Abstract
SUMMARY
Fifty years after the discovery of the mouse Mx1 gene, researchers are still trying to understand the molecular details of the antiviral mechanisms mediated by Mx proteins. Mx proteins are evolutionarily conserved dynamin-like large GTPases, and GTPase activity is required for their antiviral activity. The expression of Mx genes is controlled by type I and type III interferons. A phylogenetic analysis revealed that Mx genes are present in almost all vertebrates, usually in one to three copies. Mx proteins are best known for inhibiting negative-stranded RNA viruses, but they also inhibit other virus families. Recent structural analyses provide hints about the antiviral mechanisms of Mx proteins, but it is not known how they can suppress such a wide variety of viruses lacking an obvious common molecular pattern. Perhaps they interact with a (partially) symmetrical invading oligomeric structure, such as a viral ribonucleoprotein complex. Such an interaction may be of a fairly low affinity, in line with the broad target specificity of Mx proteins, yet it would be strong enough to instigate Mx oligomerization and ring assembly. Such a model is compatible with the broad “substrate” specificity of Mx proteins: depending on the size of the invading viral ribonucleoprotein complexes that need to be wrapped, the assembly process would consume the necessary amount of Mx precursor molecules. These Mx ring structures might then act as energy-consuming wrenches to disassemble the viral target structure.
INTRODUCTION
The interferon (IFN) system is the first line of defense against animal viruses. Binding of type I or III IFNs to their receptors (IFNAR1/2 and IL-28Rα/IL-10Rβ, respectively) induces an antiviral state within the cell by inducing the transcription of many IFN-stimulated genes. The antiviral activities of some of these genes are well understood (reviewed in reference 1), with one of the best studied being Mx1.
Mx genes exist in nearly all vertebrate genomes, from fish to primates, and they are active mainly against RNA viruses (Table 1). Mx proteins from different species possess distinct antiviral activities, and the subcellular localization of an Mx protein contributes to its antiviral specificity. In general, nuclear Mx proteins (e.g., mouse Mx1) protect against viruses that replicate in the nucleus, such as influenza virus and Thogoto virus (THOV) (2–10), whereas cytoplasmic forms (e.g., mouse Mx2) inhibit replication of vesicular stomatitis virus (VSV) and other viruses that replicate in the cytoplasm (11–13). The human MxA protein is cytoplasmic and has a broad antiviral spectrum that is seemingly unrelated to the intracellular replication site of the virus (5, 14–30). Antiviral activity has also been described for fish Mx proteins (31–37). For example, Atlantic salmon encodes multiple Mx proteins, all of which can suppress infectious salmon anemia virus (ISAV), a fish orthomyxovirus (33, 38).
Table 1.
Antiviral spectrum of Mx proteins
| Mx protein(s) | Localization | Antiviral activity | Virus family | Genome | Reference(s) |
|---|---|---|---|---|---|
| Mouse Mx1 | Nucleus | Influenza virus | Orthomyxoviridae | ssRNA (−) | 2–6, 8–10, 39, 62, 90–93, 112, 124 |
| THOV | 7, 125 | ||||
| DHOV | 126 | ||||
| BKNV | 127 | ||||
| Mouse Mx2 | Cytoplasm | VSV | Rhabdoviridae | ssRNA (−) | 11, 12 |
| HTNV | Bunyaviridae | ssRNA (−) | 13 | ||
| Human MxA | Cytoplasm | Influenza virus | Orthomyxoviridae | ssRNA (−) | 5, 10, 14, 91 |
| THOV | 14, 18, 22, 24, 105 | ||||
| VSV | Rhabdoviridae | ssRNA (−) | 14–16, 21, 68, 111, 113 | ||
| Rabies virus | 128 | ||||
| HTNV | Bunyaviridae | ssRNA (−) | 19 | ||
| LACV | 19, 23, 24, 27 | ||||
| CCHFV | 26 | ||||
| RVFV | 19, 129 | ||||
| PUUV | 20 | ||||
| TULV | 20 | ||||
| Measles virus | Paramyxoviridae | ssRNA (−) | 17, 115 | ||
| SFV | Togaviridae | ssRNA (+) | 21, 24 | ||
| CSFV | Flaviviridae | ssRNA (+) | 28 | ||
| IBDV | Birnaviridae | dsRNA | 30 | ||
| Reovirus | Reoviridae | dsRNA | 30 | ||
| HBV | Hepadnaviridae | dsDNA | 25, 120 | ||
| ASFV | Asfarviridae | dsDNA | 29 | ||
| Human MxB | Nucleus | None | 15, 18 | ||
| Rat Mx1 | Nucleus | Influenza virus | Orthomyxoviridae | ssRNA (−) | 71, 130 |
| THOV | 130 | ||||
| VSV | Rhabdoviridae | ssRNA (−) | 71 | ||
| Rat Mx2 | Cytoplasm | VSV | Rhabdoviridae | ssRNA (−) | 71, 130, 135 |
| LACV | Bunyaviridae | ssRNA (−) | 131 | ||
| RVFV | 130 | ||||
| Rat Mx3 | Cytoplasm | None | 71, 130, 131 | ||
| Chicken Mx | Cytoplasm | None | 132, 133 | ||
| Influenza virus | Orthomyxoviridae | ssRNA (−) | 134 | ||
| VSV | Rhabdoviridae | ssRNA (−) | 134–136 | ||
| NDV | Paramyxoviridae | ssRNA (−) | 136 | ||
| Duck Mx | Nucleus and cytoplasm | None | 87 | ||
| Cow Mx1 | Cytoplasm | VSV | Rhabdoviridae | ssRNA (−) | 137, 138 |
| Rabies virus | 128 | ||||
| Cow Mx2 | Cytoplasm | VSV | Rhabdoviridae | ssRNA (−) | 118, 138 |
| Pig Mx1 | Cytoplasm | Influenza virus | Orthomyxoviridae | ssRNA (−) | 107 |
| VSV | Rhabdoviridae | ssRNA (−) | 139 | ||
| Dog Mx1, Mx2 | Cytoplasm | VSV | Rhabdoviridae | ssRNA (−) | 140 |
| Atlantic salmon Mx1-Mx3 | Cytoplasm | ISAV | Orthomyxoviridae | ssRNA (−) | 33, 38 |
| IPNV | Birnaviridae | dsRNA | 34 | ||
| Japanese flounder Mx | Cytoplasm | HIRRV | Rhabdoviridae | ssRNA (−) | 31 |
| VHSV | 31 | ||||
| Grouper Mx1-Mx3 | Cytoplasm | YGNNV | Nodaviridae | ssRNA (+) | 32, 35 |
| Senegalese sole Mx | Aquabirnavirus | Birnaviridae | dsRNA | 141 | |
| Barramundi Mx | Cytoplasm | NNV | Nodaviridae | ssRNA (+) | 37 |
| IPNV-SP | Birnaviridae | dsRNA | 147 | ||
| Seabream Mx1-Mx3 | IPNV | Birnaviridae | dsRNA | 119 | |
| Rare minnow Mx | GCRV | Reoviridae | dsRNA | 36 | |
| Rainbow trout Mx1 | Cytoplasm | IPNV | Birnaviridae | dsRNA | 142 |
| SAV | Togaviridae | ssRNA (+) | 142 |
The discovery of the Mx1 gene was reported 50 years ago by Lindenmann et al. (3, 39) and was based on the resistance of an inbred mouse strain to influenza virus infection. Unlike most other mouse strains, A2G mice are highly resistant to influenza virus infection (3, 39). This resistance is inherited as a dominant autosomal trait and is dependent on a single gene (Mx1) located on chromosome 16 (40). Most inbred mouse strains carry an inactive Mx1 gene due to deletion of three exons or the presence of a nonsense mutation leading to premature termination of translation (41). IFN induces the expression of Mx1 and is required for the resistance of A2G mice to influenza A virus: treatment of these mice with an interferon-neutralizing antiserum renders them susceptible to the virus (42). The IFN inducibility of the Mx1 gene facilitated the isolation of the Mx1 protein by comparison of the proteins derived from in vitro-translated mRNAs isolated from mouse strains that are sensitive or resistant to influenza virus infection (43). However, Mx1 can also protect cells against influenza virus in the absence of other IFN-induced proteins (4, 24, 44).
Mx1 orthologs (related genes in other species that evolved from a common ancestral gene) and paralogs (related genes that originated by duplication within a genome) were later identified in many different animal species. The first homologous Mx protein was observed in human peripheral blood lymphocytes treated with type I IFN. This protein, called MxA and encoded by MX1, was isolated by immunoprecipitation with a monoclonal antibody (2C12) raised against the mouse Mx1 protein (45). By studying the structures of these Mx genes, it became clear that mice and humans carry more than one Mx gene (46). Remarkably, the second mouse Mx gene is also not functional in most inbred mouse strains, in contrast to the intact Mx2 gene in wild mouse species (11). Subsequent analysis of human cDNA clones derived from type I IFN-treated human fibroblasts led to the discovery of MxB, encoded by MX2, the second human Mx gene (47).
The strategies used to identify homologous Mx proteins in mouse and human cells were based on the assumption that the corresponding Mx genes are induced by IFN. Subsequent detailed study of IFN inducibility showed that the Mx genes are induced by type I (IFN-α and IFN-β) or type III (IFN-λ) IFNs (45, 47–50); the Mx promoter does not respond to IFN-γ (type II IFN), interleukin-1α (IL-1α), or tumor necrosis factor alpha (TNF-α). Upregulation of Mx expression in response to virus infection is indirect and depends on the production of type I or type III IFNs (47, 49–51). After the discovery of mouse and human Mx proteins, it became clear that Mx proteins are present in almost all vertebrates and that all of them are induced by IFN. For example, the chicken Mx gene can be induced by IFN, poly(I) · (C) and UV-treated Newcastle disease virus (NDV) (52). In pregnant cattle, Mx gene expression can be induced not only by the antiviral type I IFN but also by IFN-τ (a type I IFN produced by cattle during pregnancy) (53, 54). Likewise, fish Mx proteins are expressed in response to type I IFNs (e.g., induction by poly(I) · (C) or by virus infection) but not to type II IFNs or lipopolysaccharide (LPS) (55, 56). In summary, Mx genes are conserved in vertebrates and are typically induced by type I and III IFNs, and Mx proteins are effectors of the antiviral state.
STRUCTURE OF Mx PROTEINS
Mx proteins are members of the family of large GTPases, which includes dynamins (57, 58). These GTPases share an N-terminal GTPase domain, a middle domain (MD), and a C-terminal GTPase effector domain (GED) (Fig. 1A) (57, 58). Unlike classical dynamins, Mx proteins lack a pleckstrin homology domain (involved in membrane targeting) and a proline-rich domain (involved in protein-protein interactions) (58).
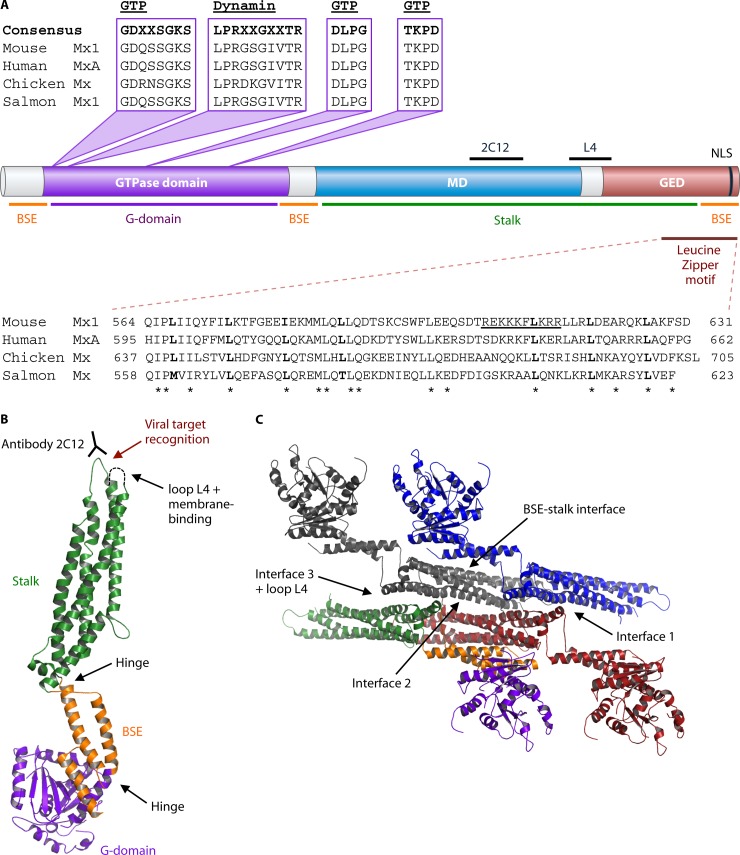
Fig 1.
Structure of Mx proteins. (A) The different domains of Mx proteins delineated by their primary sequence and their relationship to the domains determined in the 3D structure of Mx proteins. The Mx domains in the primary sequence include the GTPase domain, the middle domain (MD), and the GTPase effector domain (GED). The domains in the 3D structure include the G domain, the bundle signaling element (BSE), and the stalk domain. The amino acid sequences of domains important for the antiviral activity of Mx proteins are aligned. These domains include the GTP-binding motif, the dynamin signature, the C-terminal leucine zipper, and the nuclear localization signal (NLS). The conserved amino acids in the leucine zipper are indicated with an asterisk. The position of the epitope recognized by the monoclonal antibody 2C12, which can neutralize Mx antiviral activity, is indicated above the MD. The 2C12 epitope and loop L4 are important for viral target recognition. (B) 3D structure of Mx proteins (PDB file MxA 3SZR [73]). (C) 3D structure of MxA tetramer showing the interfaces (1 to 3 and loop L4) involved in oligomer formation (65, 73, 109). The images in panels B and C were generated with PyMOL.
GTPase Domain
The GTPase domain is the most conserved part of Mx proteins and other members of the dynamin family of large GTPases. In Mx proteins, this domain consists of a tripartite GTP-binding motif (GDXXSGKS, DLPG, and TKPD) and a dynamin signature (LPRXXGXXTR) (Fig. 1A) (59–61). The dynamin signature contains residues that coordinate the Mg2+ ion required for GTPase activity (61). The first two GTP-binding motifs, which flank the dynamin signature, bind the phosphate moiety of GTP, and the third GTP-binding motif is important for binding guanosine (62, 63). The second and third GTP-binding motifs are perfectly conserved in different species, whereas the first GTP-binding domain shows some differences between species (Fig. 1A). The dynamin signature is not completely conserved, but nearly all known Mx proteins (turkey Mx is an exception) contain the penultimate threonine residue, which is important for coordination of the Mg2+ ion. This strong conservation is probably because the biological activity of Mx proteins requires an intact GTPase domain (63, 64).
Middle Domain and GTPase Effector Domain
In the primary structure, the GTPase domain is followed by the middle domain (MD) (also called central interactive domain [CID]) and the C-terminal GTPase effector domain (GED) (65–67). Both the MD and GED are important for the conformation and activity of Mx proteins (65–67). The MD is important for oligomerization and virus target recognition (65, 68). On the other hand, the GED functions as an intramolecular GTP-activating domain: the C-terminal leucine zipper motif (65 to 70 amino acids) in the GED folds back to join the N-terminal GTP-binding domain, forming the enzymatically active center of Mx proteins. Mutants lacking the GED domain have no detectable GTPase activity (69). The C-terminal leucine zipper is evolutionarily conserved, suggesting a vital role in Mx function (Fig. 1A). The C terminus of rodent Mx1 protein is unique in that it contains a functional nuclear localization signal (NLS) (amino acids 606 to 615) (70, 71). Mouse Mx1 accumulates in the nuclei of most cell types tested, and this localization is important for its antiviral activity (70, 72).
Three-Dimensional Structure of Mx Proteins
The crystal structure of GTP-free human MxA protein was recently resolved (65, 73) and shown to resemble the three-dimensional (3D) structure of dynamin proteins (74). It is remarkable that crystallized MxA adopts a three-domain structure that does not coincide with the domains identified in the primary amino acid sequence. In the crystal structure, the GTPase domain is connected to the bundle signaling element (BSE) by a hinge, and the BSE is connected to the stalk by a second hinge (Fig. 1B) (73). The GTPase domain comprises a central β-sheet surrounded by α-helices. The BSE consists of three α-helices, each of which is derived from a different part of the MxA primary structure. One α-helix contains the amino acids just before the GTPase domain, the second one separates the GTPase domain from the MD, and the third corresponds to the C-terminal part of the GED (Fig. 1A) (73). Therefore, formation of the BSE requires backfolding of the C-terminal leucine zipper to the N-terminal GTPase domain. Concomitantly, three α-helices from the MD and one from the GED (preceding the carboxy-terminal BSE α-helix) adopt a helix-rich conformation known as the stalk (65, 73). Thus, the C-terminal leucine zipper is part of both the stalk and the BSE domains. This interaction between the C-terminal leucine zipper and the MD was originally proposed by Di Paolo et al. on the basis of two-hybrid experiments using deletion mutants of human MxA (75). The human MxA is the only Mx protein crystallized so far, but its strong structural similarity to dynamin, another large GTPase family member, and the strong conservation between Mx proteins suggest that other Mx proteins likely adopt a similar three-dimensional structure (73, 74).
Oligomerization of Mx Proteins
At low protein concentrations and physiological salt concentrations, Mx proteins form tetramers in solution. At higher protein concentrations (>1.5 mg/ml), these tetramers oligomerize further into large filaments and rings. These structures have been observed by electron microscopy and have been characterized by size exclusion chromatography and sedimentation assays (66, 67, 76–78). Mx oligomerization is mediated by three interfaces and one loop region (L4) (Fig. 1C). These interfaces mediate a crisscross interaction pattern between the stalk domains of different Mx molecules (65, 75), which ultimately results in ring formation. In these Mx rings, the stalk domains point inwards and the GTPase domains are located at the periphery. In addition to the stalk-stalk interactions, MxA oligomerization is also mediated by interactions between the BSE and the stalk of a neighboring parallel MxA monomer. These BSE-stalk interactions (Fig. 1C) are crucial for transmitting conformational changes, caused by GTPase activity, from the GTPase domain to the stalk domain of a neighboring MxA molecule. This cross talk is required for antiviral activity of Mx proteins (65, 73). Furthermore, when two Mx rings interact laterally (like two millstones), the GTPase domains of Mx molecules in adjacent rings interact with each other in the presence of GTP or its analogs (GTPγS or GDP/AlF4−) (67). This second-order oligomerization step stimulates GTPase activity by a cooperative mechanism.
The antiviral activity of MxA depends on both oligomerization and GTPase activity. However, mutations that impair oligomerization by inhibiting stalk-stalk or BSE-stalk interactions enhance the GTPase activity of MxA, probably by relieving structural constraints of Mx oligomerization on GTPase activity (65, 73). This suggests that antiviral activity requires an optimal balance between a rigid conformation (for interaction with the viral target) and mobility (allowing a stronger GTPase activity due to interactions between GTPase domains). Thus, although GTPase activity is required, stronger GTPase activity does not necessarily correlate with stronger antiviral activity (73).
Membrane Binding of Mx Proteins
Though MxA has no pleckstrin homology domain, it can associate with cellular membranes by binding to negatively charged lipids. This binding is mediated by a loop consisting of four lysine residues (MxA residues 554 to 557) located at the tip of the stalk region, distal from the G domain (Fig. 1B). MxA preferentially associates with negatively charged phospholipids, probably because of electrostatic interaction with the positively charged lysine residues in the membrane-binding loop (79). Unfortunately, this loop is unstructured and not visible in the crystal structure (65, 73). The affinity for membrane lipids supports self-assembly of dynamin and MxA, but this lipid binding leads to enhanced GTPase activity only in the case of dynamin (76, 79, 80).
In the cell, MxA associates with the smooth endoplasmic reticulum (ER) and the smooth ER-Golgi intermediate (27, 76, 80), which may contribute to antiviral activity. Membrane binding might facilitate contact with viral target structures, for example, with the nucleocapsid protein of La Crosse virus (LACV). Both MxA and the nucleocapsid protein of LACV localize to the smooth ER, which could facilitate their initial contact and interaction. This interaction leads to sequestration of the viral nucleocapsid protein to membrane-associated, large perinuclear complexes (27). Membrane binding might also lead to formation of a depot of MxA molecules that are protected from proteolytic attack: the membrane-binding loop contains a proteinase K cleavage site, which is protected from cleavage after membrane binding (79). The membrane-binding loop is fairly conserved in mammalian Mx1 proteins, which may indicate that membrane binding plays a role in antiviral activity.
PHYLOGENETIC ANALYSIS OF Mx FAMILY MEMBERS
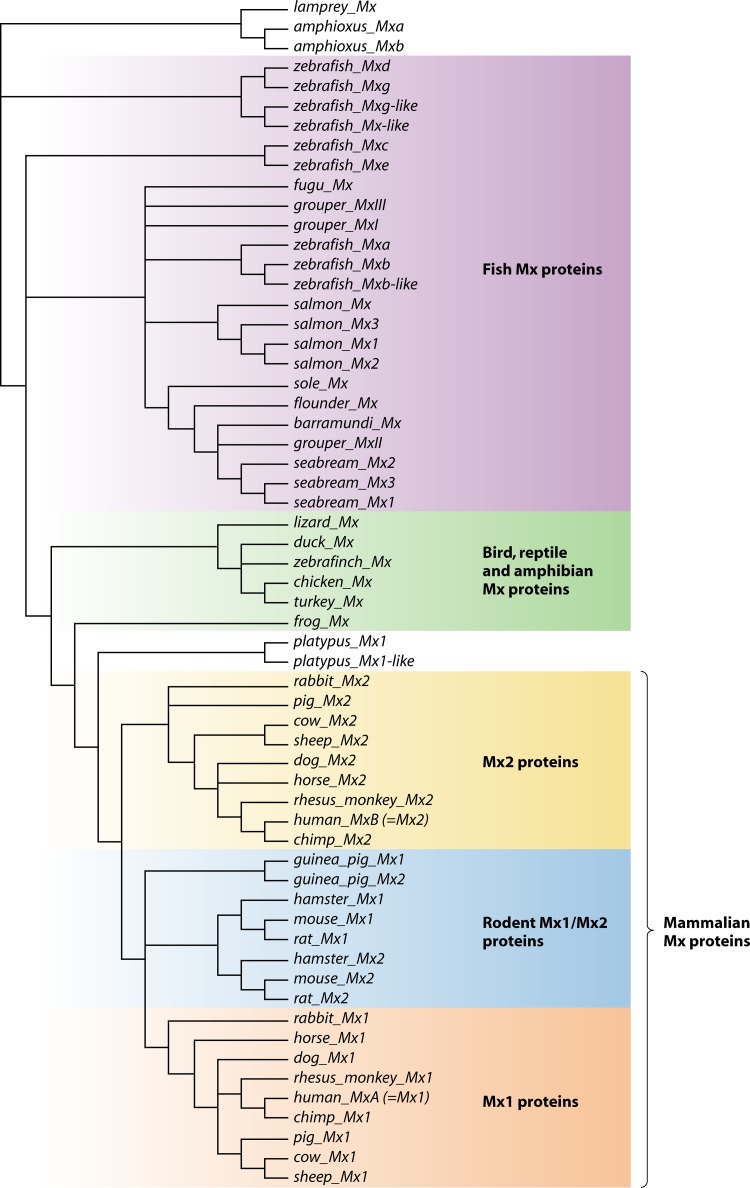
Mx genes are present in nearly all sequenced chordate genomes; a notable exception is the opossum (Monodelphis domestica) genome. We used the presence of the conserved N-terminal GTPase domain, an MD, and a C-terminal GED (Fig. 1A) in the Mx gene products to identify and compare Mx family members across a large set of vertebrate species (58). We identified Mx genes in mammals, monotremes, birds, amphibians, reptiles, and fish and determined their phylogenetic relationships. The Mx protein sequences (n = 60) were aligned in ClustalX2 (81) to estimate their phylogenetic relationships. The identified Mx protein sequences range from 620 amino acid residues (Japanese flounder [Paralichthys olivaceus]) to 721 amino acid residues (duck [Anas platyrgynchos]). Because the N-terminal region preceding the GTPase domain is highly variable, we excluded it from the alignment used to build the phylogenetic tree. The deduced phylogenetic tree reveals different subfamilies of Mx proteins that segregate into three classes: (i) fish; (ii) amphibians, reptiles, and birds; and (iii) mammals (Fig. 2).
Fig 2.
Phylogenetic tree based on primary amino acid sequences of Mx proteins. The Mx protein sequences were aligned in ClustalX2 (using the protein weight matrix PAM350), and the nonconserved N terminus was trimmed using the program Jalview (81, 143). This alignment was then used to build a bootstrap neighbor-joining tree in ClustalX2 with 1,000 bootstrap replicates. The deduced phylogenetic tree, visualized with Dendroscope (144), allows the identification of different subfamilies of Mx proteins: (i) fish Mx proteins (purple); (ii) Mx proteins of birds, reptiles, and amphibians (green); (iii) nonrodent mammalian Mx2 proteins (yellow); (iv) rodent Mx1/Mx2 proteins (blue); and (v) nonrodent mammalian Mx1 proteins (orange).
The evolutionarily most ancient Mx genes are from lamprey and amphioxus (Fig. 2, top). Depending on the species, fish have one to nine Mx genes. The multiple Mx genes in fish probably arose by gene duplications, as the encoded proteins are more closely related to their paralogs than to their orthologs in other fish species. Birds, reptiles, and amphibians have single Mx genes that presumably resemble the ancestor of mammalian Mx genes, which duplicated into two paralogous Mx genes soon after the emergence of mammals. As a result, most mammals have two Mx genes: MX1 and MX2. In humans and other primates, the gene products derived from MX1 and MX2 are usually named MxA (= Mx1) and MxB (= Mx2). Remarkably, in the phylogenetic tree the two rodent Mx proteins segregate together as a branch that is most closely linked to Mx1 proteins of other mammals (Fig. 2).
We also identified the genes flanking the Mx genes in the different species to look for conserved synteny (colocalization and sequential arrangement of related genes on a chromosome of a different species). We collected data from the UCSC, NCBI Map Viewer, and Ensembl genome browsers but excluded fish genomes because only a few fish genome sequences are available. In addition, fish Mx loci are much more complex and we found no evidence for conserved synteny to Mx genes from other vertebrates. The Mx genes from amphibians to mammals are all flanked by the FAM3B and TMPRSS2 genes, although the positions and orientations of the Mx genes relative to those of FAM3B and TMPRSS2 are occasionally shuffled (Fig. 3). The TMPRSS2 gene encodes a serine protease that can process the influenza A virus hemagglutinin into its fusion-competent state in human airway epithelial cells (82), and the FAM3B gene encodes the pancreatic secreted cytokine FAM3B (83). There is no obvious functional relationship to explain this chromosomal synteny of the Mx locus across different species ranging from amphibians to human.
Fig 3.
Genomic organization of Mx genes. The genomic organization of the different Mx genes of the different organisms was retrieved from the UCSC, NCBI Map Viewer, and Ensembl genome browsers; their organization with respect to their flanking genes FAM3B and TMPRSS2 is shown. The chromosome (Chr), scaffold (sc), or contig number is given.
The Mx locus in rodents is rearranged in comparison to the Mx loci of other mammals (Fig. 3). During evolution, rodents presumably lost their ancestral Mx2 gene. Based on sequence homology, the gene that is now named Mx2 in mouse was likely derived from an ancestral Mx1-like gene, and the mouse Mx1 evolved from that ancestral gene after duplication. This hypothesis is supported by the fact that the two rodent Mx proteins are more related to Mx1 than to Mx2 proteins from other mammals (Table 2). The mouse and rat Mx1 and FAM3B genes are oriented in opposite directions relative to the Mx2 gene, suggesting that a duplication of the ancestral Mx2 gene was accompanied by rotation of the FAM3B gene during the evolution of rodents. In rats, three Mx proteins have been reported (84). They are named Mx1, -2, and -3, but the genes encoding Mx2 and Mx3 are most likely allelic variants of the same gene, as only two Mx genes are found in the rat genome. Compared to those of mouse and rat, the guinea pig Mx locus seems to lack the orthologous Mx2 gene, and the FAM3B and TMPRSS2 genes are duplicated. However, the guinea pig Mx2 gene is located approximately 4.3 Mb further away on the same chromosome. The chromosomal arrangement of the two platypus Mx genes resembles that in other nonrodent mammals. However, the predicted gene products are more closely related to each other than to other mammalian Mx proteins. Birds, reptiles, and amphibians have one Mx gene, flanked by FAM3B and TMPRSS2 genes (Fig. 3). The lizard Anolis carolinensis has an additional Mx-like gene, but there is no evidence that this gene is transcribed, and presumably it is a pseudogene.
Table 2.
Sequence similarity between human MxA/MxB and mouse Mx1/Mx2
| Protein | % Sequence similarity with: |
||
|---|---|---|---|
| Human MxB | Mouse Mx1 | Mouse Mx2 | |
| Human MxA | 77.7 | 81.9 | 88.1 |
| Human MxB | 73.3 | 76.3 | |
| Mouse Mx1 | 83.5 | ||
The genetic relationship between the Mx proteins of different organisms can provide clues about the antiviral spectrum and the presence of specific functions in different subclasses of Mx proteins. For example, the Mx1 proteins from rodents are the only ones known to have an NLS close to the C terminus. This NLS is important for their antiviral activity against different viruses, including orthomyxoviruses (72). Another example is the variable and sometimes extended N-terminal domain that precedes the GTPase domain in Mx2 proteins in mammals (except rodents) and Mx proteins in birds. This N-terminal part of human MxB contains a functional NLS and a proline-rich region, which are absent in human MxA. Human MxB localizes in the heterochromatin region beneath the nuclear envelope, unlike its paralog MxA, which is cytoplasmic (85). This extended N terminus was probably lost during the evolution of mammalian Mx1 proteins. Though the expression of human MxB depends on type I IFN, no antiviral activity has yet been described for human MxB. Instead, this protein is involved in cellular processes, such as nuclear import and cell cycle progression (86). Related Mx2 proteins without known antiviral activity might be involved in similar cellular processes in other mammals. Duck Mx localizes in both the cytoplasm and the nucleus, but no nuclear localization motifs are apparent in the primary sequence. The N terminus of duck Mx might also harbor an NLS, but the importance of the N terminus for nuclear localization needs to be determined (87).
ANTIVIRAL MECHANISMS OF Mx PROTEINS
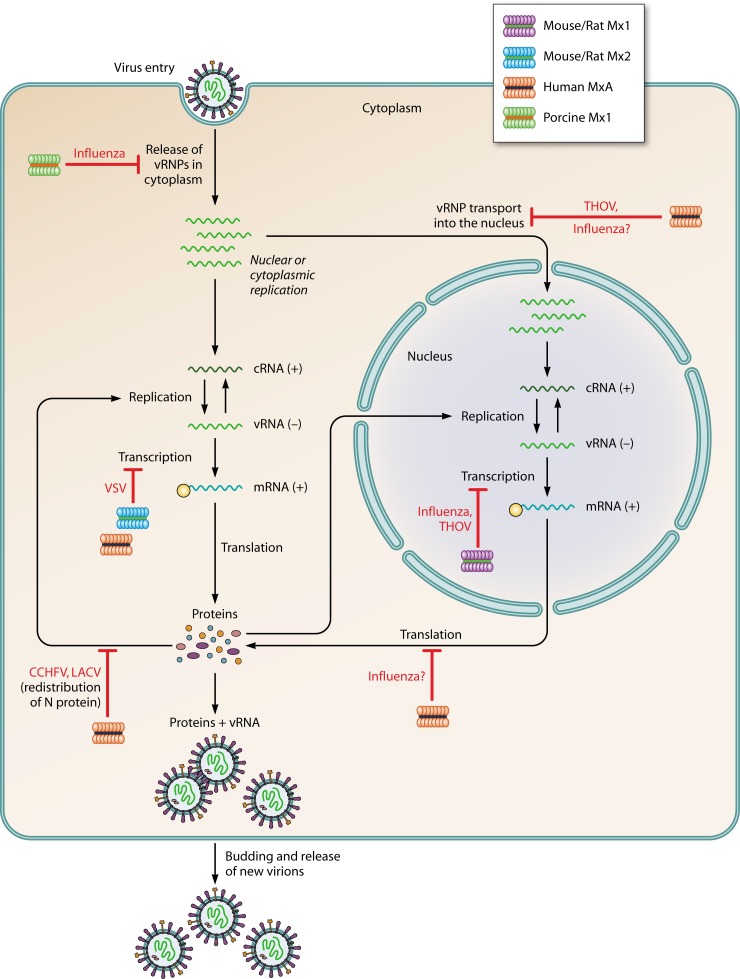
The antiviral activities of Mx proteins of different species have been studied for many RNA viruses (Table 1). Figure 4 depicts examples of different steps in the life cycles of a selection of viruses that are repressed by Mx proteins. How Mx protein family members are thought to hinder viral replication is amazingly diverse. We provide an overview of these proposed mechanisms of action, according to the virus family that is targeted.
Fig 4.
Antiviral mechanisms of Mx proteins. Steps in the life cycles of different viruses that are inhibited by Mx proteins are shown. Human MxA, mouse Mx2, and porcine Mx1 inhibit Thogoto virus (THOV), vesicular stomatitis virus (VSV), La Crosse virus (LACV), and influenza viruses in the cytoplasm. Mouse Mx1 inhibits THOV and influenza viruses in the nucleus.
Antiviral Activity against Orthomyxoviridae
Influenza virus.
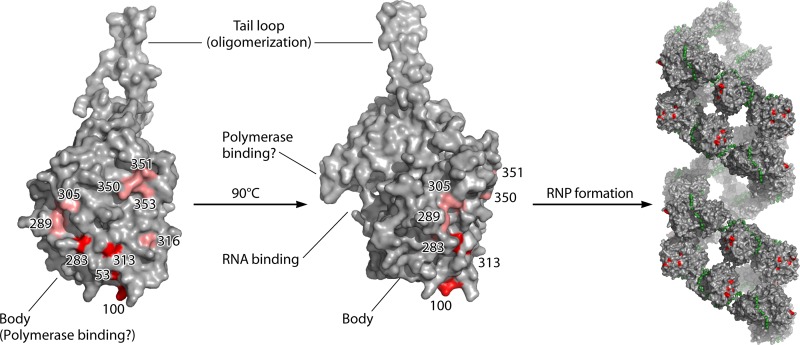
The reference virus for the antiviral activity of the Mx proteins is influenza virus, and different Mx proteins can inhibit influenza virus replication. The mouse Mx1 was the first (ortho)myxovirus resistance gene to be discovered (3, 39). The effect of mouse Mx1 expression on different steps of influenza virus replication has been studied extensively, yet the exact antiviral mechanism remains unclear. Mouse Mx1 does not seem to affect the uncoating of the virus or the transport of viral ribonucleoproteins (vRNPs) into the nucleus (88–90). Infection of IFN-stimulated cells, and therefore Mx1-expressing cells, resulted in reduced primary viral transcription and viral mRNA translation (2, 90). A caveat of these two studies is that the antiviral activity of the mouse Mx1 protein was combined with other IFN-dependent antiviral effects. Although both studies showed that IFN treatment inhibits transcription and translation, the part attributed to the Mx1 protein was assigned differently: in IFN-treated macrophages the presence of Mx1 affected mainly viral translation, whereas in IFN-treated embryonic fibroblasts it affected mostly primary transcription (2, 90). Pavlovic and colleagues confirmed that mouse Mx1 suppresses primary transcription of influenza A virus genes by using a stable fibroblast cell line that constitutively expresses mouse Mx1, thereby excluding IFN-dependent effects on viral replication (91). Mx1 differentially inhibits the transcription of the different influenza virus segments. It strongly affects the largest transcripts (encoding PB1, PB2, and PA) and moderately affects the hemagglutinin (HA) and NP transcripts, but it hardly affects the transcripts generated from the M and NS segments. This correlation between the degree of inhibition and the length of the segment suggests that Mx1 hampers transcriptional elongation rather than initiation (2, 91). The inhibitory effect of mouse Mx1 has often been studied by using a minireplicon system with a reporter gene as a readout (5, 10, 92). This assay indicates that mouse Mx1 targets the influenza vRNPs. Two subunits of the ribonucleoprotein complex have been suggested as targets for mouse Mx1 activity. The first is the PB2 protein, as its overexpression can outcompete the activity of Mx1 (92, 93). The second is the NP protein, because the sensitivities of different influenza A virus strains to mouse Mx1 and human MxA depend on their NP proteins: virus strains of human origin are more resistant to the effects of Mx1 and MxA than strains of avian origin (5, 10, 94). Recently, Mänz and colleagues (95) identified the parts in NP that are responsible for Mx sensitivity in the 1918 and 2009 pandemic virus strains (A/Brevig Mission/1/1918 and A/Hamburg/4/2009, respectively). In both cases, a minimal set of three residues was important for increased Mx1 (and MxA) resistance (positions 100/283/313 and 53/100/313 for the 1918 and 2009 pandemic viruses, respectively). These residues are grouped in separate but overlapping regions in the body domain of the NP protein (Fig. 5). These residues are solvent exposed and therefore readily accessible for cellular factors, such as the Mx1 protein (95). We recently demonstrated that Mx1 interacts with both the NP and PB2 proteins and disturbs their interaction by a mechanism that depends on GTP binding and/or GTPase activity. Surprisingly, the interactions of Mx1 with NP and PB2 do not depend on GTPase activity, which is essential for its antiviral activity (94). In the future, it will be interesting to determine if a higher Mx resistance of specific NP variants correlates with a weaker binding to Mx1 (and MxA) to confirm the crucial role of influenza virus NP in Mx1 sensitivity. Taken together, these results suggest that Mx1 targets vRNPs, possibly at the NP-polymerase interface, leading to disruption of the vRNPs and inhibition of influenza virus infection. It will be interesting to confirm these interactions in a cell-free system using purified, enzymatically active vRNPs and Mx1 and to demonstrate that in this system Mx1 can disrupt the PB2-NP interaction.
Fig 5.
Amino acid residues in influenza A virus nucleoprotein associated with sensitivity to Mx. Left, 3D structure of an NP monomer (PDB file 2Q06 [145]). The residues important for Mx1 and MxA resistance are highlighted in red. Residues that contribute in only a minor way to Mx1 resistance are indicated in light red (95). Right, these important residues are also exposed on the surface of viral ribonucleoprotein (RNP) complexes (PDB file 4BBL [146]). The RNA strand is depicted in green. The images were generated with PyMOL.
Human MxA localizes to the cytoplasm, and, like Mx1, it has been extensively studied as a suppressor of influenza virus infection. For MxA, this suppression occurs at an ill-defined step after viral primary transcription but before viral protein expression (15, 91). When the MxA protein is directed to the nucleus by making a fusion with a heterologous NLS (e.g., that of the simian virus 40 [SV40] large T antigen), its mode of action becomes similar to that of the mouse Mx1 protein in that it also blocks primary transcription of influenza virus genes (96). In contrast to the human MxA protein, which can inhibit influenza virus replication inside the cytoplasm and the nucleus, the mouse Mx1 protein is active only inside the nucleus. This restriction for mouse Mx1 was demonstrated by using a cytoplasmic mutant of Mx1 (R614E, mutated to resemble MxA) that has no anti-influenza virus (or anti-VSV) activity. When this protein was translocated back to the nucleus by a heterologous NLS, it regained its antiviral activity (72).
Why does mouse Mx1 have to be in the nucleus to exert its antiviral activity? It is possible that mouse Mx1 needs another cellular factor that is present only in the nucleus or that it is posttranslationally modified by a nuclear protein. For example, the RNA helicase UAP56 (or URH49) might act as a bridging protein, as it interacts with NP and with mouse Mx1 and human MxA (97, 98). However, this helicase is not restricted to the nucleus and partially redistributes to the cytoplasm in the presence of human MxA (98), which argues against the hypothesis that it could be responsible for the strict nuclear activity of the mouse Mx1 protein. Other potential bridging factors that bind to influenza virus vRNP components include nucleophosmin (99), CDK9 (100), USP11 (101), and Hsp90 (102, 103). It is also possible that mouse Mx1 recognizes viral targets that adopt the right conformation only in the nucleus. These viral targets are probably the incoming vRNPs of orthomyxoviruses, which are transported to the nucleus following their release in the cytoplasm after viral entry (2, 91). Apparently, these vRNPs cannot be recognized by the cytoplasmic mouse Mx1 R614E mutant when they are transported to the nuclear pore. Recognition of these vRNPs by Mx1 might require conformational or structural changes during or after their transport to the nucleus. Although experimental evidence is missing, the recently resolved crystal structure of MxA led to the suggestion that human MxA blocks influenza virus by recognizing the incoming vRNPs, followed by self-assembly of MxA rings around these vRNPs (73, 104). This mechanism is comparable to that for the recognition and inhibition of THOV (22, 105).
Nuclear transport of orthomyxovirus vRNPs is dependent on an NLS on their main structural component, the NP protein. The NP protein is not the only viral protein that interacts with the nuclear import machinery. The interaction between the viral PB2 and NP proteins and different importin alpha isoforms is also important for virus replication (106). If, as proposed, NP and PB2 are targets for the Mx1 protein, their colocalization at nuclear pores could be important for the antiviral activity of the Mx1 protein. Similarly, MxA could recognize both proteins at the cytoplasmic face of the nuclear pores. In this way, Mx proteins could function as antiviral gatekeepers at the nuclear membrane, blocking vRNPs just before or after nuclear entry.
The similarity in anti-influenza virus mechanisms of mouse Mx1 and MxA targeted to the nucleus, together with the similar Mx resistance phenotypes of certain virus strains, indicates that both Mx proteins recognize the same viral target, most likely the NP protein in vRNP structures. This is in line with the demonstrated interaction between (nuclear) MxA and influenza virus NP after cross-linking (96).
In contrast to mouse Mx1, which does not recognize the influenza virus vRNPs inside the cytoplasm, the pig Mx1 protein recognizes these vRNPs and somehow prevents their transport into the nucleus (107). However, pig Mx1 works indirectly by influencing the endocytic pathway, leading to a delayed and centripetal movement of viral particles to the nucleus (107).
THOV.
Another member of the Orthomyxoviridae that is inhibited by Mx proteins is Thogoto virus (THOV), a natural pathogen of mice (7, 14, 18, 22, 105, 108). THOV is very sensitive to human MxA activity, and the antiviral mechanism was resolved by Kochs and colleagues (22, 105). Human MxA blocks a very early step in the viral life cycle by preventing the transport of THOV vRNPs into the nucleus. This blockage is mediated by the interaction of MxA with RNA-bound NP; this interaction probably shields the signals responsible for nuclear translocation of the vRNPs (22, 105). The NP interaction interface in MxA was mapped to its C terminus, in a domain that includes loop L4 (105, 109). The interaction between MxA and THOV NP was also blocked by the 2C12 antibody, which recognizes an epitope in the C terminus, confirming the importance of the C terminus for NP binding (22, 68, 105). MxA had no effect on other steps in the viral life cycle, such as protein synthesis or transport of newly produced proteins into the nucleus. In contrast, the nuclear forms of human MxA and probably also mouse Mx1 block THOV polymerase activity in the nucleus without affecting vRNP transport (108). This demonstrates that the antiviral mechanism of Mx proteins is determined at least in part by their subcellular localization. It is possible that the mouse Mx1 protein inhibits the THOV polymerase complex like it inhibits the influenza virus polymerase complex, but this has not been demonstrated.
Common features of Mx antiviral activity against Orthomyxoviridae.
Inhibition of orthomyxoviruses by Mx proteins generally requires an intact GTPase domain. This has been demonstrated by examining the antiviral activities of different GTPase-inactive mutants (63, 64, 110). For example, the K49A mutation in the first GTP-binding motif of mouse Mx1 prevents GTP binding and subsequent GTPase activity. This mutant Mx1 protein lacks antiviral activity against influenza A virus, as determined by viral protein expression and by an influenza virus minireplicon assay (5, 63). The importance of an intact GTPase domain was further demonstrated by the observation that human MxA interacts with THOV NP in a cosedimentation assay only in the presence of GTPγS. This nonhydrolyzable GTP analog presumably stabilized the MxA-NP interaction (105). It is not clear if GTPase activity is required or if GTP binding alone is sufficient for antiviral activity. GTP binding and GTPase activity are intimately connected, and there are no reports on Mx mutants that lack GTPase but still have GTP-binding activity. However, one mutant of human MxA (L612K) lacks detectable GTPase activity but has antiviral activity against THOV in infection and in a minireplicon system (111). Since the L612K mutation is located near the C terminus of MxA, far from the GTP-binding domain, this mutant can probably bind GTP, indicating that GTP binding is sufficient for the antiviral activity of MxA (111).
The activity of Mx proteins against orthomyxoviruses is neutralized by the 2C12 antibody (68, 71, 84, 112). This indicates a similar virus recognition region in three Mx homologs: MxA (residues 432 to 471), mouse Mx1, and rat Mx1. This 2C12 region is well conserved in all Mx proteins, which suggests that this region is important for its biological function. Besides the 2C12 epitope region, a second loop in the stalk domain of Mx proteins was recently found to be a genetic determinant of the species-specific antiviral activity of primate MxA proteins against members of the Orthomyxoviridae family (109). This loop, called L4 (MxA residues 533 to 572 [Fig. 1]), is essential for the antiviral activity of primate MxA against THOV and is also essential in the interaction between active MxA proteins and THOV NP protein. In addition, replacing loop L4 in mouse Mx1 with the human variant enhances its antiviral activity against THOV and allows the coimmunoprecipitation of THOV NP. However, replacing this loop in inactive primate MxA proteins by the active human MxA L4 loop is not enough to provide full protection against THOV infection. This suggests that other determinants are also important for MxA activity. Finally, loop L4, and residue F561 in particular, is essential for the antiviral activity of human MxA against influenza viruses, which means that loop L4 is important in the protection against orthomoxoviruses in general (109). This loop is less conserved than the 2C12 region. Particularly the second part of the loop, which contains residue F561, differs strongly among Mx proteins. It is tempting to speculate that this part of Mx is a determinant of its viral target specificity. The position of both loops at the tip of the Mx stalk suggests that this region is important for virus target recognition (Fig. 1B). This similarity in virus recognition provides support for the recently proposed model of a general antiviral activity against Orthomyxoviridae, in which MxA oligomerizes around incoming vRNPs and prevents their nuclear import (73, 104). However, subtle differences in Mx proteins, in particular their subcellular localization, are involved in antiviral specificity.
Antiviral Activity against Rhabdoviridae
Vesicular stomatitis virus (VSV) is inhibited by most cytoplasmic Mx proteins (references are given in Table 1). MxA inhibits VSV mRNA synthesis, probably by affecting elongation of the viral RNA chain, and requires an intact GTPase domain (16, 63, 110). Inhibition of viral transcription was confirmed by an in vitro study using purified MxA and viral vRNPs (113). Purified MxA inhibits VSV transcription in the presence of GTP as well as in the presence of nonhydrolyzable GTP analogs (GMP-PNP or GTPγS) (69, 113). This suggests that GTP binding is sufficient for the antiviral activity of MxA against VSV and that GTPase activity is not needed. Although this in vitro study suggests that MxA targets the vRNPs of VSV, an interaction between human MxA and VSV proteins has not yet been demonstrated (16). The inhibition of VSV mRNA synthesis resembles the inhibition of influenza virus transcription by mouse Mx1. However, it is not known if and how human MxA interferes with the interaction between the VSV nucleoprotein and the polymerase complex.
Antiviral Activity against Bunyaviridae
Human MxA and mouse and rat Mx2 proteins inhibit different members of the Bunyaviridae (references are given in Table 1). The best-studied example is the inhibition of La Crosse virus (LACV) by human MxA (19, 23, 24, 27). MxA inhibits LACV replication by sequestering the newly produced viral nucleocapsid protein at large, membrane-associated perinuclear complexes. This missorting prevents the nucleocapsid protein from performing its function in viral replication (23, 27). In contrast, Frese and colleagues suggested that the active viral RNA polymerase complex is the target for MxA. They demonstrated that human MxA protein inhibits the accumulation of viral transcripts, and particularly the longer ones, suggesting an effect on elongation (19). However, this effect on elongation could be secondary to sequestration of the nucleocapsid protein. This sequestration would probably also affect mostly the large segments, as they need more nucleocapsid protein for their replication.
MxA also seems to inhibit Crimean-Congo hemorrhagic fever virus (CCHFV) by a similar mechanism (26), indicating that it has a similar mode of action against other viruses of this family, including Rift Valley fever virus (RVFV), Hantaan virus (HTNV), Puumala virus (PUUV), and Tula virus (TULV) (19, 20). However, an argument against a general antiviral mechanism is the observation that the nucleocapsid protein of Dugbe virus (DUGV) appears to avoid MxA activity and is not sequestered to the perinuclear region by human MxA (114).
Antiviral Activity against Paramyxoviridae
Human MxA has antiviral activity against some viruses from the Paramyxoviridae family (17, 115). These antiviral activities are dependent on cell type and virus strain. For example, MxA can inhibit measles virus in the human mononuclear cell line U937 and in the glioblastoma cell line U87 but not in Vero or Hep-2 cells (17, 21, 115). Moreover, MxA can inhibit primary transcription in U87 cells but not in U937 cells (17), in which it inhibits viral glycoprotein synthesis instead (115). Human respiratory syncytial virus (HRSV) is not inhibited by MxA in U87 or Vero cells (116), but its close relative murine pneumovirus is inhibited by transgenic mouse cells expressing bovine (Bos taurus) Mx1 protein (117).
Antiviral Activity against Other Viruses
The antiviral activities of the different Mx proteins described so far are all directed against negative-stranded RNA viruses. Nevertheless, human MxA and different fish Mx proteins have antiviral activity also against other RNA and even some DNA viruses. Human MxA has antiviral activity against the positive-stranded RNA viruses Semliki Forest virus (SFV) and classical swine fever virus (CSFV) (21, 24, 28). Also, Mx proteins of fish confer resistance against positive-stranded RNA viruses (32, 35, 37, 118). For example, grouper Mx can inhibit yellow grouper nervous necrosis virus (YGNNV) by inhibiting viral mRNA synthesis. The inhibition is likely mediated by the observed interaction between Mx and the viral coat protein, which probably disturbs the localization of the coat protein (32). In addition, the barramundi Mx protein inhibits nervous necrosis virus (NNV) by redistribution of the viral RNA-dependent RNA polymerase to the perinuclear area, where it is degraded (37). Human MxA and fish Mx proteins can also inhibit double-stranded RNA (dsRNA) viruses. For example, salmon Mx and seabream Mx proteins can inhibit infectious pancreatic necrosis virus (IPNV) (30, 34, 119).
Mx proteins also have antiviral activity against dsDNA viruses. Human MxA can inhibit hepatitis B virus (HBV) (25, 120, 121) and African swine fever virus (ASFV) (29). MxA inhibits HBV replication at several steps during the viral life cycle. For example, MxA can inhibit the nucleocytoplasmic export of viral mRNAs, probably by being redistributed partly to the nucleus during HBV replication (25, 120). It was also demonstrated that MxA can interact with the hepatitis B virus core antigen, leading to its sequestration and immobilization in perinuclear compartments (121). This antiviral mechanism resembles the antiviral activity of MxA against LACV, an RNA virus (23, 27). Remarkably, inhibition of HBV by MxA is retained by MxA mutants with K83A or T103A substitutions. These mutations abolish the GTPase activity of MxA and are detrimental for the antiviral activity of MxA against RNA viruses (63, 120). This demonstrates that GTPase activity of MxA is dispensable for the inhibition of HBV (120). Finally, MxA protein complexes inhibit ASFV replication by encircling the virus factories (29).
CONCLUSIONS
The studies described here collectively demonstrate that Mx proteins have a broad range of antiviral activity. It is unclear if all the sensitive viruses are inhibited by a common mechanism, but in general, Mx proteins recognize the nucleoproteins or (nucleo)capsid proteins of different viruses, perhaps concurrently with the viral polymerase. The C termini of Mx proteins are responsible for recognition of the viral targets and for their differential antiviral activities. Identifying the common molecular pattern in the viral target proteins that is recognized by the Mx proteins is an exciting research objective. One possible common feature is the nucleotide-binding viral proteins. Moreover, the arrangement of multiple nucleoprotein or (nucleo)capsid protein molecules, e.g., in vRNPs or virus factories, allows extensive, repetitive contacts between Mx complexes and the viral targets. Consequently, even a weak-affinity contact could result in a biologically significant interaction.
Recent studies indicate ongoing coevolution between viruses and antiviral Mx proteins. For example, the stronger resistance of human influenza virus strains (relative to that of avian strains) against MxA activity illustrates the evolution of seasonal influenza virus strains to cope with MxA. This phenomenon is also seen for swine influenza viruses, although less prominently due to the lower antiviral activity of the swine Mx1 protein (95). The higher sensitivity of avian influenza virus strains suggests that MxA operates as an important barrier against the zoonotic introduction of avian influenza viruses in the human population. Understanding the molecular determinants of Mx resistance will help to estimate the pandemic potential of new influenza strains. In addition, a recent study also highlights the importance of this coevolution by revealing specific residues in primate MxA proteins which are under positive selection. These residues (mainly in loop L4) are predicted to be important in viral target recognition (109) (the evolution of MxA antiviral diversity was recently reviewed in reference 122).
How do Mx proteins perform their antiviral functions? An intact GTPase domain is required for activity against RNA viruses, but it is not clear if GTP binding is sufficient or GTPase activity is also needed. As the crystal structure of MxA is based on nucleotide-free MxA, it will be interesting to determine the conformational changes caused by GTP binding and GTP hydrolysis in an enzymatically active MxA protein. The information obtained from the crystal structure of MxA, together with mutational studies, led to the proposition of an antiviral mechanism that depends on the multimeric assembly of Mx proteins. In this model, Mx forms tetramers that further oligomerize into large Mx rings at higher Mx concentrations. These rings contain GTPase domains on the outside of the ring and stalks pointing inwards (123). This assembly and oligomerization allow the Mx stalk to interact with viral target structures, e.g., vRNPs, possibly leading to multiple Mx rings wrapping around these structures. Adjacent Mx rings can interact through their GTPase domains, leading to enhanced GTPase activity. This conformation allows cross talk between the GTPase domains and the stalks of neighboring Mx molecules and causes conformational changes leading to constriction of Mx rings and disruption of functional viral vRNPs by a mechano-chemical cycle. In this way, Mx rings could disrupt the association of nucleoprotein complexes with the viral polymerase, thereby inhibiting virus transcription or replication. These Mx rings could also act to relocalize viral (ribo)nucleoproteins or (nucleo)capsid proteins to perinuclear complexes in a GTPase-dependent way, leading to their sequestration and/or degradation. Although this ring-forming model is very appealing, there is no proof that ring formation of Mx around vRNPs or other viral structures actually occurs.
To summarize, Mx proteins are evolutionarily conserved, and they inhibit a wide range of viruses, probably by recognizing large viral structures possessing a degree of symmetry and/or iteration. In this sense, Mx proteins could be considered pattern recognition molecules with a direct antiviral effector function.
ACKNOWLEDGMENTS
We thank Amin Bredan for critically reading the manuscript.
Research related to Mx in the group of Xavier Saelens was supported by research grant G.0.375.10 and IOF project IOF10/StarTT/027 from Ghent University. Judith Verhelst was a research fellow of FWO-Vlaanderen.
Biographies

Judith Verhelst obtained her Ph.D. in Sciences-Biotechnology in 2012 from Ghent University, Belgium. She currently works as a postdoctoral researcher in the Molecular Virology Unit in the Department for Molecular Biomedical Research at Ghent University and VIB. Her research focuses on the interaction between mammalian Mx and the influenza A virus proteins to understand the molecular details of the antiviral activity of these dynamin-like GTPases.

Paco Hulpiau is a bioinformatician in the Department for Molecular Biomedical Research at VIB. He is a guest lecturer in structural bioinformatics at Howest, University College, West Flanders, from which he graduated magna cum laude in pharmaceutical and biological laboratory technology.

Xavier Saelens obtained his Ph.D. in Sciences-Biotechnology in 1990 at Ghent University. His postdoctoral research included the development of novel vaccines against influenza viruses and the elucidation of signal transduction pathways that control the fate of protein synthesis during cell death. In 2004 he became the head of the Molecular Virology Unit in the Department for Molecular Biomedical Research at Ghent University and VIB. In 2008 he became an Associate Professor in Molecular Virology at Ghent University. His team is studying innate and adaptive immune responses against influenza A and B viruses and human respiratory syncytial virus, with a focus on Mx proteins and the development of novel vaccines and antivirals against these respiratory viruses.
REFERENCES
- 1.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krug RM, Shaw M, Broni B, Shapiro G, Haller O. 1985. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. J. Virol. 56:201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindenmann J, Lane CA, Hobson D. 1963. The resistance of A2G mice to myxoviruses. J. Immunol. 90:942–951 [PubMed] [Google Scholar]
- 4.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147–158 [DOI] [PubMed] [Google Scholar]
- 5.Dittmann J, Stertz S, Grimm D, Steel J, Garcia-Sastre A, Haller O, Kochs G. 2008. Influenza A virus strains differ in sensitivity to the antiviral action of the Mx-GTPase. J. Virol. 82:3624–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm D, Staeheli P, Hufbauer M, Koerner I, Martinez-Sobrido L, Solorzano A, Garcia-Sastre A, Haller O, Kochs G. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:6806–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haller O, Frese M, Rost D, Nuttall PA, Kochs G. 1995. Tick-borne thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J. Virol. 69:2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salomon R, Staeheli P, Kochs G, Yen HL, Franks J, Rehg JE, Webster RG, Hoffmann E. 2007. Mx1 gene protects mice against the highly lethal human H5N1 influenza virus. Cell Cycle 6:2417–2421 [DOI] [PubMed] [Google Scholar]
- 9.Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 81:10818–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza a viruses. J. Virol. 85:8133–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin HK, Takada A, Kon Y, Haller O, Watanabe T. 1999. Identification of the murine Mx2 gene: interferon-induced expression of the Mx2 protein from the feral mouse gene confers resistance to vesicular stomatitis virus. J. Virol. 73:4925–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zurcher T, Pavlovic J, Staeheli P. 1992. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology 187:796–800 [DOI] [PubMed] [Google Scholar]
- 13.Jin HK, Yoshimatsu K, Takada A, Ogino M, Asano A, Arikawa J, Watanabe T. 2001. Mouse Mx2 protein inhibits hantavirus but not influenza virus replication. Arch. Virol. 146:41–49 [DOI] [PubMed] [Google Scholar]
- 14.Pavlovic J, Arzet HA, Hefti HP, Frese M, Rost D, Ernst B, Kolb E, Staeheli P, Haller O. 1995. Enhanced virus resistance of transgenic mice expressing the human MxA protein. J. Virol. 69:4506–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlovic J, Zurcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64:3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staeheli P, Pavlovic J. 1991. Inhibition of vesicular stomatitis virus mRNA synthesis by human MxA protein. J. Virol. 65:4498–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider-Schaulies S, Schneider-Schaulies J, Schuster A, Bayer M, Pavlovic J, ter Meulen V. 1994. Cell type-specific MxA-mediated inhibition of measles virus transcription in human brain cells. J. Virol. 68:6910–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frese M, Kochs G, Meier-Dieter U, Siebler J, Haller O. 1995. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J. Virol. 69:3904–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frese M, Kochs G, Feldmann H, Hertkorn C, Haller O. 1996. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J. Virol. 70:915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanerva M, Melen K, Vaheri A, Julkunen I. 1996. Inhibition of puumala and tula hantaviruses in Vero cells by MxA protein. Virology 224:55–62 [DOI] [PubMed] [Google Scholar]
- 21.Landis H, Simon-Jodicke A, Kloti A, Di Paolo C, Schnorr JJ, Schneider-Schaulies S, Hefti HP, Pavlovic J. 1998. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J. Virol. 72:1516–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochs G, Haller O. 1999. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 96:2082–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kochs G, Janzen C, Hohenberg H, Haller O. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc. Natl. Acad. Sci. U. S. A. 99:3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hefti HP, Frese M, Landis H, Di Paolo C, Aguzzi A, Haller O, Pavlovic J. 1999. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La crosse virus and other lethal viral infections. J. Virol. 73:6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordien E, Rosmorduc O, Peltekian C, Garreau F, Brechot C, Kremsdorf D. 2001. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 75:2684–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson I, Bladh L, Mousavi-Jazi M, Magnusson KE, Lundkvist A, Haller O, Mirazimi A. 2004. Human MxA protein inhibits the replication of Crimean-Congo hemorrhagic fever virus. J. Virol. 78:4323–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichelt M, Stertz S, Krijnse-Locker J, Haller O, Kochs G. 2004. Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic 5:772–784 [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Pang D, Wang T, Yang X, Wu R, Ren L, Yuan T, Huang Y, Ouyang H. 2011. Human MxA protein inhibits the replication of classical swine fever virus. Virus Res. 156:151–155 [DOI] [PubMed] [Google Scholar]
- 29.Netherton CL, Simpson J, Haller O, Wileman TE, Takamatsu HH, Monaghan P, Taylor G. 2009. Inhibition of a large double-stranded DNA virus by MxA protein. J. Virol. 83:2310–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mundt E. 2007. Human MxA protein confers resistance to double-stranded RNA viruses of two virus families. J. Gen. Virol. 88:1319–1323 [DOI] [PubMed] [Google Scholar]
- 31.Caipang CM, Hirono I, Aoki T. 2003. In vitro inhibition of fish rhabdoviruses by Japanese flounder, Paralichthys olivaceus Mx. Virology 317:373–382 [DOI] [PubMed] [Google Scholar]
- 32.Chen YM, Su YL, Shie PS, Huang SL, Yang HL, Chen TY. 2008. Grouper Mx confers resistance to nodavirus and interacts with coat protein. Dev. Comp. Immunol. 32:825–836 [DOI] [PubMed] [Google Scholar]
- 33.Kibenge MJ, Munir K, Kibenge FS. 2005. Constitutive expression of Atlantic salmon Mx1 protein in CHSE-214 cells confers resistance to infectious salmon anaemia virus. Virol. J. 2:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen R, Rokenes TP, Robertsen B. 2004. Inhibition of infectious pancreatic necrosis virus replication by atlantic salmon Mx1 protein. J. Virol. 78:7938–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin CH, Christopher John JA, Chang CY. 2006. Inhibition of nervous necrosis virus propagation by fish Mx proteins. Biochem. Biophys. Res. Commun. 351:534–539 [DOI] [PubMed] [Google Scholar]
- 36.Su J, Yang C, Zhu Z, Wang Y, Jang S, Liao L. 2009. Enhanced grass carp reovirus resistance of Mx-transgenic rare minnow (Gobiocypris rarus). Fish Shellfish Immunol. 26:828–835 [DOI] [PubMed] [Google Scholar]
- 37.Wu YC, Lu YF, Chi SC. 2010. Anti-viral mechanism of barramundi Mx against betanodavirus involves the inhibition of viral RNA synthesis through the interference of RdRp. Fish Shellfish Immunol. 28:467–475 [DOI] [PubMed] [Google Scholar]
- 38.Jensen I, Robertsen B. 2002. Effect of double-stranded RNA and interferon on the antiviral activity of Atlantic salmon cells against infectious salmon anemia virus and infectious pancreatic necrosis virus. Fish Shellfish Immunol. 13:221–241 [DOI] [PubMed] [Google Scholar]
- 39.Lindenmann J. 1962. Resistance of mice to mouse-adapted influenza A virus. Virology 16:203–204 [DOI] [PubMed] [Google Scholar]
- 40.Staeheli P, Pravtcheva D, Lundin LG, Acklin M, Ruddle F, Lindenmann J, Haller O. 1986. Interferon-regulated influenza virus resistance gene Mx is localized on mouse chromosome 16. J. Virol. 58:967–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haller O, Arnheiter H, Gresser I, Lindenmann J. 1979. Genetically determined, interferon-dependent resistance to influenza virus in mice. J. Exp. Med. 149:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staeheli P, Colonno RJ, Cheng YS. 1983. Different mRNAs induced by interferon in cells from inbred mouse strains A/J. and A2G. J. Virol. 47:563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horisberger MA, Staeheli P, Haller O. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc. Natl. Acad. Sci. U. S. A. 80:1910–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staeheli P, Haller O. 1985. Interferon-induced human protein with homology to protein Mx of influenza virus-resistant mice. Mol. Cell. Biol. 5:2150–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staeheli P, Sutcliffe JG. 1988. Identification of a second interferon-regulated murine Mx gene. Mol. Cell. Biol. 8:4524–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aebi M, Fah J, Hurt N, Samuel CE, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staeheli P, Danielson P, Haller O, Sutcliffe JG. 1986. Transcriptional activation of the mouse Mx gene by type I interferon. Mol. Cell. Biol. 6:4770–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hug H, Costas M, Staeheli P, Aebi M, Weissmann C. 1988. Organization of the murine Mx gene and characterization of its interferon- and virus-inducible promoter. Mol. Cell. Biol. 8:3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holzinger D, Jorns C, Stertz S, Boisson-Dupuis S, Thimme R, Weidmann M, Casanova JL, Haller O, Kochs G. 2007. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J. Virol. 81:7776–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon A, Fah J, Haller O, Staeheli P. 1991. Interferon-regulated Mx genes are not responsive to interleukin-1, tumor necrosis factor, and other cytokines. J. Virol. 65:968–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schumacher B, Bernasconi D, Schultz U, Staeheli P. 1994. The chicken Mx promoter contains an ISRE motif and confers interferon inducibility to a reporter gene in chick and monkey cells. Virology 203:144–148 [DOI] [PubMed] [Google Scholar]
- 53.Yankey SJ, Hicks BA, Carnahan KG, Assiri AM, Sinor SJ, Kodali K, Stellflug JN, Ott TL. 2001. Expression of the antiviral protein Mx in peripheral blood mononuclear cells of pregnant and bred, non-pregnant ewes. J. Endocrinol. 170:R7–R11 [DOI] [PubMed] [Google Scholar]
- 54.Gerardin JA, Baise EA, Pire GA, Leroy MP, Desmecht DJ. 2004. Genomic structure, organisation, and promoter analysis of the bovine (Bos taurus) Mx1 gene. Gene 326:67–75 [DOI] [PubMed] [Google Scholar]
- 55.Collet B, Secombes CJ. 2001. The rainbow trout (Oncorhynchus mykiss) Mx1 promoter. Structural and functional characterization. Eur. J. Biochem. 268:1577–1584 [PubMed] [Google Scholar]
- 56.Ooi EL, Hirono I, Aoki T. 2006. Functional characterisation of the Japanese flounder, Paralichthys olivaceus, Mx promoter. Fish Shellfish Immunol. 21:293–304 [DOI] [PubMed] [Google Scholar]
- 57.Haller O, Kochs G. 2002. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710–717 [DOI] [PubMed] [Google Scholar]
- 58.Praefcke GJ, McMahon HT. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147 [DOI] [PubMed] [Google Scholar]
- 59.Nakayama M, Nagata K, Kato A, Ishihama A. 1991. Interferon-inducible mouse Mx1 protein that confers resistance to influenza virus is GTPase. J. Biol. Chem. 266:21404–21408 [PubMed] [Google Scholar]
- 60.Leong JC, Trobridge GD, Kim CH, Johnson M, Simon B. 1998. Interferon-inducible Mx proteins in fish. Immunol. Rev. 166:349–363 [DOI] [PubMed] [Google Scholar]
- 61.Song BD, Leonard M, Schmid SL. 2004. Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J. Biol. Chem. 279:40431–40436 [DOI] [PubMed] [Google Scholar]
- 62.Melen K, Ronni T, Lotta T, Julkunen I. 1994. Enzymatic characterization of interferon-induced antiviral GTPases murine Mx1 and human MxA proteins. J. Biol. Chem. 269:2009–2015 [PubMed] [Google Scholar]
- 63.Pitossi F, Blank A, Schroder A, Schwarz A, Hussi P, Schwemmle M, Pavlovic J, Staeheli P. 1993. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 67:6726–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melen K, Julkunen I. 1994. Mutational analysis of murine Mx1 protein: GTP binding core domain is essential for anti-influenza A activity. Virology 205:269–279 [DOI] [PubMed] [Google Scholar]
- 65.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465:502–506 [DOI] [PubMed] [Google Scholar]
- 66.Melen K, Ronni T, Broni B, Krug RM, von Bonsdorff CH, Julkunen I. 1992. Interferon-induced Mx proteins form oligomers and contain a putative leucine zipper. J. Biol. Chem. 267:25898–25907 [PubMed] [Google Scholar]
- 67.Nakayama M, Yazaki K, Kusano A, Nagata K, Hanai N, Ishihama A. 1993. Structure of mouse Mx1 protein. Molecular assembly and GTP-dependent conformational change. J. Biol. Chem. 268:15033–15038 [PubMed] [Google Scholar]
- 68.Flohr F, Schneider-Schaulies S, Haller O, Kochs G. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett. 463:24–28 [DOI] [PubMed] [Google Scholar]
- 69.Schwemmle M, Richter MF, Herrmann C, Nassar N, Staeheli P. 1995. Unexpected structural requirements for GTPase activity of the interferon-induced MxA protein. J. Biol. Chem. 270:13518–13523 [PubMed] [Google Scholar]
- 70.Noteborn M, Arnheiter H, Richter-Mann L, Browning H, Weissmann C. 1987. Transport of the murine Mx protein into the nucleus is dependent on a basic carboxy-terminal sequence. J. Interferon Res. 7:657–669 [DOI] [PubMed] [Google Scholar]
- 71.Meier E, Kunz G, Haller O, Arnheiter H. 1990. Activity of rat Mx proteins against a rhabdovirus. J. Virol. 64:6263–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zurcher T, Pavlovic J, Staeheli P. 1992. Nuclear localization of mouse Mx1 protein is necessary for inhibition of influenza virus. J. Virol. 66:5059–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, Kochs G, Daumke O. 2011. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525 [DOI] [PubMed] [Google Scholar]
- 74.Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. 2011. Crystal structure of nucleotide-free dynamin. Nature 477:556–560 [DOI] [PubMed] [Google Scholar]
- 75.Di Paolo C, Hefti HP, Meli M, Landis H, Pavlovic J. 1999. Intramolecular backfolding of the carboxyl-terminal end of MxA protein is a prerequisite for its oligomerization. J. Biol. Chem. 274:32071–32078 [DOI] [PubMed] [Google Scholar]
- 76.Accola MA, Huang B, Al Masri A, McNiven MA. 2002. The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 277:21829–21835 [DOI] [PubMed] [Google Scholar]
- 77.Kochs G, Haener M, Aebi U, Haller O. 2002. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 277:14172–14176 [DOI] [PubMed] [Google Scholar]
- 78.Richter MF, Schwemmle M, Herrmann C, Wittinghofer A, Staeheli P. 1995. Interferon-induced MxA protein. GTP binding and GTP hydrolysis properties. J. Biol. Chem. 270:13512–13517 [PubMed] [Google Scholar]
- 79.von der Malsburg A, Abutbul-Ionita I, Haller O, Kochs G, Danino D. 2011. Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop. J. Biol. Chem. 286:37858–37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stertz S, Reichelt M, Krijnse-Locker J, Mackenzie J, Simpson JC, Haller O, Kochs G. 2006. Interferon-induced, antiviral human MxA protein localizes to a distinct subcompartment of the smooth endoplasmic reticulum. J. Interferon Cytokine Res. 26:650–660 [DOI] [PubMed] [Google Scholar]
- 81.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 82.Bottcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y, Xu G, Patel A, McLaughlin MM, Silverman C, Knecht K, Sweitzer S, Li X, McDonnell P, Mirabile R, Zimmerman D, Boyce R, Tierney LA, Hu E, Livi GP, Wolf B, Abdel-Meguid SS, Rose GD, Aurora R, Hensley P, Briggs M, Young PR. 2002. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics 80:144–150 [DOI] [PubMed] [Google Scholar]
- 84.Meier E, Fah J, Grob MS, End R, Staeheli P, Haller O. 1988. A family of interferon-induced Mx-related mRNAs encodes cytoplasmic and nuclear proteins in rat cells. J. Virol. 62:2386–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melen K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, Julkunen I. 1996. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 271:23478–23486 [DOI] [PubMed] [Google Scholar]
- 86.King MC, Raposo G, Lemmon MA. 2004. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. U. S. A. 101:8957–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bazzigher L, Schwarz A, Staeheli P. 1993. No enhanced influenza virus resistance of murine and avian cells expressing cloned duck Mx protein. Virology 195:100–112 [DOI] [PubMed] [Google Scholar]
- 88.Broni B, Julkunen I, Condra JH, Davies ME, Berry MJ, Krug RM. 1990. Parental influenza virion nucleocapsids are efficiently transported into the nuclei of murine cells expressing the nuclear interferon-induced Mx protein. J. Virol. 64:6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horisberger MA, Haller O, Arnheiter H. 1980. Interferon-dependent genetic resistance to influenza virus in mice: virus replication in macrophages is inhibited at an early step. J. Gen. Virol. 50:205–210 [DOI] [PubMed] [Google Scholar]
- 90.Meyer T, Horisberger MA. 1984. Combined action of mouse alpha and beta interferons in influenza virus-infected macrophages carrying the resistance gene Mx. J. Virol. 49:709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pavlovic J, Haller O, Staeheli P. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 66:2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang T, Pavlovic J, Staeheli P, Krystal M. 1992. Overexpression of the influenza virus polymerase can titrate out inhibition by the murine Mx1 protein. J. Virol. 66:4154–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stranden AM, Staeheli P, Pavlovic J. 1993. Function of the mouse Mx1 protein is inhibited by overexpression of the PB2 protein of influenza virus. Virology 197:642–651 [DOI] [PubMed] [Google Scholar]
- 94.Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. 2012. Interferon-inducible Mx1 protein inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 86:13445–13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Manz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, Kochs G, Schwemmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 9:e1003279. 10.1371/journal.ppat.1003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turan K, Mibayashi M, Sugiyama K, Saito S, Numajiri A, Nagata K. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res. 32:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wisskirchen C, Ludersdorfer TH, Muller DA, Moritz E, Pavlovic J. 2011. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 85:8646–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wisskirchen C, Ludersdorfer TH, Muller DA, Moritz E, Pavlovic J. 2011. Interferon-induced antiviral protein MxA interacts with the cellular RNA helicases UAP56 and URH49. J. Biol. Chem. 286:34743–34751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Li G, Ye X. 2010. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J. Virol. 84:12619–12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. 2010. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 29:3879–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Naito T, Momose F, Kawaguchi A, Nagata K. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81:1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306–45314 [DOI] [PubMed] [Google Scholar]
- 104.Daumke O, Gao S, von der Malsburg A, Haller O, Kochs G. 2010. Structure of the MxA stalk elucidates the assembly of ring-like units of an antiviral module. Small GTPases 1:62–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kochs G, Haller O. 1999. GTP-bound human MxA protein interacts with the nucleocapsids of Thogoto virus (Orthomyxoviridae). J. Biol. Chem. 274:4370–4376 [DOI] [PubMed] [Google Scholar]
- 106.Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11. 10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Palm M, Garigliany MM, Cornet F, Desmecht D. 2010. Interferon-induced Sus scrofa Mx1 blocks endocytic traffic of incoming influenza A virus particles. Vet. Res. 41:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weber F, Haller O, Kochs G. 2000. MxA GTPase blocks reporter gene expression of reconstituted Thogoto virus ribonucleoprotein complexes. J. Virol. 74:560–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mitchell PS, Patzina C, Emerman M, Haller O, Malik HS, Kochs G. 2012. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12:598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ponten A, Sick C, Weeber M, Haller O, Kochs G. 1997. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 71:2591–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Janzen C, Kochs G, Haller O. 2000. A monomeric GTPase-negative MxA mutant with antiviral activity. J. Virol. 74:8202–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arnheiter H, Haller O. 1988. Antiviral state against influenza virus neutralized by microinjection of antibodies to interferon-induced Mx proteins. EMBO J. 7:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwemmle M, Weining KC, Richter MF, Schumacher B, Staeheli P. 1995. Vesicular stomatitis virus transcription inhibited by purified MxA protein. Virology 206:545–554 [DOI] [PubMed] [Google Scholar]
- 114.Bridgen A, Dalrymple DA, Weber F, Elliott RM. 2004. Inhibition of Dugbe nairovirus replication by human MxA protein. Virus Res. 99:47–50 [DOI] [PubMed] [Google Scholar]
- 115.Schnorr JJ, Schneider-Schaulies S, Simon-Jodicke A, Pavlovic J, Horisberger MA, ter Meulen V. 1993. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J. Virol. 67:4760–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Atreya PL, Kulkarni S. 1999. Respiratory syncytial virus strain A2 is resistant to the antiviral effects of type I interferons and human MxA. Virology 261:227–241 [DOI] [PubMed] [Google Scholar]
- 117.Dermine M, Desmecht D. 2012. In vivo modulation of the innate response to pneumovirus by type-I and -III interferon-induced Bos taurus Mx1. J. Interferon Cytokine Res. 32:332–337 [DOI] [PubMed] [Google Scholar]
- 118.Babiker HA, Nakatsu Y, Yamada K, Yoneda A, Takada A, Ueda J, Hata H, Watanabe T. 2007. Bovine and water buffalo Mx2 genes: polymorphism and antiviral activity. Immunogenetics 59:59–67 [DOI] [PubMed] [Google Scholar]
- 119.Fernandez-Trujillo MA, Novel P, Manchado M, Sepulcre MP, Mulero V, Borrego JJ, Alvarez MC, Bejar J. 2011. Three Mx genes with differential response to VNNV infection have been identified in Gilthead seabream (Sparus aurata). Mol. Immunol. 48:1216–1223 [DOI] [PubMed] [Google Scholar]
- 120.Yu Z, Wang Z, Chen J, Li H, Lin Z, Zhang F, Zhou Y, Hou J. 2008. GTPase activity is not essential for the interferon-inducible MxA protein to inhibit the replication of hepatitis B virus. Arch. Virol. 153:1677–1684 [DOI] [PubMed] [Google Scholar]
- 121.Li N, Zhang L, Chen L, Feng W, Xu Y, Chen F, Liu X, Chen Z, Liu W. 2012. MxA inhibits hepatitis B virus replication by interaction with hepatitis B core antigen. Hepatology 56:803–811 [DOI] [PubMed] [Google Scholar]
- 122.Mitchell PS, Emerman M, Malik HS. 15 May 2013. An evolutionary perspective on the broad antiviral specificity of MxA. Curr. Opin. Microbiol. 10.1016/j.mib.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. 2010. Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity. J. Biol. Chem. 285:28419–28424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scornavacca C, Zickmann F, Huson DH. 2011. Tanglegrams for rooted phylogenetic trees and networks. Bioinformatics 27:i248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ng AK, Zhang H, Tan K, Li Z, Liu JH, Chan PK, Li SM, Chan WY, Au SW, Joachimiak A, Walz T, Wang JH, Shaw PC. 2008. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 22:3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637 [DOI] [PubMed] [Google Scholar]
- 128.Haller O, Arnheiter H, Gresser I, Lindenmann J. 1981. Virus-specific interferon action. Protection of newborn Mx carriers against lethal infection with influenza virus. J. Exp. Med. 154:199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pichlmair A, Buse J, Jennings S, Haller O, Kochs G, Staeheli P. 2004. Thogoto virus lacking interferon-antagonistic protein ML is strongly attenuated in newborn Mx1-positive but not Mx1-negative mice. J. Virol. 78:11422–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thimme R, Frese M, Kochs G, Haller O. 1995. Mx1 but not MxA confers resistance against tick-borne Dhori virus in mice. Virology 211:296–301 [DOI] [PubMed] [Google Scholar]
- 131.Frese M, Weeber M, Weber F, Speth V, Haller O. 1997. Mx1 sensitivity: Batken virus is an orthomyxovirus closely related to Dhori virus. J. Gen. Virol. 78:2453–2458 [DOI] [PubMed] [Google Scholar]
- 132.Leroy M, Pire G, Baise E, Desmecht D. 2006. Expression of the interferon-alpha/beta-inducible bovine Mx1 dynamin interferes with replication of rabies virus. Neurobiol. Dis. 21:515–521 [DOI] [PubMed] [Google Scholar]
- 133.Habjan M, Penski N, Wagner V, Spiegel M, Overby AK, Kochs G, Huiskonen JT, Weber F. 2009. Efficient production of Rift Valley fever virus-like particles: the antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology 385:400–408 [DOI] [PubMed] [Google Scholar]
- 134.Sandrock M, Frese M, Haller O, Kochs G. 2001. Interferon-induced rat Mx proteins confer resistance to Rift Valley fever virus and other arthropod-borne viruses. J. Interferon Cytokine Res. 21:663–668 [DOI] [PubMed] [Google Scholar]
- 135.Johannes L, Kambadur R, Lee-Hellmich H, Hodgkinson CA, Arnheiter H, Meier E. 1997. Antiviral determinants of rat Mx GTPases map to the carboxy-terminal half. J. Virol. 71:9792–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Benfield CT, Lyall JW, Kochs G, Tiley LS. 2008. Asparagine 631 variants of the Chicken Mx protein do not inhibit influenza replication in primary chicken embryo fibroblast cells or in vitro surrogate assays. J. Virol. 82:7533–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bernasconi D, Schultz U, Staeheli P. 1995. The interferon-induced Mx protein of chickens lacks antiviral activity. J. Interferon Cytokine Res. 15:47–53 [DOI] [PubMed] [Google Scholar]
- 138.Ko JH, Jin HK, Asano A, Takada A, Ninomiya A, Kida H, Hokiyama H, Ohara M, Tsuzuki M, Nishibori M, Mizutani M, Watanabe T. 2002. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 12:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ko JH, Takada A, Mitsuhashi T, Agui T, Watanabe T. 2004. Native antiviral specificity of chicken Mx protein depends on amino acid variation at position 631. Anim. Genet. 35:119–122 [DOI] [PubMed] [Google Scholar]
- 140.Li B, Fu D, Zhang Y, Xu Q, Ni L, Chang G, Zheng M, Gao B, Sun H, Chen G. 2012. Partial antiviral activities of the Asn631 chicken Mx against Newcastle disease virus and vesicular stomatitis virus. Mol. Biol. Rep. 39:8415–8424 [DOI] [PubMed] [Google Scholar]
- 141.Baise E, Pire G, Leroy M, Gerardin J, Goris N, De Clercq K, Kerkhofs P, Desmecht D. 2004. Conditional expression of type I interferon-induced bovine Mx1 GTPase in a stable transgenic vero cell line interferes with replication of vesicular stomatitis virus. J. Interferon Cytokine Res. 24:513–521 [DOI] [PubMed] [Google Scholar]
- 142.Garigliany MM, Cloquette K, Leroy M, Decreux A, Goris N, De Clercq K, Desmecht D. 2009. Modulating mouse innate immunity to RNA viruses by expressing the Bos taurus Mx system. Transgenic Res. 18:719–732 [DOI] [PubMed] [Google Scholar]
- 143.Asano A, Ko JH, Morozumi T, Hamashima N, Watanabe T. 2002. Polymorphisms and the antiviral property of porcine Mx1 protein. J. Vet. Med. Sci. 64:1085–1089 [DOI] [PubMed] [Google Scholar]
- 144.Nakamura T, Asano A, Okano S, Ko JH, Kon Y, Watanabe T, Agui T. 2005. Intracellular localization and antiviral property of canine Mx proteins. J. Interferon Cytokine Res. 25:169–173 [DOI] [PubMed] [Google Scholar]
- 145.Fernandez-Trujillo MA, Garcia-Rosado E, Alonso MC, Borrego JJ, Alvarez MC, Bejar J. 2008. In vitro inhibition of sole aquabirnavirus by Senegalese sole Mx. Fish Shellfish Immunol. 24:187–193 [DOI] [PubMed] [Google Scholar]
- 146.Lester K, Hall M, Urquhart K, Gahlawat S, Collet B. 2012. Development of an in vitro system to measure the sensitivity to the antiviral Mx protein of fish viruses. J. Virol. Methods 182:1–8 [DOI] [PubMed] [Google Scholar]
- 147.Wu SC, Chi SC. 2007. Cloning and analysis of antiviral activity of a barramundi (Lates calcarifer) Mx gene. Fish Shellfish Immunol. 23:97–108 [DOI] [PubMed] [Google Scholar]