Abstract
Carl Woese developed a unique research program, based on rRNA, for discerning bacterial relationships and constructing a universal tree of life. Woese's interest in the evolution of the genetic code led to him to investigate the deep roots of evolution, develop the concept of the progenote, and conceive of the Archaea. In so doing, he and his colleagues at the University of Illinois in Urbana revolutionized microbiology and brought the classification of microbes into an evolutionary framework. Woese also provided definitive evidence for the role of symbiosis in the evolution of the eukaryotic cell while underscoring the importance of lateral gene transfer in microbial evolution. Woese and colleagues' proposal of three fundamental domains of life was brought forward in direct conflict with the prokaryote-eukaryote dichotomy. Together with several colleagues and associates, he brought together diverse evidence to support the rRNA evidence for the fundamentally tripartite nature of life. This paper aims to provide insight into his accomplishments, how he achieved them, and his place in the history of biology.
TEXT
Carl Woese challenged doctrines at the core of 20th-century biology. When microbiologists had declared that a phylogenetic classification of all the bacteria was impossible, Woese began a research program based on comparisons of rRNA to reveal a universal tree of life. His methods and concepts revitalized the study of microbial evolution and taxonomy. Woese proposed, in direct conflict with the prokaryote-eukaryote dichotomy, three fundamental lineages or domains, i.e., the Archaea, the Bacteria, and the Eucarya, each arising from a “progenote” phase of evolution, in which precellular entities were in the throes of developing the modern genetic system.
Woese's research program has a remarkably singular quality in the history of science. Great scientific thinkers are typically on the same path. Alfred Russel Wallace conceived of natural selection independently of Charles Darwin. Had James Watson and Francis Crick not discovered the structure of DNA, Linus Pauling was on track to do so. However, there was no one else who would have discovered the Archaea in the 1970s and 1980s. Indeed, one can wonder if the Archaea would even have been recognized after the development of microbial genomics. As Otto Kandler commented, when Woese was nominated to the National Academy of Science, “He opened a door which nobody expected to exist” (unpublished). The path to that door originated by posing a question few others did and approaching it in a methodical scientific manner. How did the genetic code evolve? Why does CCC encode proline? For Woese, understanding the evolution of molecular genetic machinery would unearth the base of the tree of life, and a deep phylogeny would in turn help reveal the evolution of the genetic code and of cells. That unique perspective, and the methods he and his colleagues developed, led to the discovery of the Archaea and the construction of a universal tree of life, hitherto deemed to be impossible.
Molecular biology was established without an evolutionary foundation. For the first generation of molecular biologists, the genetic code was a meaningless accident, “a frozen accident,” as Francis Crick called it (1). Of course evolution was axiomatic, but molecular biology largely developed as an engineering discipline, far removed from evolutionary biology. James Watson's classic Molecular Biology of the Gene (2) is strikingly different from Woese's book in 1967, The Genetic Code (3), the latter being steeped in evolutionary inquiry. Molecular biologists feared that questions in regard to the genetic code would lead to metaphysics (4), but Woese was an outsider; he was not part of the Watson and Crick “RNA Tie Club.”
Similarly, microbiologists of the 1970s had come to a consensus that an evolutionary understanding of microbial relationships was impossible to achieve, but Woese was not imbued with the doctrines of the classical microbiology, and there too he challenged its basic canons.
MICROBIOLOGY WITHOUT AN EVOLUTIONARY FRAMEWORK
In On the Origin of Species, Darwin wrote, “All true classification is genealogical; that community of descent is the hidden bond which naturalists have been unconsciously seeking, and not some unknown plan of creation, or … the mere putting together and separating objects more or less alike” (5). To construct a deep classification, Darwin argued, required comparisons of “essential” “characters” far removed from the vicissitudes of life. “Embryological characters,” he said, “are the most valuable of all.” As Darwin opined to his friend Thomas Henry Huxley on 26 September 1857, “The time will come I believe, though I shall not live to see it, when we shall have very fairly true genealogical trees of each great kingdom of nature” (Darwin Correspondence Project, www.darwinproject.ac.uk/).
From the late 19th century to the late 20th century, an evolutionary classification was based on principles of homology, through comparative morphology, comparative embryology, and the fossil record. Bacteria lacked morphological complexity and a detailed fossil record. Bacteriology for the most part developed as an applied science, and bacteria were not put into groups based on principles of homology and evolution but arbitrarily on as many characteristics as possible and on utility for industry and medicine. Microbial classification remained a world apart from evolutionary biology until the rise of molecular methods.
Some microbiologists had attempted to bring microbes into the evolutionary fold on the basis of morphology, as had been done for plants and animals. In 1941, Roger Stanier and C. B. van Niel proposed an evolutionary course based on ever-increasing morphological complexity, and on that basis, they suggested that “the simplest sphere” was the original shape of all bacteria and thus placed “the hypothetical coccus type” at the foot of the microbial tree (6). By the middle of the century, they recognized their speculations to be worthless and, like others, abandoned an evolutionary understanding of microbial relationships (7). As Stanier, Michael Doudoroff, and Edward Adelberg declared in the first edition of their popular text, The Microbial World (1957), “… it is a waste of time to attempt a natural system of classification for bacteria” (8).
Still, a classification based on morphological complexity remained in the prokaryote-eukaryote dichotomy, which Stanier and van Niel articulated in 1962. It had long been assumed that bacteria were different from other kinds of microbes. They seemed to lack sex, they divided by simple fission and not by mitosis, and they seemed to lack a nucleus like true cells. Ernst Haeckel (in 1866) grouped all the bacteria and blue-green algae as the Monera, a division within his newly proposed kingdom, Protista (9). However, throughout most of the 20th century, it remained uncertain whether the blue-green algae lacked a nucleus and divided by simple fission and how one could distinguish small bacteria, such as Rickettsia, from larger viruses (7).
In his 1957 paper “The Concept of a Virus,” André Lwoff drew a distinction between a virus and a bacterium on the basis of electron microscopy and biochemistry, arguing that the virus contained only one kind of nucleic acid, either RNA or DNA, enclosed in a coat of protein; it possessed few if any enzymes, and it did not reproduce by division like a cell (10). Five years later (in 1962), Stanier and van Niel wrote a sister paper, “The Concept of a Bacterium,” which aimed to resolve issues about the anatomy of the bacterium in the same way (11). Employing terms briefly mentioned by Edouard Chatton decades earlier (12, 13), they distinguished prokaryotic cells from eukaryotic cells on the basis that the former did not divide by mitosis, lacked a membrane-bound nucleus, and lacked mitochondria and chloroplasts.
Stanier and his colleagues understood that they could not create a natural classification of microbes based on morphology (14), but it seemed self-evident that all bacteria had a common ancestry. It would be counterintuitive to think otherwise. As they commented in the second edition of The Microbial World (1963), “All these organisms share the distinctive structural properties associated with the procaryotic cell … and we can therefore safely infer a common origin for the whole group in the remote evolutionary past” (15). “In fact,” they said, “this basic divergence in cellular structure, which separates the bacteria and blue-green algae from all other cellular organisms, probably represents the greatest single evolutionary discontinuity to be found in the present-day world” (15).
For some biologists, it made sense to assign the prokaryotes to a kingdom of their own: the Monera. That was the basis of the five-kingdom proposal of Richard Whittaker (16) and Lynn Margulis (17), which embellished the Haeckel scheme by the addition of a kingdom of fungi. There had long been prior suggestions of other kingdoms for protists, protozoa, monera, fungi, and viruses (7), but classical evolutionists throughout most of the 20th century had considered none of them. The Darwinian evolutionary synthesis of the 20th century was situated in a two-kingdom world of plants and animals. When classical evolutionist Ernst Mayr responded to proposals of microbial kingdoms, he said that accepting some of them was merely “a matter of taste” and “convenience” (18). He also said, “The classification by me is based on the traditional principles of classification, which biology shares with all fields in which items are classified, as are books in a library or goods in a warehouse” (52). Thus, Mayr argued that all bacteria should be classified as prokaryotes, which he defined as cells “lacking an organized cell nucleus and complex chromosomes” (18).
INNOVATING METHODS
That was the world of morphology Woese found himself in at the University of Illinois in 1969, when Sol Spiegelman left for a position at the Columbia College of Physicians and Surgeons. Woese inherited Spiegelman's electrophoresis equipment and wanted to put it to good use by comparing the rRNA sequences of various bacteria in order to establish a bacterial phylogeny. The choice of rRNA reflected both Carl's interest in the translation machinery and the realization that the equivalent rRNAs could be unambiguously isolated from essentially every organism of interest, whereas this would be problematic with most proteins. Initially, Carl considered using the 120-nucleotide 5S rRNA, but initial studies convinced him that the information content would be insufficient, and so he settled on the 16S rRNA found in the small ribosomal subunit.
Photograph of Carl working on a fingerprint, circa 1976. (Courtesy of Ken Luehrsen.)
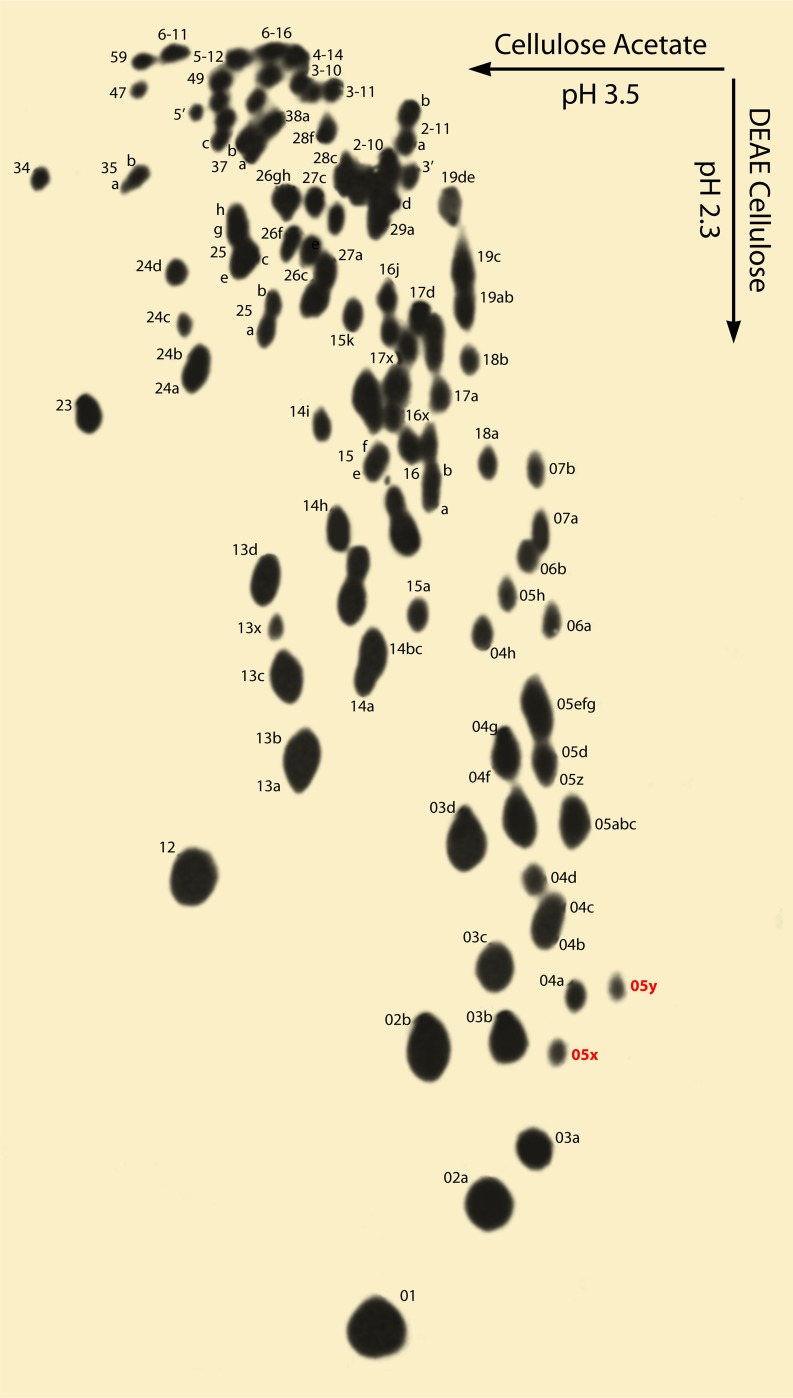
To establish a phylogeny, he would utilize the Sanger RNA sequencing technique (19). The basic idea was to digest radioactive rRNA with RNase T1, which cuts RNA almost exclusively after guanosine residues. The mixture of fragments was then separated by two-dimensional (2D) electrophoresis on paper. A picture of the pattern was obtained by exposing the finished chromatogram to X-ray film. The result is a “fingerprint” (Fig. 1). Each spot on the fingerprint corresponds to a specific sequence that ends in G but contains no other Gs. In addition, the position of each spot indicated the length and number of uracils contained therein. Thus, if one compared a fingerprint from organism 1 with fingerprints from organisms 2 and 3, one might see that there was more similarity between organisms 1 and 2 than between organisms 1 and 3 and thereby deduce organisms 1 and 2 to be more closely related.
Fig 1.
The fingerprint produced by ribosome T1 digestion of E. coli 16S rRNA is shown. Each spot contains one or more individual digestion products. They congregate in groups based on uracil content that are referred to as isopliths. Within each isoplith, the spots are further separated by the number of residues they contain. Thus, spot 24d is one of several tetramers with 2 uracil residues and 18b is an 8-mer with one uracil. The spots labeled 05x and 05y (in red) contain posttranscriptionally modified nucleotides that cause them to have aberrant mobility. The absence of these highly characteristic spots on the methanogen 16S rRNA fingerprints was the first indication of the uniqueness of the Archaea. (Adapted from reference 20 with permission of the publisher.)
Carl at a light table with various fingerprints in the background. (Courtesy of the Institute for Genomic Biology, University of Illinois at Urbana-Champaign.)
However, direct comparison of the images was not realistic, as the separation of the larger, more-informative fragments was not sufficient to distinguish matches from mismatches. In addition, as the spots get closer together, they run together such that more than one sequence is frequently present in each spot. Thus, one would instead sequence the oligonucleotide(s) (oligo[s]) associated with every spot on the primary fingerprint. The result would be a list of fragments, a catalog, found in each organism. One would then compare the catalogs rather than the images. The sequencing was accomplished by treating the RNA from each spot with additional enzymes to fragment it further to produce products of known sequence. Remaining unidentified digestion products in this secondary analysis were then subjected to further digestions in tertiary analysis. Ultimately, all the information pertaining to a single spot would be assembled and the sequence of the RNA fragment was deduced. With the core plan in mind, Woese wrote to Francis Crick in 1968, explaining his intentions and seeking advice on finding someone to help with the technical aspects of the project. Woese did not then view himself as an experimentalist and was well aware that as good as the plan was, the technology as it existed was simply not up to the task. Such help was not directly forthcoming, but as it turned out, it was not needed. David Bishop, a Spiegelman postdoc from the Sanger lab, taught graduate student Mitchell Sogin in Woese's lab the basics. From that core knowledge, Sogin and Woese led a revolution in the fingerprinting technology (7).
In order to sequence the various spots on the fingerprints, the traditional approach was to cut each spot out of the primary fingerprint and then elute the RNA before performing the secondary digestions. Although not fully published until 1976, five major improvements were quickly forthcoming (20, 21) and were fully in place by 1973. By increasing the salt concentration and pH of the second-dimension buffer, the core separation was greatly improved, thereby allowing access to the large fragments containing multiple uracil residues. This was accompanied by the use of maceration of RNA-containing spots directly onto the DEAE cellulose paper for secondary and tertiary analysis. This improvement eliminated the need for elution and greatly speeded up the process. Improved enzyme choices provided better accuracy in sequence determination and the ability to sequence oligomers of most any length. RNase U2, which cleaved the guanosine-ending RNA fragments primarily after adenine, was added to the mix along with full and partial digestions with the traditional RNase A that cleaves after cytosine and uracil. Despite reviewer skepticism, it was found that RNase T1 itself in high concentrations could be convinced to cleave at non-G locations. This activity was especially useful for C stretches, as one would produce a complete set of products defining the number of Cs in the stretch. For example, if the oligomer were CCCCU, then CU, CCU, CCCU, and the undigested CCCCU would all be seen on the secondaries. With the experimental procedures well established, it was possible to sequence all the informative oligos (five or more residues in length). The final step was development of a primitive algorithm to produce trees from comparisons of the catalogs (22). Initially, single-linkage clustering was used, but this was quickly replaced with the more appropriate average-linkage clustering.
With the technology in place, Woese quickly moved to establish a pipeline to determine complete catalogs for both 16S rRNA and 5S rRNA from large numbers of organisms. On every other Monday, the lab would receive 50 millicuries of 32P. This would be divvied up in 10-mC aliquots between various lab members who would then grow cells in a low-phosphate medium with 32P added and then recover the radioactive rRNAs. The group's then-technician, Linda Magrum, would run most of the fingerprints as well as the subsequent secondary and tertiary analyses in the electrophoresis system. After exposure of the chromatograms to X-ray film, Woese would analyze the spots to design secondary and tertiary digestions that were then carried out by Linda. All the analysis information was recorded on the relevant X-ray films with Sharpie markers that allowed easy correction as the process proceeded. Once the process was completed, George Fox recorded the data in the “oligo book” and transferred it to punch cards for construction of trees. The entire process would typically take 3 to 4 weeks per organism. Much of this was spent waiting for the tertiary digestion products to be developed, as the short half-life of 32P coupled with the decreasing amounts of material required long exposures. However, time was not lost, as multiple fingerprints were always under way and Carl would spend most every hour of every day analyzing them. It should be noted that the analysis of each spot on the primary pattern was essentially a puzzle. Thus, as Woese analyzed the spots, he would quickly realize that he had seen certain spots before. Hence, he quickly learned which sequences were highly conserved.
Organism choice quickly became a priority. In the first experiments, local laboratory strains were used, but it was soon realized that one needed to use literature strains. Multiple members of the scientific community were approached regarding their recommendations and to provide cultures. Here the Woese group was somewhat limited, as many of the more interesting strains were not readily cultured by standard techniques and most collaborators were unprepared to culture them with 32P. Chief among the organisms of interest were the methanogens. Ralph Wolfe had educated Woese about the many unique features of these organisms, and Woese was convinced of the importance of including this group. Wolfe was in fact listed as a consultant on Woese's NASA Exobiology grant. This was on hold, however, due to the culturing difficulties that precluded growth in the Woese lab and the unsuitability of the Wolfe lab for work with high levels of 32P.
This problem was overcome when Bill Balch, then a graduate student in the Wolfe lab, developed a vastly improved culturing technique for these organisms (23) that allowed 32P labeling in the Woese lab, where high radiation levels were routine. Balch and Fox had taken the 1973 Woods Hole microbial ecology course together and now worked to obtain the first labeled methanogen 16S rRNA. Thus, in 1976, the first effort was made with the delta H strain. This failed, as the incorporation of 32P was not sufficient. However, Ken Luehrsen recovered the 5S rRNA, and subsequent analysis of his fingerprints revealed mainly novel oligos, including UUAAG, which had previously been found in eukaryotic 5S rRNAs. This heightened expectations. A week or two later, success was had, and when Woese saw the primary fingerprint and initial secondary results, that was the eureka moment.
Woese immediately went to everybody in the group proclaiming that a new form of life had been discovered. How could this be with virtually no analysis? The answer lies with the posttranscriptionally modified bases, a few of which occur in 16S rRNA. The presence of the modification changes the mobility. There are two short sequences containing modifications that migrate in unexpected areas of the fingerprint (Fig. 1). These two spots had been seen on every previous fingerprint and were now missing. In addition, the initial secondary results made it clear that the most universal sequences were also missing. After a few calm-downs and shared euphoria, Carl posed the alternative explanation that Fox had somehow screwed up the RNA isolations and that the fingerprint was not from a 16S rRNA. It was decided that while the analysis of the fingerprint preceded, a second methanogen would be processed on the expectation that nothing was wrong. As the secondary and tertiary analyses came in, again and again the known conserved sequences were missing, and this was soon confirmed with the second methanogen, too. Carl was thus able to share his euphoria with Wolfe the day he returned from a European trip a few weeks later as if it had happened that very day (7).
Eureka moment over, serious discussion followed. If a new “urkingdom” of life had been discovered, it should satisfy a few minor conditions. For example, there would most likely be more than one genus therein. It would also be crucial to know that the methanogens were as distinct from the eukaryotes as they were from normal bacteria. To address the latter point, Woese undertook a heroic effort, the fingerprinting of several eukaryotic 18S rRNAs. Unlike the bacterial rRNAs, the eukaryotic small-subunit (SSU) rRNAs were heavily modified and were larger and thereby produced more-complex fingerprints. It was not readily possible to determine complete catalogs. Rather, Woese sought to determine if each oligo was present in the bacteria or the emerging archaebacterial group. As documented in detail elsewhere (7), the discussions continued as more data were collected. Knowledge gained from Otto Kandler in Munich, Germany, that the methanogens lacked true peptidoglycan, led to the search for other organisms of that ilk. The result was the rapid discovery of the extreme halophiles, Sulfolobus, and Thermoplasma as candidates for inclusion in what was initially referred to as a new urkingdom. Woese sought a name for what had been discovered. When protozoologist David Nanney and Linda Magrum independently suggested “archaebacteria,” that was adopted. The initial publications came in late 1977, accompanied by a press release from NASA's Exobiology program. As a result, the archaebacteria were immediately highlighted on 3 November with a front-page article in the New York Times accompanied by a picture of Carl relaxing at his desk (53). A week of frenzy followed, as reporters from CBS News, Good Morning America, and others descended on the lab.
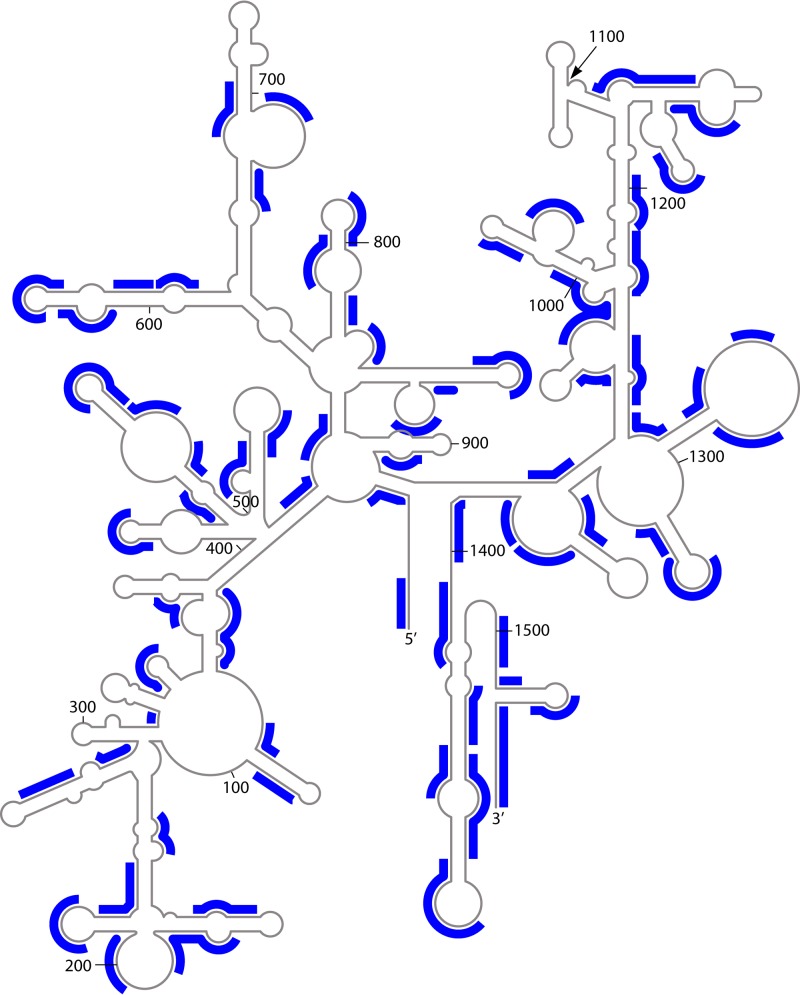
In retrospect, the fingerprinting technology had two major advantages. First, it turns out that the large oligomers (six residues and up) generated by RNase T1 are an incredibly good sample of the 16S rRNA sequence (Fig. 2) (24). Not only are they distributed rather uniformly throughout the 16S sequence, but they frequently cover only one side of a base pair, thereby largely avoiding redundant information. The second aspect is that the catalog comparison is much more discrete than continuous sequences. When comparing aligned sequences, one sees differences in percent identity as something of a continuum. Thus, if two sequences have 50% identity when everything else is 70% or more, one tends to propose a deep branch. In terms of a catalog, however, such a level of change quickly translates into essentially no similarity and a new urkingdom or domain can come to mind.
Fig 2.
The E. coli RNase T1 oligomers ≥6 residues in length are mapped onto the secondary structure of 16S rRNA. It is immediately seen that they are an extremely good sample of the entire sequence. In addition to being distributed throughout the RNA, they frequently cover only one side of helical regions. (Adapted from reference 24 with permission from Elsevier.)
Microbiologists were startled, and indeed, many were incredulous when Woese and Fox introduced the archaebacteria as an ancient lineage of bacteria (25). They were a phenotypically diverse group of little-studied organisms: methanogens that live in coal mine refuse, swamps, and the guts of rumens, extreme halophiles which live in brines five times as salty as the ocean, and thermophiles which live in geothermal environments that would cook other organisms (26). Previously believed to be wholly unrelated, they were reconceptualized by Woese and colleagues as an ancient lineage of organisms from the dawn of life, an urkingdom as phylogenetically distinct from the typical bacteria as they and the typical bacteria were from eukaryotes.
The archaebacterial concept received great support from the work of several biochemists and microbiologists and especially from Otto Kandler, Karl Stetter, and Wolfram Zillig in Germany, who contributed vital data and hosted the first archaebacterial conferences (7). The archaebacterial group was thus shown to possess many common characteristics: their walls lacked peptidoglycan, a defining feature of “prokaryotes”; the lipids in their cell membranes were unique, as were their transfer rRNAs, transcription enzymes, introns, and viruses. To underscore the deep trifurcation of life and the great diversity in the microbial world, Woese, Otto Kandler, and Mark Wheelis (27) formally proposed three “domains” of life (representing a rank above kingdoms), the Archaea, the Bacteria, and the Eucarya, to replace the classical bipartite division of life into prokaryotes and eukaryotes.
The three-domain proposal drew fire from the biological establishment imbued with the cytologically based prokaryote-eukaryote dichotomy (28, 29, 30). Molecular methods of classification were worlds apart from those of classical biology based on comparative morphology, whether microscopic or macroscopic. Certainly, Woese was not alone in seeing the great importance of molecular methods for phylogenetics. Classifications based on GC content, DNA-RNA hybridization, and amino acid sequencing of proteins had begun to revitalize the aim for a microbial phylogeny in the 1960s and 1970s. However, none of those methods would be used to construct a universal tree of life. Woese's molecular genetic perspective on the evolution of the genetic code allowed him to think beyond the accepted world of morphology and to question the untested assumptions that prokaryotes had a common prokaryotic ancestry and that they preceded and gave rise to eukaryotes.

Cropped color photograph of Carl taken by Tom Roberts in July 2002. (Courtesy of the Institute for Genomic Biology, University of Illinois at Urbana-Champaign.)
SYMBIOSIS AND THE PROGENOTE AT THE ROOTS
It would be false to understand Woese's program to be only the completion of the Darwinian program through the creation of new molecular methods. The evolutionary concepts of his program were as novel to classical evolutionary biologists as were his methods. Classical evolutionists assumed that natural selection operating on gene mutations and recombination within species was the sole basis of evolutionary change. In Woese's program, lateral gene transfer between “species” and symbiosis were central features of the evolutionary process beginning with the evolution of “the progenote,” in the throes of developing the cellular translation machinery (31, 32, 33, 34). Woese explained the thinking that shaped his program in a letter to Emile Zuckerkandl, one of the founders of molecular evolution, as well as the editor of the Journal of Molecular Evolution, on 29 March 1977 (personal communication):
The problem with the current conventional view, outside of its being muddled, is that the prokaryote is given phylogenetic connotation … In the proper view, one could have in principle some prokaryotes more closely related to eucaryotes than they are to other prokaryotes. With such a view the evolutionary questions become more clear. Was the (most recent) “common ancestor” a prokaryote; i.e., did all life arise from a common prokaryotic ancestor? If not, how many times did the prokaryotic state evolve (from the progenote level)? How many times, by analogy then, did the eucaryotic level arise (from the prokaryotic level)? These questions are not generally asked explicitly.
You see, Emile, it is a very different thing if you feel obliged (and don't know it) to derive your eucaryote from species all of which are phylogenetically related prokaryotes, than if you don't feel so obliged. In the former instance you are stuck with a whole spectrum of properties, phylogenetic tethers, that change the basic quality of your thinking. The chief examples here are how we treat endosymbiosis and how we treat genome organization. If we are obliged to start with typical bacteria … then you have to remove the wall and develop phagocytosis. This makes endosymbiotic interactions: (1) late on the scene (i.e., evolving after the grand plan of the bacteria has begun to unfold) and (2) a special occurrence, as the capacity evolved (in a rather unlikely way) in only one group of bacteria. In other words, endosymbiosis seems ad hoc; it needs to be excused. Now, when progenote vs. prokaryote is recognized, as an organizational distinction, then one no longer takes this narrow and misleading view of endosymbiosis. Endosymbiosis becomes an aboriginal property of progenotes, not an acquired property of prokaryotes. The “ancestor” has no cell wall; this evolved separately in typical bacteria (peptidoglycan walls) and eucaryotes (plant and fungal walls). Methanogens also have their own versions of the cell wall. Endosymbiosis then suddenly becomes an interaction that is widespread and diverse. What we now take as endosymbiosis is only the tip of the iceberg. Therefore, what we now define as eucaryotic is only a restricted segment of the real class thereof. Endosymbioses have been a major force in evolution for over three billion years …
It always astounds me how little attention is paid by the usual writers in this area to the evolution of translation. They totally ignore it, including the facts that are known.
With the progenote concept at the root of his thinking, Woese contradicted two central assumptions of the prokaryote concepts: that all prokaryotes were derived from a universal ancestor itself a prokaryote and that prokaryotes preceded and gave rise to eukaryotes. Woese articulated the fundamental assumptions and concepts of his research program in his paper “Bacterial Evolution” of 1987, a citation classic (35). As the translation machinery became optimized and fixed, one could then follow different lineages through the neutral changes in rRNA, the “ultimate molecular chronometer.” Not just any genes of course could be used to follow vertical evolution. Horizontal gene transfer, which some bacteriologists of the 1960s and 1970s thought to be the pervasive mode of evolutionary change in the bacterial (sensu lato) world, was indeed shown to be so with the rise of genomics in the 1990s. Once the first whole bacterial genome was sequenced, the first archaeal genome soon followed. Analyses of some gene histories conflicted with one another. Genes for protein A might not have the same history as genes for protein B. Horizontal gene transfer was thus deduced; its importance in evolution, like hereditary symbiosis, had been underestimated (7). Still, as long had been hypothesized, nonadaptive “informational genes” involved in DNA replication, transcription, and translation were shown to be more resilient to lateral gene transfer. A small set of informational genes representing the genetic core could be traced back to the universal ancestor (36).
The SSU rRNA methodology also confirmed and refined proposals made in the late 19th century and early 20th century, that mitochondria and chloroplasts originated exogenously as symbionts eons ago (37). Such speculations arose anew in the early 1960s when it was discovered that those organelles each contained DNA and a translation apparatus. However, proof of origin was lacking. Stanier spoke for many cell biologists when he commented in 1970 (38):
It might have happened thus; but we shall surely never know with certainty. Evolutionary speculation constitutes a kind of metascience, which has the same intellectual fascination for some biologists that metaphysical speculation possessed for some medieval scholastics. It can be considered a relatively harmless habit, like eating peanuts, unless it assumes the form of an obsession; then it becomes a vice.
Lynn Margulis, who did so much to champion symbiosis theories of organelles, agreed that theories of organellar origin could “never be directly tested” (39). Evolutionary biologists, she said, were in the same logical predicament as historians and “can only present arguments based on the assumption that of all the plausible historical sequences one is more likely to be a correct description of the past events than another.”
Woese's research program belied those statements. Mitochondria were derived from alphaproteobacteria, and chloroplasts were derived from cyanobacteria (40). Indeed, the SSU rRNA methods were remarkably predictive. They also contradicted many classifications of classical bacteriologists. Indeed, all the bacterial taxa above the level of genus had to be reorganized according to the SSU rRNA phylogenies.
In the midst of 16S rRNA cataloging, Woese maintained his interest in the genetic code and the ribosome. The 5S rRNA sequencing studies in his lab supported his efforts to understand the structure of the rRNAs. Holley and colleagues had drawn tRNA secondary structure in a “cloverleaf” arrangement (41). In his Nobel Lecture, Holley pointed out that “the strongest evidence for the ‘cloverleaf' arrangement of the secondary structure of tRNA's came from the finding that all of the tRNA sequences that have been determined since 1965 fit the same type of base-pairing arrangement” (42). It was also shown that when the nucleotides at positions 15 and 48 in the canonical tRNA structure varied, they did so in a coordinated way such that standard base pairing appears to be preserved (43). (The 15-48 interaction is actually a reverse Watson-Crick pair.) Fox and Woese (44) turned these observations around to use patterns of base variation that preserved possible helical segments as a tool to predict secondary structure rather than to just verify it.
Thus, “comparative analysis” of RNA structure was born, and soon, by collaboration with Harry Noller, the first model of 16S rRNA (45) (and later 23S rRNA [46]) secondary structure was at hand. As the large rRNA sequence database grew, these models were refined by Robin Gutell and others and in their final form are in extremely good agreement with actual structures seen by crystallography (47). Indeed, all large RNA secondary structures were initially determined by comparative sequence analysis. Woese was also the first to observe that tetraloops with the sequences GNRA, UNCG, and CUUG occurred with high frequency in the rRNAs (48). Common features of many RNAs, these tetraloops are widely used to stabilize artificial RNAs by synthetic biologists. Ultimately, Woese returned to his focus on translation and the origin of the genetic code, writing several important papers on the tRNA synthetases (49), the translation initiation factors (50), and other aspects.
With Gary Olsen, Woese also established the Ribosomal Database Project, which compiles ribosomal sequence information (51). It continues under the management of the Center for Microbial Ecology at Michigan State University (http://rdp.cme.msu.edu/). As the genomics initiative unfolded, Woese ensured that the Archaea were included from the very beginning.
EVOLUTIONARY RELATIONS AMONG ALL THE BACTERIA
Woese's discovery of the Archaea is certainly a career highlight, but from some perspectives, his contributions to unraveling the evolutionary relations among all bacteria was even more significant and, unlike the concept of the Archaea, largely not disputed. By the 1960s, the microbiology community had given up hope of obtaining an overall classification based on true phylogenetic relationship but nevertheless saw its merits. On a more local scale, microbiologists frequently had ongoing debates about relationship. For example, the organism Sporosarcina ureae was said by some to be related to Bacillus because it made endospores. Others argued that because it had the sarcina morphology, it should be considered a member of the genus Sarcina. Fingerprints were made, trees were drawn, and it was shown that the first explanation was to be preferred. Another case was the photosynthetic bacteria: all in a single large grouping or dispersed in different parts of the tree of life? Again, fingerprints were obtained from key organisms and it was shown that they are dispersed in multiple lineages. Thus, as the number of catalogs grew from a crude 27 in 1974 to over 170 solid examples in the first overview paper in 1980 (26), the recognition of 16S rRNA as a premier tool for the study of bacterial relations grew rapidly.
Unlike the discovery of the Archaea, the key to the success of the phylogeny project was the analysis of more and more fingerprints. There would be no eureka moment, just years of hard work. Kandler's enthusiasm for the Archaea and the 16S rRNA approach in general resulted in his encouraging Erko Stackebrandt to do a postdoctoral stint with Woese. Stackebrandt immediately brought a whole new level of activity to the cataloging enterprise. With his incredible work ethic and knowledge of microbiology, the number of high-quality catalogs rapidly increased. Upon his return to Germany, Erko was able to perfect a thin-layer chromatography fingerprinting methodology that avoided the requirement of growing cells in the presence of 32P. This allowed 16S rRNA fingerprint data to be obtained from almost any organism that could be cultivated and thus contributed greatly to the growing database that would eventually peak at around 375 organisms when modern sequencing technology finally began to take over.
Once readily accessible sequencing technologies did become available, 16S was unstoppable and has contributed in a major way to our understanding of microbial ecosystems, including the human microbiome. Indeed, the argument that Woese was not an attractive candidate for the Nobel Prize because the Archaea lack medical relevance is not true and in any event completely overlooks the fact that his pioneering efforts with 16S rRNA in the end revolutionized microbial ecology and medical microbiology. He did of course receive many awards, including the Crafoord Prize and the MacArthur “genius” award.
LIFE IN CARL'S LAB—1974 THROUGH 1977
Carl had established routines that allowed him to be with the fingerprints 8 hours a day, 5 days a week. He went to great lengths to avoid interruptions and non-research-related activities. He dismissed mail, especially internal, as a scourge. During his tenure in the lab, Fox recalls taking on the task of getting the mail and screening it. It was essential that someone do this, if only to hear from editors regarding manuscript decisions. When the mail was brought in and Carl informed of it, he would say “Thanks” and indicate that it should be left on a table near the door to his office where he would look at it “later.” As months went by, the pile got taller and taller until one day, as the mail was placed on top, it fell and landed in a wastebasket that “happened” to be next to the table. As Fox started to retrieve it, Carl announced that it was okay, “Just leave it.” A minor downside occurred when Carl's suitability for an annual salary increase was questioned because he did not respond to a request for an activity report.
When fingerprinting was in its heyday, the Woese space consisted of three main labs with some accessory rooms (dark room, etc.) across the hall. Carl analyzed the fingerprints in the center lab. His office immediately adjoined the main lab but, except for occasional efforts at transcendental meditation, was seldom used. He ran his lab in a relaxed manner, never having group meetings.
Carl was a master at piecing together the puzzle of each oligo to determine its sequence. This was, however, extremely tedious, and thus he would regularly recruit people from the lab to analyze fingerprints with him. Typically, this would be the person who generated the rRNA to begin with. By the time final sequence assignments were being made, the relevant data relating to each spot on the primary fingerprint were scattered among several pieces of X-ray film. Thus, it was helpful to have someone else monitoring one of the easier-to-interpret films and relay what was there to Carl. This was a serious activity with very little if any side conversation. Carl wanted every oligo completely and correctly sequenced.
During the day, when Carl would sometimes take breaks, he would visit one of the two adjoining labs to find someone who would talk science with him. Alternatively, lab members might go to him to discuss some results or ideas or to improve procedures and possible side projects. Visiting with Carl was risky, as there was a good chance you would end up helping analyze fingerprints for two to three hours.
For Linda Magrum, working as Carl's technician was far more intellectually rewarding and inclusive than she expected:
Although I did my share of the grunt work (like washing lab ware), Carl treated me more like a student and colleague than a technician. I too got to analyze endless fingerprints when I wasn't running countless chromatograms. I had the chance to sit and participate in Carl's ruminations on science. Working in Carl's lab is probably what made me decide to go for a Ph.D.—I figured I was at least as smart as some of his students. My most vivid memories involve radioactive doorknobs, sinks, and the cockroaches that came out of the sinks at night. I recall this being the case in the little lab off the main lab and on the opposite side from the room where we ran the chromatograms. I think that was also the room in which I ate my lunch!
If Carl was especially excited about an idea, he would actively try to recruit a group member to work on it. He wouldn't care who, he simply wanted the science done. The result was that on more than one occasion, two or more lab members would discover they were working on the same project. This even occasionally extended to external collaborators, as when the Fox and Pace labs were separately asked to help sequence a particularly difficult region of an archaeal 5S rRNA when the new sequencing ladder technology was just emerging, The Pace lab finished first by a few days and so got the joint authorship. In some contexts, this behavior would have been frowned upon, but in Carl's world, it was just normal and nobody took offense at it. It was simply getting the science done!
Carl was adept at finding ways to avoid nonresearch interruptions and willingly shared this information. One of his sage admonitions was that lab members, if asked, should inform others that the Woese group works on bacterial taxonomy. This would cause the inquirer to rapidly go elsewhere to discuss “real” science. The centerpiece of his efforts to create time for research was his practice of “dynamic incompetence,” a secret he revealed to a selected few. The essence of the idea is to always be the good guy who will try to do what is asked but then always manage to mess it up. This was at times a bit ugly, as for example when his rare trips to the library in the adjoining building happened to correspond with a graduate student's qualifying exam that Woese had “forgotten.” On the whole, though, the approach was amazingly successful.
When asked to run the departmental seminar program, Woese accepted gladly, and as it turned out, all the speakers “happened” to work on the ribosome, thereby ensuring Carl would never be asked again. Among these speakers was Harry Noller. When Fox asked Carl if this was the same Noller that had written the organic chemistry text he had used as an undergraduate, Carl was very concerned that he had invited some irritating big shot. He was greatly relieved when the early-1970s version of Harry showed up. They quickly became collaborators and friends.
As a professor, Carl was required at times to teach, which was not his favorite activity. Then-graduate student Ken Luehrsen recalls Woese's antics in his seminar course on the ribosome:
He was giving us a summary of past research done to elucidate ribosome structure and function. As he summarized the researcher names, he wrote them on the board starting first with those who he thought made the bigger contribution. The names were written large, but as he continued with people he thought were mediocre or poor scientists, he made the letters smaller and smaller. When he got to the end he was writing in very tiny letters, unreadable if you were more than a foot away from the board. Very funny at the time and quintessentially Carl!
Carl was great to work with. He was always willing to discuss science with anyone who would seriously listen and respond with real arguments rather than “That is crazy.” All ideas were allowed, and in the end, the good ones would survive. Very few biologists have had a greater impact on science than he.
ACKNOWLEDGMENTS
We are grateful to Norman Pace, who encouraged us to write this paper and kindly reviewed it. We also thank Ken Luehrsen and Linda Magrum for helpful comments and suggestions. Most of all, we are grateful to Carl R. Woese, who encouraged us to question untested assumptions in evolutionary biology.
This work was supported in part by the Social Science and Humanities Research Council of Canada (J.S.) and by the Center for Ribosomal Evolution and Adaptation at the Georgia Institute of Technology (NASA cooperative agreement NNA09DA78A) (G.E.F.).
Biographies

Jan Sapp received his Ph.D. in the history of science from the University of Montréal in 1984. He subsequently held an appointment at the University of Melbourne for 8 years. He was Andrew Mellon Fellow at the Rockefeller University, 1991-1992. He held the Canada Research Chair in the History of the Biological Sciences at the University of Québec at Montréal from 2001 to 2003 before returning to York University, where he has been a professor since 1992. His writings focus on evolutionary biology and ecology, emphasizing the importance of symbiosis and horizontal gene transfer. He has authored numerous papers and several books, including Beyond the Gene (1987), Where the Truth Lies (1990), Evolution by Association: a History of Symbiosis (1994), What is Natural? Coral Reef Crisis (1999), Genesis: the Evolution of Biology (2003), and The New Foundations of Evolution: on the Tree of Life (2009).

George E. Fox received a B.S. and Ph.D. in chemical engineering at Syracuse University. Subsequently, he worked with Carl Woese as a postdoctoral scientist at the University of Illinois at Urbana-Champaign from 1973 until 1977. During that time, Woese and Fox described the Archaea, introduced the concept of the progenote, and developed the comparative approach to RNA secondary structure prediction. Together they ultimately published 27 papers, including the first overview of bacterial evolutionary relations based on 16S rRNA in 1980. Dr. Fox joined the University of Houston faculty in September of 1977 and is now John and Rebecca Moores Professor of Biology and Biochemistry. His current research continues the focus on the origin and evolution of the translation machinery that he began while in the Woese lab. As Woese said in 2001, “Translation not only defines gene expression, but it is the sine qua non without which modern cells would not have come into existence.”
Footnotes
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Crick FH. 1968. The origin of the genetic code. J. Mol. Biol. 38:367–379 [DOI] [PubMed] [Google Scholar]
- 2.Watson J. 1965. Molecular biology of the gene. Benjamin Cummings, New York, NY [Google Scholar]
- 3.Woese CR. 1967. The genetic code. Harper and Row, New York, NY [Google Scholar]
- 4.Jacob F. 1974. The logic of living systems: a history of heredity. Translated from the French by Betty Spillmann. Allen Lane, London, United Kingdom [Google Scholar]
- 5.Darwin C. 1964. On the origin of species. (Facsimile of 1859 edition.) Harvard University Press, Cambridge, MA [Google Scholar]
- 6.Stanier RY, van Niel CB. 1941. The main outlines of bacterial classification. J. Bacteriol. 42:437–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sapp J. 2009. The new foundations of evolution: on the tree of life. Oxford University Press, New York, NY [Google Scholar]
- 8.Stanier RY, Doudoroff M, Adelberg EA. 1957. The microbial world. Prentice-Hall, Englewood Cliffs, NJ [Google Scholar]
- 9.Haeckel E. 1866. Generelle Morphologie der Organismen, vol 2 George Reimer, Berlin, Germany [Google Scholar]
- 10.Lwoff A. 1957. The concept of virus. J. Gen. Microbiol. 17:239–253 [DOI] [PubMed] [Google Scholar]
- 11.Stanier RY, van Niel CB. 1962. The concept of a bacterium. Arch. Mikrobiol. 42:17–35 [DOI] [PubMed] [Google Scholar]
- 12.Chatton E. 1925. Pansporella perplexa, amoebien á spores protégées parasite des Daphnies, reflexions sur la biologie et la phylogenie des protozoairies. Ann. Sci. Nat. Zool. 10th Ser. 8:1–84 [Google Scholar]
- 13.Chatton E. 1938. Titres et travaux scientifiques (1906–1937). Sottano, Italy [Google Scholar]
- 14.Sapp J. 2005. The prokaryote-eukaryote dichotomy: meanings and mythology. Microbiol. Mol. Biol. Rev. 69:292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanier RY, Doudoroff M, Adelberg E. 1963. The microbial world, 2nd ed. Prentice-Hall, Englewood Cliffs, NJ [Google Scholar]
- 16.Whittaker RH. 1969. New concepts of kingdoms of organisms. Science 163:150–163 [DOI] [PubMed] [Google Scholar]
- 17.Whittaker RH, Margulis L. 1978. Protist classification and the kingdoms of organisms. Biosystems 10:3–18 [DOI] [PubMed] [Google Scholar]
- 18.Mayr E. 1982. The growth of biological thought, p 244 Harvard University Press, Cambridge, MA [Google Scholar]
- 19.Sanger F, Brownlee GG, Barrell BG. 1965. A two-dimensional fractionation procedure for radioactive nucleotides. J. Mol. Biol. 13:373–398 [DOI] [PubMed] [Google Scholar]
- 20.Uchida T, Bonen L, Schaup HW, Lewis BJ, Zablen L, Woese C. 1974. The use of ribonuclease U2 in RNA sequence determination. Some corrections in the catalog of oligomers produced by ribonuclease T1 digestion of Escherichia coli 16S ribosomal RNA. J. Mol. Evol. 3:63–77 [DOI] [PubMed] [Google Scholar]
- 21.Woese CR, Sogin M, Stahl D, Lewis BJ, Bonen L. 1976. A comparison of the 16S ribosomal RNAs from mesophilic and thermophilic bacilli: some modifications in the Sanger method for RNA sequencing. J. Mol. Evol. 7:197–213 [DOI] [PubMed] [Google Scholar]
- 22.Fox GE, Pechman KR, Woese CR. 1977. Comparative cataloging of 16S ribosomal ribonucleic acid: molecular approach to prokaryotic systematics. Int. J. Syst. Bacteriol. 27:44–57 [Google Scholar]
- 23.Balch WE, Wolfe RS. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stackebrandt E, Ludwig W, Fox GE. 1985. 16 S ribosomal RNA oligonucleotide cataloguing. Methods Microbiol. 18:75–107 [Google Scholar]
- 25.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryote domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 75:5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, Zablen LB, Blakemore R, Gupta R, Bonen L, Lewis BJ, Stahl DA, Luehrsen KR, Chen KN, Woese CR. 1980. The phylogeny of prokaryotes. Science 209:457–463 [DOI] [PubMed] [Google Scholar]
- 27.Woese CR, Kandler O, Wheelis M. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87:4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr E. 1990. A natural system of organisms. Nature 348:4911701032 [Google Scholar]
- 29.Margulis L, Guerrero R. 1991. Kingdoms in turmoil. New Sci. 129:46–50 [PubMed] [Google Scholar]
- 30.Cavalier-Smith T. 1992. Bacteria and eukaryotes. Nature 356:570 [Google Scholar]
- 31.Woese CR, Fox GE. 1977. The concept of cellular evolution. J. Mol. Evol. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 32.Woese CR. 1982. Archaebacteria and cellular origins: an overview. Zentralbl. Bakteriol. Hyg. I Abt. Orig. C 3:1–17 [Google Scholar]
- 33.Fox GE, Luehrsen KR, Woese CR. 1982. Archaebacterial 5S ribosomal RNA. Zentralbl. Bakteriol. Hyg. I Abt. Orig. C 3:330–345 [Google Scholar]
- 34.Woese CR. 1998. The universal ancestor. Proc. Natl. Acad. Sci. U. S. A. 95:6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese CR. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris JK, Kelley S, Spiegelman GB, Pace NR. 2003. The genetic core of the universal ancestor. Genome Res. 13:407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sapp J. 1994. Evolution by association: a history of symbiosis. Oxford University Press, New York, NY [Google Scholar]
- 38.Stanier RY. 1970. Some aspects of the biology of cells and their possible evolutionary significance, p 1–38 In Charles HP, Knight BC. (ed), Organization and control in prokaryotic cells. 20th Symp. Soc. Gen. Microbiol. Cambridge University Press, London, United Kingdom [Google Scholar]
- 39.Margulis L. 1975. Symbiotic theory of the origin of eukaryotic organelles: criteria for proof. Symp. Soc. Exp. Biol. 29:21–38 [PubMed] [Google Scholar]
- 40.Gray MW, Doolittle WF. 1982. Has the endosymbiont hypothesis been proven? Microbiol. Rev. 46:1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. 1965. The structure of a ribonucleic acid. Science 147:1462–1465 [DOI] [PubMed] [Google Scholar]
- 42.Holley RW. 1972. Alanine transfer RNA (1968). Nobel lectures, physiology or medicine 1963–1970. Elsevier Publishing Company, Amsterdam, The Netherlands [Google Scholar]
- 43.Levitt M. 1969. Detailed molecular model for transfer ribonucleic acid. Nature 224:759–763 [DOI] [PubMed] [Google Scholar]
- 44.Fox GE, Woese CR. 1975. 5S rRNA secondary structure. Nature 256:505–507 [DOI] [PubMed] [Google Scholar]
- 45.Noller HF, Woese CR. 1981. Secondary structure of 16S ribosomal RNA. Science 212:403–411 [DOI] [PubMed] [Google Scholar]
- 46.Kop J, Wheaton V, Gupta R, Woese CR, Noller HF. 1984. Complete nucleotide sequence of a 23S ribosomal RNA gene from Bacillus stearothermophilus. DNA 3:347–357 [DOI] [PubMed] [Google Scholar]
- 47.Petrov AS, Bernier CR, Hershkovits E, Xue Y, Waterbury CC, Hsiao C, Stepanov VG, Gaucher EA, Grover MA, Harvey SC, Hud NV, Wartell RM, Fox GE, Williams LD. 14 June 2013. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. [Epub ahead of print.] 10.1093/nar/gkt513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woese CR, Winker S, Gutell RR. 1990. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops.” Proc. Natl. Acad. Sci. U. S. A. 87:8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woese CR, Olsen GJ, Ibba M, Soll D. 2000. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64:202–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kyrpides NC, Woese CR. 1998. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. U. S. A. 95:224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen GJ, Overbeek R, Larsen N, Marsh TL, McCaughey MJ, Maciukenas MA, Kuan WM, Macke TJ, Xing Y, Woese CR. 1992. The Ribosome Database Project. Nucleic Acids Res. 20(Suppl):2199–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayr E. 1991. More natural classification. Nature 353:122 [Google Scholar]
- 53.Lyons RD. 3 November 1977. Scientists discover a form of life that predates higher organisms. The New York Times, New York, NY [Google Scholar]