K)-specific demethylase 6A | Exon exclusion | A + U |
| MDM4 | Mdm4 p53 binding protein homolog (mouse) | Exon exclusion | A + U |
| CDK13 | Cyclin-dependent kinase 13 | Alt 3′ SS | A + U |
| SLTM | SAFB-like transcription modulator | Alt 5′ SS | A + U |
| BAP1 | BRCA1-associated protein-1 | Alt 5′ SS | A + U |
| RBBP5 | Retinoblastoma binding protein 5 | Alt 5′ SS | A + U |
Alt; alternative: SS; splice site.
aBased on chicken NCBI and Ensemble databases, ‘U’ stands for unannotated splice transcripts, whereas A stands for annotated transcripts.
Figure 2.
SRSF10 can promote both exon inclusion and exclusion in vivo. SRSF10-activated cassette exons (A), SRSF10-repressed cassette exons (B), SRSF10-activated alternative 5′ exons (C) and SRSF10-activated alternative 3′ exons (D) were identified by RNA-seq analysis and validated by RT-PCR. Each case is schematically diagrammed with mapped RNA-seq reads that cover the corresponding exons (left panels). Black boxes, alternative exons; black lines, introns; white boxes, flanking constitutive exons. The primer pairs used for RT-PCR are indicated by arrows. SRSF10-regulated AS changes were analyzed by RT-PCR (right bottom panels). Quantification of the RNA products was measured as the inclusion/exclusion (In/Ex) ratio, as shown in the top right histogram. Values shown are the mean ± SD, n = 3. Significance for each detected change was evaluated by Student’s t-test.
These results are in agreement with previous findings that SRSF10 functions as a splicing activator to promote exon inclusion in vivo. However, the increased inclusion of exon 31 of the EPRS pre-mRNA and exon 13 of the KDM6A pre-mRNA was observed in the SRSF10-deficient cells (Figure 2B), suggesting that SRSF10 can also activate exon exclusion in vivo. In addition, the selection of alternative 3′ and 5′ splice sites was similarly affected by SRSF10. This was illustrated by two cases in which the inclusion of the BAP1 alternative 5′ exon and the CDK13 alternative 3′ exon were significantly decreased on SRSF10 depletion (Figure 2C and D). Taken together, these results provide strong evidence that SRSF10 can promote both exon inclusion and exclusion in vivo.
Restoration of WT-splicing patterns by exogenously expressed SRSF10 in KO cells
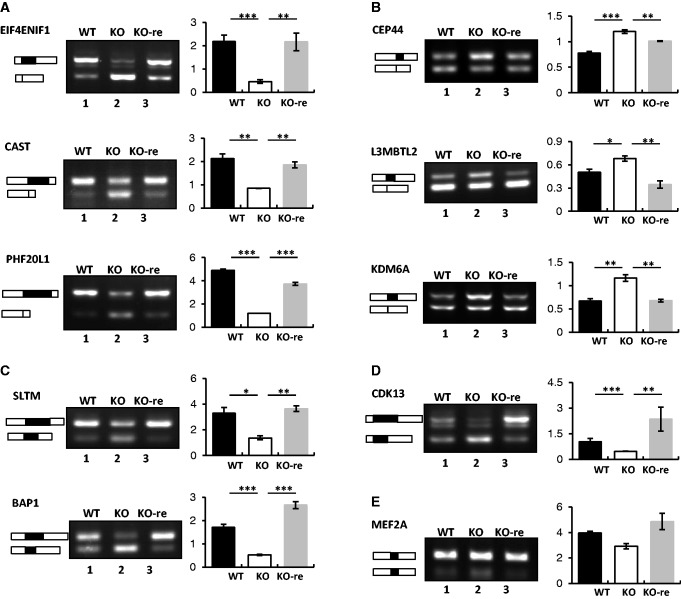
We next tested whether exogenously expressed SRSF10 could rescue the splicing variation observed in the KO cells. To this end, we first examined the splicing patterns of 32 validated transcripts in KO-re cells, which highly express flu-tagged SRSF10 (12), along with those obtained from WT and KO cells for comparison. Strikingly, the induction of SRSF10 restored the WT splicing patterns of all of the examined transcripts with one exception: the inclusion of EPRS exon 11 was not decreased to the WT level on SRSF10 re-expression (data not shown). Representative RT-PCR results of the examined transcripts and quantification of their RNA products are shown in Figure 3.
Figure 3.
Exogenous expression of SRSF10 in KO cells restores WT splicing patterns of SRSF10-targeted transcripts. SRSF10-activated cassette exons (A), SRSF10-repressed cassette exons (B), alternative 5′ exons (C), alternative 3′ exons (D) and mutually exclusive exons (E) were analyzed for splicing changes in WT, KO and KO-re cells by RT-PCR (left panels). The primers used for RT-PCR are the same as in Figure 3. Bar graphs show the ratios of inclusion versus exclusion (A–D) or exon a versus exon b (E) (right panels). Values shown are the mean ± SD, n = 3. The significance of each detected change was evaluated by Student’s t-test.
The levels of SRSF10-activated exons were all increased to nearly the WT levels for the EIF4ENIF1, CAST and PHF20L1 transcripts on SRSF10 re-expression (Figure 3A, compare lane 3 with lanes 1 and 2 for each target). In contrast, a decreased inclusion ratio was observed for the SRSF10-repressed exons, such as for the transcripts encoding CEP44, L3MBTL2 and KDM6A (Figure 3B, compare lane 3 with lanes 1 and 2 for each target). Similar restoration was reproduced for other types of alternative exons, such as the alternative 5′ exons of SLAM and BAP1 (Figure 3C, compare lane 3 with lanes 1 and 2 for each) and the alternative 3′ exon of CDK13 (Figure 3D, compare lane 3 with lanes 1 and 2). These results strongly suggest that SRSF10 is directly involved in the AS regulation of these significant transcripts in vivo.
We then determined whether the minor changes that were observed in several transcripts on SRSF10 deficiency could be rescued by the re-expression of SRSF10. Surprisingly, we found that these minor changes reverted back to the normal WT splicing levels in KO-re cells, as was observed for the MEF2A mutually exclusive exon (Figure 3E, compare lane 3 with lanes 1 and 2). This result provides evidence that SRSF10 also regulates the AS of pre-mRNAs, such as MEF2A, in vivo, although the output effect can be negligible. Together, these findings demonstrate that SRSF10 exerts differential activation or repression effects on the various splicing events in vivo.
Position-dependent activity revealed by SRSF10 binding motif analysis
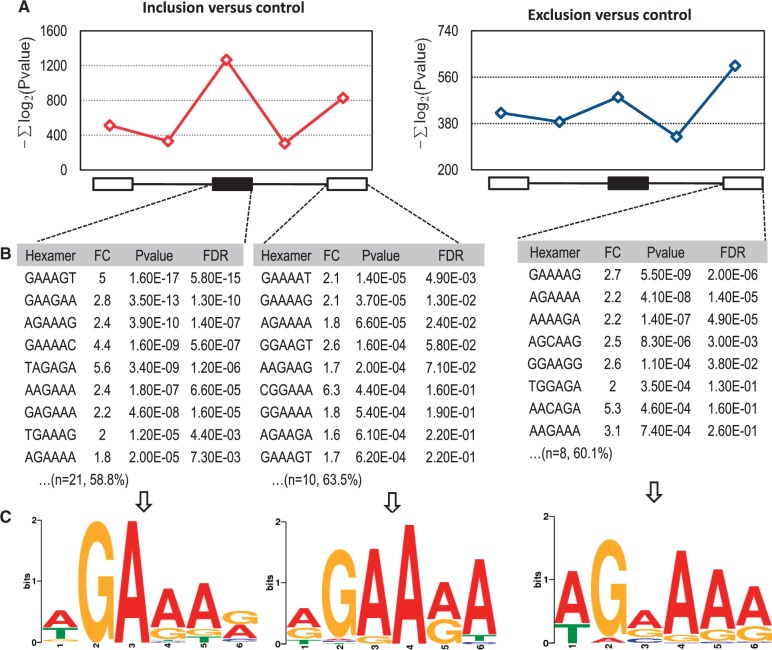
Next, we investigated whether the distribution of SRSF10 binding motifs differs between SRSF10-activated and -repressed cassette exons. To this end, we simply defined GA-rich hexamers as potential SRSF10 binding sites based on the SELEX-identified (11) and RNAcompete-predicted (16) sequences for SRSF10 (see ‘Materials and Methods’ Section). We then searched five transcript regions, including the cassette exon, the two flanking constitutive exons and the two flanking introns, for GA-rich hexamer enrichments among 78 transcripts with SRSF10-activated exons and 52 transcripts with SRSF10-repressed cassette exons. Another 80 SRSF10-unaffected events served as controls. As shown in Figure 4A, no GA-rich hexamers were found in the upstream exon or in the two flanking introns for both groups. However, some overrepresented GA-rich hexamers (e.g. GAAAGT GAAGAA and AGAAAG) were only found within the SRSF10-activated exons, whereas other overrepresented GA-rich hexamers (e.g., GAAAAG AGAAA and GAAAAT) were found in the flanking downstream constitutive exons for both groups (Figure 4B). These overrepresented GA-rich 6-mers was further analyzed by MEME for possible SRSF10 binding motifs (Figure 4C; see ‘Materials and Methods’ Section).
Figure 4.
Hexamer and MEME analysis of SRSF10-regulated exon inclusion and exclusion. (A) The sum of the minus log2-transformed P-values of GA-rich hexamers within the five regions around regulated cassette exons was compared with control cassette exons that are not affected by SRSF10. The red line represents SRSF10-mediated exon inclusion compared with the control (left), and the blue line represents SRSF10-mediated exon exclusion compared with the control (right). There is no obvious peak in the upstream exon or the two flanking introns around the cassette exons. (B) A representative list of top GA-rich hexamers was shown for SRSF10-activated cassette exons, -activated downstream constitutive exons and -repressed downstream constitutive exons, respectively. FC is fold change, P-value stands for statistically significance of GA-rich hexamers between SRSF10-regulated group and controls and FDR indicates false discovery rate. Shown in brackets are the number of overrepresented GA-rich 6-mers and the percentage of exons containing at least one overrepresented 6-mer. (C) Potential GA-rich motifs derived from the aforementioned overrepresented GA-rich 6-mers using MEME (P < 0.0001).
In summary, SRSF10-activated exons showed a predominant enrichment of GA-rich hexamers within the exons themselves over the flanking downstream constitutive exon (Figure 4A). However, SRSF10-repressed exons were associated with the enrichment of GA-rich hexamers only in the downstream constitutive exon (Figure 4A). Thus, the binding of SRSF10 to the alternative exon was associated with exon inclusion, whereas the binding of SRSF10 within the downstream constitutive exon was associated with exon exclusion. This positional effect as to inducing exon inclusion on the cassette exon and skipping on the downstream exon was only previously demonstrated with typical SR protein SRSF1 using MS2 tethering approach (19).
Mechanistic insights into SRSF10-regulated exon inclusion and exclusion
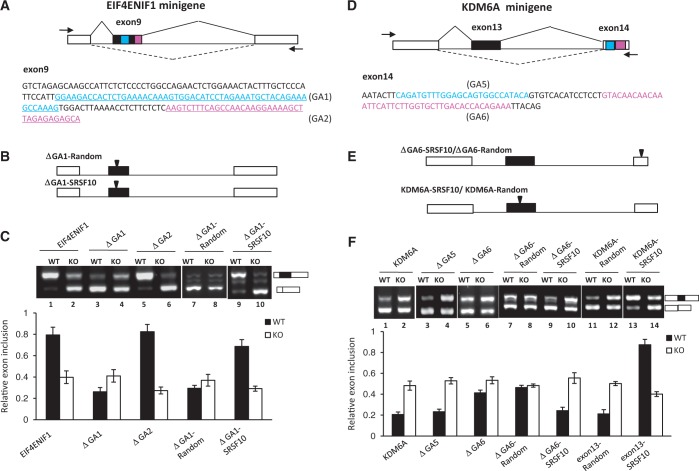
For SRSF10-activated exons, potential SRSF10 binding motifs were observed predominantly within cassette exons over downstream exons. We thus reasoned that the binding of SRSF10 to cassette exons is associated with exon inclusion. To test this, we analyzed individual cases that displayed SRSF10-dependent exon inclusion, such as EIF4ENIF1 exon 9. We constructed a minigene plasmid in which a genomic DNA fragment spanning EIF4ENIF1 exons 8–10 was preceded by the CMV promoter and followed by an SV(40) polyA site (Figure 5A, top). Splicing was assayed following transient expression in DT40 cells. Total RNA was extracted from the transfected cells, and exon inclusion/exclusion was analyzed by RT-PCR (Figure 5B). Consistent with its endogenous splicing pattern, the EIF4ENIF1 exon 9 was almost fully included in the WT cells (lane 1), whereas SRSF10 depletion significantly decreased the inclusion of exon 9 to nearly 30–40% (lane 2), indicating that inclusion of EIF4ENIF1 exon 9 was SRSF10 dependent. We identified four GA-rich clusters: GA1 and GA2 within exon 9 (Figure 5A, bottom), whereas GA3 and GA4 within exon 10 (Supplementary Figure S3A).
Figure 5.
Characteristics of SRSF10-regulated exon inclusion and exclusion. (A) Schematic representation of the EIF4ENIF1 minigene construct (top). The intact genomic region of EIF4ENIF1 from exon 8–10 was cloned into the CMV-Tag2b vector. The exon 9 sequences are shown at the bottom, with two GA-rich motifs (GA1 and GA2) marked with a different color. The PCR primers used to detect the alternatively spliced products are indicated by arrows. (B) Three copies of the SRSF10 consensus binding motif or random sequences were inserted within the middle exon of the GA1-deletion construct. (C) RT-PCR analysis of the EIF4ENIF1 minigene or each of the indicated mutants transfected in WT or KO cells, respectively. The percentage of exon inclusion is shown at the bottom of the gel. Values shown are the mean ± SD, n = 3. (D) Schematic representation of the KDM6A minigene construct (top). The intact genomic region of KDM6A from exon 12–14 was cloned into the CMV-Tag2b vector. The exon 14 sequences are shown at the bottom, with two potential GA-rich motifs (GA5 and GA6). The PCR primers are indicated by arrows. (E) Three copies of the SRSF10 consensus binding motif or random sequences were either inserted within exon 14 using the GA6-deletion construct (top) or within exon 13 of the KDM6A minigene (bottom). (F) RT-PCR analysis of the KDM6A minigene or each of indicated mutant transfected in WT or KO cells, respectively. The histogram below shows mean percentage of exon inclusion ± SD, n = 3.
We next examined, in more detail, the role of the individual GA elements in exon inclusion. To this end, we introduced a single GA deletion into the minigene construct and analyzed the effects of each mutant on the exon inclusion in vivo. As shown in Figure 5C, deletion of GA1 led to nearly 80% exon exclusion, which is similar to the effects caused by SRSF10 deficiency (compare lanes 3 and 4 with lanes 1 and 2). In contrast, the other three mutants displayed no effect on exon exclusion and still remained responsive to SRSF10 deficiency (compare lanes 5–6 and Supplementary Figure S3B). Taken together, these results provide strong evidence that the GA1 element has SRSF10-dependent enhancer activity.
To provide direct evidence that SRSF10-dependent enhancer activity within the GA1 fragment can lead to exon inclusion, we made two other constructs in which three copies of the SRSF10 consensus motif or random sequences were inserted in the middle exon of the ΔGA1 plasmid (Figure 5B). When SRSF10 binding motifs were present in the ΔGA1 plasmid, exon inclusion was restored nearly to the WT level, compared with the control sequence (Figure 5C, compare lane 9, lane 7 and lane 1). However, the differential effects on exon inclusion between the two inserted sequences were completely diminished on SRSF10 depletion (Figure 5C, compare lane 10 and lane 8).
For SRSF10-repressed exons, a key insight from the motif analysis was the enrichment of SRSF10 binding motifs within the downstream constitutive exon. To investigate the possible role of downstream SRSF10 binding motifs in the regulation of exon exclusion, we selected an exon from the KDM6A gene, which displayed exon exclusion in the WT cells but showed increased inclusion on SRSF10 depletion (Figure 2B). This effect was reproduced in the transient transfection of the WT or KO cells with a KDM6A minigene construct (Figure 5F, lanes 1 and 2). Although potential SRSF10 binding motifs were not observed within cassette exon 13, two GA-rich elements (named GA5 and GA6) were identified within downstream constitutive exon 14 (Figure 5D, bottom). Although the deletion of GA5 had little effect on exon inclusion, the deletion of GA6 increased exon inclusion from 20% to nearly 40% (Figure 5F, compare lanes 5 and 6 with lanes 3 and 4). These results suggest that the GA6 motif partially contributes to exon 13 exclusion. Importantly, insertion of SRSF10 consensus motifs in the exon 14 restored the WT splicing pattern of ΔGA6 substrate (Figure 5E, and F, compare lanes 7–10 with lanes 1–2), further demonstrating that SRSF10-dependent binding motifs within the constitutive exon resulted in exon skipping.
Induction of exon inclusion by the manipulation of SRSF10 binding motifs
We next considered whether the introduction of SRSF10 binding sequences in a naturally excluded exon would be sufficient to induce inclusion. To this end, mutations were made in the KDM6A minigene construct to allow for the insertion of three copies of the SRSF10 consensus motif or random sequences just downstream of the 3′ splice site in the cassette exon (Figure 5E). As shown in Figure 5F, ∼90% of the mutated exon with the inserted SRSF10 binding motifs was included, compared with the ∼20% exon inclusion observed with the control sequence (Figure 5F, compare lane 13 and 11). Moreover, the effect on exon inclusion critically depended on the motifs that were directly bound by SRSF10, as it was diminished in response to SRSF10 depletion (Figure 5F, lane 14). Taken together, these results suggest that the activation activity of SRSF10 binding sequences within the cassette exon is dominant over the repression activity of the downstream exon.
SRSF10 protects DT40 cells from ER stress-induced apoptosis
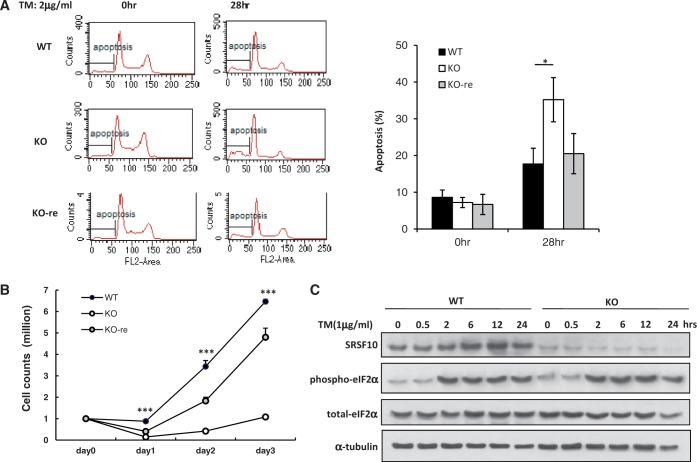
Through an extensive literature review, we learned that a large subset of genes have functional annotations related to stress and apoptosis among SRSF10-regulated AS events, such as genes of HSF2 (20), CASP1 (21), CAST (22), SLTM (23), EIF4ENIF1 (24,25), FKBP14 (26), BAP1 (27), SEC16A (28) or MDM4 (29). Because SRSF10 is not required for cell survival (12), we questioned whether SRSF10 favors cell growth under stress conditions by regulating these splicing events. To this end, we treated WT, KO and KO-re cells with different stress inducers and observed the responses of these cell lines to each treatment. Although there were no obvious differences observed among these cell lines in response to most stresses, ER stress induced by either thapsigargin or TM exerted greater deleterious effects on the KO cells compared with the WT and KO-re cells. As shown in Figure 6A, significant apoptosis was observed in the KO cells after 28 h of treatment with TM, as measured by propidium iodide staining and flow cytometry.
Figure 6.
SRp38 protects DT40 cells from ER stress-induced apoptosis. (A) WT, KO and KO-re cells were treated with the ER stress-inducer tunicamycin (TM, 2 ug/ml) for 28 h. Apoptosis was measured by propidium iodide staining and flow cytometry (right panel). (B) Cells were treated with TM (2 ug/ml) for 16 h and allowed to recover in medium for 1–3 days. The number of surviving cells was determined by trypan blue exclusion. The error bars represent the SD. (C) WT and KO cells were subjected to TM-induced ER stress for the indicated times. Immunoblotting was performed with whole cell extracts using antibodies against SRSF10, phospho-eIF2A and β-actin.
We further measured the ability of these cell lines to recover after ER stress. Whereas both the WT and KO-re cells resumed rapid growth after an ∼1-day lag, the KO cells showed considerably delayed recovery, resuming growth only after at least a 3-day lag (Figure 6B). These results reflect the loss of SRSF10 rather than an unrelated property of the cells because (i) another independent SRSF10-KO cell line behaved identically to the first (data not shown) and (ii) stable expression of exogenous SRSF10 (KO-re cells) considerably rescued the ER stress-related defects. In addition, in contrast to heat shock-induced rapid dephosphorylation (12), SRSF10 remained phosphorylated and dephosphorylated SRSF10 was not detected, regardless of short-term or long-term treatment of ER stress (Figure 6C). Consistent with this finding, none of SRSF10-dependent exon inclusion or skipping was reversed under ER stress conditions (Supplementary Figure S4). Therefore, we concluded that it is phosphorylated SRSF10 that plays a critical role in cell survival after ER stress, perhaps reflecting its ability to regulate many stress- and apoptosis-related AS events.
DISCUSSION
In this study, we globally analyzed SRSF10-regulated AS events based on deep sequencing of the chicken transcriptome. We found that SRSF10 is involved in all of the common modes of AS, with cassette exons being the most frequent targets for SRSF10 regulation. The most striking finding of our analysis is that SRSF10 shows position-dependent activity. Later, we discuss the possible mechanisms by which SRSF10 regulates exon inclusion and exclusion compared with the previously reported mechanisms of splicing regulation by other RNA regulatory proteins.
We previously demonstrated that SRSF10 functions as a sequence-specific splicing activator by directly binding to exonic splicing enhancers. The proposed mechanism for SRSF10 activation involves the stabilization of the binding of U1 and U2 small nuclear ribonucleoproteins (snRNPs) to the 5′ splice site and branch site, respectively (13). Consistent with this positive regulation, in this study we identified that SRSF10 activates a large subset of exon inclusions in chicken cell lines, including cassette exons, alternative 5′ exons and alternative 3′ exons. Motif analysis and functional studies conducted on a model minigene provided strong evidence that when binding within an alternative exon, SRSF10 acts as an activator to promote exon inclusion. This was further corroborated by previous findings in which SRSF10 was shown to enhance the inclusion of a triadin alternative 3′ exon by directly binding to the exon (14).
However, we also observed that SRSF10 can repress exon inclusion in vivo, and thus, sought to determine the mechanism underlying this SRSF10-dependent exon exclusion. It is possible that when binding occurs within flanking constitutive exons, SRSF10 may stabilize the interactions of U1 and/or U2 snRNPs with the nearby constitutive splice sites, thereby decreasing the competitiveness of the alternative sites. Kinetic analysis of splice site competition has demonstrated that a minor modulation of splice site recognition might be converted into a major functional consequence (30). This suggests that repression by SRSF10 results from its usual splicing activator activity, with partial activation of the flanking constitutive sites, preventing the cassette exon from inclusion. A similar principle has only been previously demonstrated for the SR protein SRSF1 (19). It is important to note that the manipulation of SRSF10 binding sequences could significantly switch an excluded exon into an included exon, indicating that SRSF10-dependent activation activity predominates over its repression activity.
Recent genome-wide studies of splicing regulators, including the SR proteins, hnRNPs and other RNA regulatory proteins, have revealed that they can all function both positively and negatively in regulating exon inclusion, regardless of their predominant splicing activator or repressor activity, which has been previously demonstrated using model genes (7–10,31). The opposite splicing activity could be attributed to their different binding locations in the pre-mRNA, although the positional effect exerted by individual splicing regulators appears to be fundamentally distinct. However, in some cases, the final splicing outcomes are not directly linked to the positional effect of individual protein binding in vivo, thus emphasizing the combinatory control of AS in vivo. Most AS events are likely subjected to regulation by multiple different splicing regulators, which may act synergistically or antagonistically. In this study, we observed the differential effects exerted by SRSF10 in modulating exon inclusion. Whether this reflects differences in the affinity of SRSF10 for target exons or complex mechanisms involving SRSF10 and other regulatory proteins remains to be determined
Our data also revealed that ∼70% of the validated splicing variants caused by the loss of SRSF10 have not been annotated, which indicates that the current chicken transcriptome is incomplete. Importantly, these newly identified variants do not result from aberrant splicing events, as they are also observed to be normally expressed in WT cells, albeit at much lower levels than their corresponding dominant isoforms. As WT cells are more resistant to ER stress-induced apoptosis than KO cells, this raises the possibility that SRSF10 protects cells from stress-induced apoptosis by maintaining the normal levels of these major isoforms in cells. By contrast, SRSF10 deletion results in isoform switching in these events and renders cells susceptible to specific apoptosis triggers. This possibility was further demonstrated by the observation that KO cells with reconstituted SRSF10 recovered the WT splicing patterns for the majority of SRSF10-verified splicing events and considerably rescued the ER stress-related defects. How these novel splice variants resulting from the loss of SRSF10 contribute to ER stress-induced apoptosis is currently unclear and will be an important focus of future studies.
Together, our findings provide mechanistic insights into SRSF10-regulated exon inclusion and exclusion, which depend on its binding location with respect to the regulated exons within the pre-mRNA. By regulating many stress- and apoptosis-related splicing events, our findings demonstrate that SRSF10 is required for cell viability under conditions of stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2012CB524900 and 2012BAK01B00]; National Natural Science Foundation [31170753 and 31070704] and the One Hundred Talents Program of the Chinese Academy of Sciences. China Postdoctoral Science Foundation [2013M541564 to W.W.]. Funding for open access charge: National Natural Science Foundation [31170753].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JJ, Lin KK, Huque M, Arani RB. Weighted p-value adjustments for animal carcinogenicity trend test. Biometrics. 2000;56:586–592. doi: 10.1111/j.0006-341x.2000.00586.x. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Chen M, Manley JL. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 2008;15:1040–1048. doi: 10.1038/nsmb.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vethantham V, Rao N, Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22:499–511. doi: 10.1101/gad.1628208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Biondo PD, Brindley DN, Sawyer MB, Field CJ. The potential for treatment with dietary long-chain polyunsaturated n-3 fatty acids during chemotherapy. J. Nutr. Biochem. 2008;19:787–796. doi: 10.1016/j.jnutbio.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 13.Nunez E, Kwon YS, Hutt KR, Hu Q, Cardamone MD, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell. 2008;132:996–1010. doi: 10.1016/j.cell.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev. Cell. 2009;16:528–538. doi: 10.1016/j.devcel.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves G, Yu YK. Combining independent, weighted P-values: achieving computational stability by a systematic expansion with controllable accuracy. PLoS One. 2011;6:e22647. doi: 10.1371/journal.pone.0022647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Wang Z, Zhou X, Cheng Y, Xie Z, Manley JL, Feng Y. Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression. Proc. Natl Acad. Sci. USA. 2013;110:E2687–E2695. doi: 10.1073/pnas.1310607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol. Cell. Biol. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Ann. N. Y. Acad. Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- 21.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 22.Porn-Ares MI, Samali A, Orrenius S. Cleavage of the calpain inhibitor, calpastatin, during apoptosis. Cell Death Differ. 1998;5:1028–1033. doi: 10.1038/sj.cdd.4400424. [DOI] [PubMed] [Google Scholar]
- 23.Chan CW, Lee YB, Uney J, Flynn A, Tobias JH, Norman M. A novel member of the SAF (scaffold attachment factor)-box protein family inhibits gene expression and induces apoptosis. Biochem. J. 2007;407:355–362. doi: 10.1042/BJ20070170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladomery M. Aberrant Alternative Splicing Is Another Hallmark of Cancer. Int. J. Cell Biol. 2013;2013:463786. doi: 10.1155/2013/463786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penkert RR, Kalejta RF. Tale of a tegument transactivator: the past, present and future of human CMV pp71. Future Virol. 2012;7:855–869. doi: 10.2217/fvl.12.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Temme N, Tudzynski P. Does botrytis cinerea Ignore H(2)O(2)-induced oxidative stress during infection? Characterization of botrytis activator protein 1. Mol. Plant Microbe Interact. 2009;22:987–998. doi: 10.1094/MPMI-22-8-0987. [DOI] [PubMed] [Google Scholar]
- 28.Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 2008;27:2043–2054. doi: 10.1038/emboj.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry ME. The regulation of the p53-mediated stress response by MDM2 and MDM4. Cold Spring Harb. Perspect. Biol. 2010;2:a000968. doi: 10.1101/cshperspect.a000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.