Abstract
SUMMARY
Clostridium difficile is a Gram-positive, spore-forming organism which infects and colonizes the large intestine, produces potent toxins, triggers inflammation, and causes significant systemic complications. Treating C. difficile infection (CDI) has always been difficult, because the disease is both caused and resolved by antibiotic treatment. For three and a half decades, C. difficile has presented a treatment challenge to clinicians, and the situation took a turn for the worse about 10 years ago. An increase in epidemic outbreaks related to CDI was first noticed around 2003, and these outbreaks correlated with a sudden increase in the mortality rate of this illness. Further studies discovered that these changes in CDI epidemiology were associated with the rapid emergence of hypervirulent strains of C. difficile, now collectively referred to as NAP1/BI/027 strains. The discovery of new epidemic strains of C. difficile has provided a unique opportunity for retrospective and prospective studies that have sought to understand how these strains have essentially replaced more historical strains as a major cause of CDI. Moreover, detailed studies on the pathogenesis of NAP1/BI/027 strains are leading to new hypotheses on how this emerging strain causes severe disease and is more commonly associated with epidemics. In this review, we provide an overview of CDI, discuss critical mechanisms of C. difficile virulence, and explain how differences in virulence-associated factors between historical and newly emerging strains might explain the hypervirulence exhibited by this pathogen during the past decade.
INTRODUCTION
Clostridium difficile was first isolated from infant stool in 1935, by Hall and O'Toole (1), and was further characterized by Snyder in 1937 (2). Initially, this organism was classified as Bacillus difficilis. There were no other documented reports of B. difficilis until 1960, when the renamed organism, Clostridium difficile, was cultured from the intestinal contents of a Weddell seal (3). Smith and King first suggested that C. difficile was a human pathogen when they isolated the organism from wounds, abscesses, blood, and pleural fluid of patients with a variety of illnesses (4). These human isolates of C. difficile were found to be virulent, and the authors noted that injection of the isolates into guinea pigs did not result in growth of the bacterium, yet the animals succumbed to the illness. From this observation, the authors suggested that death could occur from a toxin produced by C. difficile, a concept widely accepted today.
After the initial report by Smith and King, C. difficile was not reported as a human pathogen again until 1978, when it was discovered to cause pseudomembranous colitis (PMC). The first association between C. difficile and PMC was reported by George et al., in a study that described the isolation of C. difficile from patients with the illness or experiencing postoperative diarrhea (5). The authors also found that isolates of C. difficile produced a toxin that was detected in patients' stools. This observation correlated with an earlier finding indicating that toxin is produced in the intestinal tract in patients with antibiotic-associated diarrhea, although the source of this toxin had not been established (6, 7). Other studies soon found that C. difficile causes PMC in hamsters if the animals are pretreated with clindamycin (8). Collectively, studies in the late 1970s and early 1980s identified C. difficile as the cause of PMC in patients undergoing antibiotic treatment and suggested that a toxin(s) is involved with this illness.
During initial characterizations of C. difficile's virulence factors, two toxins were identified, one of which exhibited enterotoxic activity (TcdA) and one of which functioned as a cytotoxin (TcdB) (9, 10). Vaccination against the toxins or administration of toxin-neutralizing antibodies was found to protect against C. difficile infection (CDI), demonstrating that TcdA and TcdB are important to this illness (11, 12).
During the 1990s, C. difficile continued to be a significant cause of nosocomial illness. Until the late 1990s, CDI was a troublesome illness that was difficult to manage, but the mortality rate was low. This changed dramatically when new reports of serious hospital outbreaks began, around the year 2000. Most of these reports documented an increase in the mortality rate of CDI and an association with a strain of C. difficile rarely found before that time (13–16). This C. difficile strain has many designations, including North American pulsovar 1 (NAP1), PCR ribotype 027, and restriction endonuclease analysis group BI (NAP1/027/BI). NAP1/027/BI strains have predominated in many hospitals and have changed perspectives on the severity of CDI. What was once considered to be a manageable illness is now a more substantial challenge to clinicians, and this appears to be due to the emergence of NAP1/027/BI-related strains.
CLINICAL SIGNIFICANCE
C. difficile has been a major cause of hospital-acquired diarrhea for many years and rivals Staphylococcus aureus as the leading cause of nosocomial infections in developed countries (17). In 2010, CDI was the 18th leading cause of death in all individuals over the age of 65 (18). In addition, there is a severe economic cost to the inpatient treatment of CDI, as the most recent report in the United States estimates this figure at $8.2 billion (19). Age (>65 years), exposure to two or more antibiotics, surgical intervention, and infection with a hypervirulent strain increase the likelihood of death from CDI (20, 21). Because hospitalized patients are the most susceptible to CDI, it is not surprising that there is a strong comorbidity with this illness (22).
The clinical aspects of the disease involve, with increasing severity, diarrhea, pseudomembranous colitis, and toxic megacolon (23). In a typical disease scenario, antibiotics alter the intestinal flora, and this provides a niche for C. difficile to colonize the large intestine (Fig. 1). Although alteration of the normal flora is associated with CDI, the mechanism by which this occurs and how this initiates disease remain unclear. The fact that virtually every class of antibiotic has been associated with inciting CDI makes it unlikely that a single nutrient-niche competitor is associated with the illness. More recent work suggests that the normal flora primes a Myd88- and Toll-like receptor 5 (TLR5)-dependent innate immune response which protects against CDI (24). When the organisms providing this stimulus are eliminated by antibiotics, immune modulation subsides and C. difficile establishes infection.
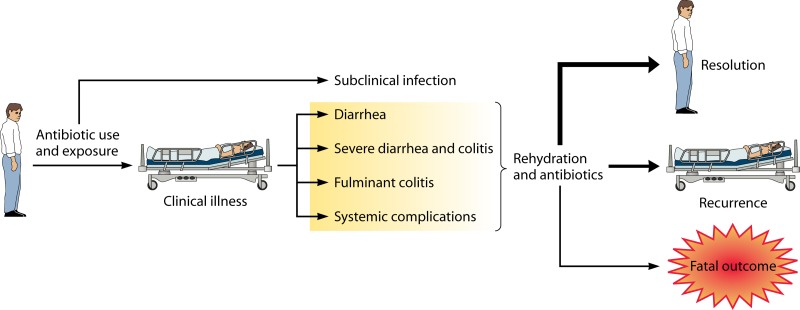
Fig 1.
Overview of C. difficile infection and outcomes. In a typical scenario, a patient is exposed to C. difficile spores, although a small number of cases appear to be community acquired. Alterations in the normal flora by antibiotic therapy likely support a niche for the bacterium in the gastrointestinal tract. The order of exposure to spores and antibiotic use required for clinical illness has not been established. The disease can range in severity from subclinical to clinical disease with systemic complications. Typically, rehydration and appropriate antibiotic therapy result in resolution of symptoms. The boldness of the arrows reflects the fact that most patients recover fully from the disease, while some will relapse, and death is the least frequent outcome.
Following ingestion, spores survive transit to the colon, and exposure to bile salts in the small intestine triggers germination (25). Vegetative C. difficile colonizes the large intestine and releases TcdA and TcdB. A series of inflammatory events occur, with penetration of macrophages, mast cells, and neutrophils into the site of infection (26). The epithelial barrier is compromised in this setting, although bacteremia involving C. difficile is extremely rare (27). Leukocytosis is common in CDI patients, and white blood cell (WBC) counts can be as high as 40,000 cells/mm3. WBC counts above 16,000 cells/mm3 are predictive of severe disease (28, 29). Interestingly, the cause of death in this illness is still not known; however, severely ill patients exhibit symptoms of systemic complications. Major organs are affected in life-threatening cases of CDI. Acute respiratory distress syndrome (30, 31), multiple-organ dysfunction (32), cardiopulmonary arrest (33), and chronic renal failure (34) have been described for CDI. In some cases, a fatal outcome occurs despite total colectomy and eradication of the organism (35, 36), further suggesting systemic effects. TcdA and TcdB have been detected in the blood of animals experimentally infected with C. difficile (37) and in human cases of C. difficile infection (38). TcdB has been shown to be a cardiotoxin (39). Thus, as is the case for many infections involving toxin-producing bacteria, toxemia is a likely contributor to the severity of C. difficile disease.
Soon after the discovery of C. difficile as the cause of PMC, a series of reports revealed a high frequency of relapse in CDI patients undergoing treatment with vancomycin (40, 41). Relapse or recurrence is a major clinical complication of CDI. Approximately 25% of CDI patients relapse at least one time following recovery, and patients relapsing once have a 40% chance of relapsing again (42–44). Multiple relapses over a long period are not unheard of for CDI patients and can dramatically exacerbate this illness. Interestingly, reports indicate that up to 50% of patients relapse with a strain that differs from the one causing the initial case of CDI (45–47), which conflicts with the earlier thought that relapse occurs due to residual C. difficile in the gut (45, 48). Thus, it is unlikely that all cases of relapse are simply due to inadequate clearance of C. difficile after initial infection. Multiple factors contribute to relapse, and the host immune response figures prominently among these factors. Patients who generate antibody to C. difficile toxins are less likely to experience relapse than patients exhibiting an undetectable response to the toxins (49–51).
Diagnosing and Predicting CDI
The gold standard of CDI diagnosis is culture of the bacterium from loose stool and testing for production of toxin (toxigenic culture) (52). However, due to the time-consuming nature of toxigenic culture, more rapid diagnostic procedures are used. The most widely adopted techniques for rapid diagnosis of CDI are enzyme immunoassays (EIA) (53) that test for the presence of TcdA and/or TcdB as well as glutamate dehydrogenase (GDH). These tests have been reported to have a wide range of false-positive and false-negative results and are thus often combined with a PCR assay for the TcdA, TcdB, or 16S rRNA gene (54–56). These molecular techniques to detect toxins by PCR or GDH by EIA are also combined with toxigenic culture in two- and three-step methods to reduce error (54–56). While EIA may not be the most sensitive assay available, there is evidence which suggests that, during outbreaks, continued surveillance by repeat EIA testing may be beneficial (57).
As mentioned above, rapid diagnostic methods using PCR are becoming more common in clinical laboratories as reports emerge that these molecular methods are highly accurate and more sensitive and specific than EIA alone (56, 58, 59). The increased sensitivity of PCR testing may reduce the need for repeat testing, as Nistico et al. have reported that repeat testing of negative patient samples by PCR rarely produced subsequent positive results (0.05%) (60).
There are several risk factors associated with CDI, including age, antibiotic use, and hospitalization. Cooper et al. additionally identified stool history, admission from another health care facility, and prior positive CDI as risk factors in a retrospective study of over 29,000 patients to develop a model to predict the predisposition of a given patient to CDI with a high accuracy (area under the curve of 0.929) (61). This is promising, but Cooper et al. acknowledge that their observed efficacy may be locale specific and would potentially need tailoring to each health care facility. It is nonetheless clear that there is no agreed-upon standard protocol for the rapid detection of CDI. Without such standardization, comparisons of statistics for various diagnostic techniques from site to site are difficult. It appears that, in most cases, PCR typing techniques are more likely to provide an accurate diagnosis in a timely manner, which is critical to selecting the appropriate intervention to prevent serious disease and/or complications due to CDI.
Treatment of C. difficile
CDI is associated with some of the most commonly prescribed antibiotics, including second- and third-generation cephalosporins and carbapenems (62). However, there is a type-specific association with emerging NAP1/027/BI strains and the use of fluoroquinolones (62–65). Several studies indicate that patients on antibiotic therapy are most at risk for CDI within 90 days of discontinuing treatment (62, 66).
Currently, metronidazole, vancomycin, and fidaxomicin are used to treat CDI (52). Therapy with oral metronidazole has been shown to be noninferior to vancomycin during primary and first recurrences of CDI (67, 68). Fidaxomicin has been FDA approved for the treatment of CDI in the United States since May 2011 and has proved to be noninferior to oral vancomycin for the treatment of CDI. Fidaxomicin has been reserved for cases of severe CDI where metronidazole or vancomycin has failed or where their use is contraindicated, in no small part due to the high cost of fidaxomicin therapy. A review of studies by Drekonja et al. has shown that all three of these antibiotics are equally effective in treating the primary occurrence of CDI (69), although fidaxomicin is more effective at preventing relapse (66, 70). The decrease in relapse rates for patients treated with fidaxomicin may be related to reports that fidaxomicin causes less disruption of the normal colonic flora than that with vancomycin (71, 72).
The recognition that normal flora maintenance is important for prevention of relapse of CDI has led to the development of fecal microbiota transplantation (FMT). This procedure restores the normal flora of the gut by introducing stool from healthy donors by enema, nasogastric tube, or colonoscopy. The efficacy of this procedure varies but is typically high (>75%), and several groups are using it to successfully treat recurrent CDI in patients unresponsive to traditional therapy (73–75). Recently, a long-term follow-up study of 77 FMT recipients reported a secondary cure rate of 98% (76). In addition, several clinical trials to assess FMT prospectively are currently recruiting individuals in the United States (77), and it will be exciting to see the results of these studies.
There has been recent interest in development of monoclonal antibodies directed toward TcdA and/or TcdB to treat recurrent CDI. Several groups have reported promising data showing that anti-TcdA and anti-TcdB therapy can prevent mortality independent of antibiotic therapy (11, 78, 79). Lowy et al. reported in a placebo-controlled trial that only 7% of patients receiving traditional antibiotic therapy plus anti-TcdA and anti-TcdB monoclonal antibodies relapsed with CDI by 84 days posttreatment, compared to 25% of those receiving antibiotics plus placebo (80). They reported no difference in the duration of hospitalization between the groups; however, only 9% of the treatment group required hospitalization after infusion of antibodies, compared to 20% of the placebo group. In the study of Lowy et al., placebo groups received normal saline instead of intravenous immunoglobulin (IVIG). However, these results are promising considering a review of clinical reports showing that there is no benefit to IVIG therapy in the CDI population (81).
In regard to the antibody-based treatments for CDI, it is important that TcdB amino acid sequences vary at the protein level between different ribotypes and that there is evidence to support that targeting TcdB, but not TcdA, is important for preventing disease in a gnotobiotic piglet model of CDI (82). In addition, Marozsan et al. have shown that monoclonal antibodies vary in the ability to neutralize toxins from diverse C. difficile strains in vitro (83). These neutralization data suggest that, going forward, targeted epitopes will have to be selected carefully to have the most effect on diverse strains, especially considering the emergence of hypervirulent non-NAP1/027/BI strains.
FACTORS IMPORTANT TO C. DIFFICILE VIRULENCE
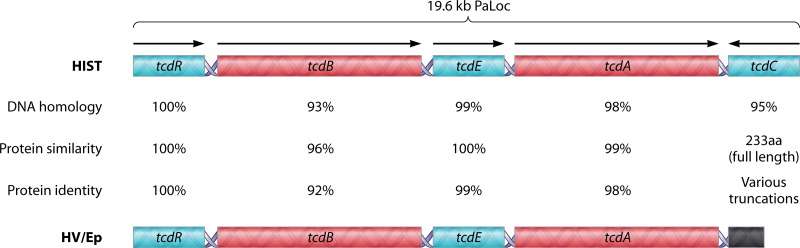
The C. difficile pathogenicity locus (PaLoc) is a 19.6-kb genomic region which carries five genes: tcdR (552 nucleotides [nt]), tcdB (7,098 nt), tcdE (498 nt), tcdA (8,130 nt) and tcdC (696 nt) (85) (Fig. 2).
Fig 2.
Comparison of C. difficile pathogenicity loci between historical and hypervirulent strains. The schematics depict the 19.6-kb PaLoc of historical C. difficile (HIST) (top) and hypervirulent C. difficile (HV/Ep) (bottom). DNA homology, protein similarity, and protein identity are based upon reference strains CD630 (GenBank accession no. NC_009089.1) and BI1 (GenBank accession no. NC_017179.1). TcdC protein similarity and identity calculations are not trivial, as truncations of tcdC vary among hypervirulent/epidemic (HV/Ep) isolates, resulting in final products ranging from 61 to 226 amino acids (aa).
TcdR (∼22.1 kDa) is an RNA polymerase sigma factor that is expressed in stationary phase, is repressed in high glucose, and autoregulates its own expression (86). TcdR is a member of a group of RNA polymerase sigma-70 sigma factors that includes BotR, TetR, and UviA, which regulate toxin and bacteriocin production in Clostridium botulinum, Clostridium tetani, and Clostridium perfringens, respectively (87, 88). TcdR binds to promoter regions upstream of tcdA and tcdB, leading to transcription of these toxin-encoding genes.
TcdE (∼18.8 kDa) is a holin-like molecule often suggested to be important for release of TcdA and TcdB from the organism. Given tcdE's close localization to tcdB and tcdA on the PaLoc and the reported ability of TcdE to lyse Escherichia coli when expressed as a recombinant protein, the concept has merit. Yet this idea has come under more scrutiny recently with the publication of two diametrically opposed reports, one of which found no role for TcdE in toxin release (89), while the other showed TcdE to be absolutely essential for toxin release from the organism (90). Both studies relied on in vitro analyses. Therefore, whether TcdE plays a role in pathogenesis has not been determined. In all, the current understanding of TcdE, especially regarding its mechanism of action, is limited.
TcdC (∼25.5 kDa) has been examined extensively during the past few years, mostly due to its association with emerging hypervirulent strains of C. difficile. TcdC is an anti-sigma factor which disrupts TcdR interaction with RNA polymerase in the holoenzyme complex (91). TcdC appears to be part of a negative regulator system that represses TcdR and toxin expression during exponential growth. This idea is further supported by the fact that TcdC is the only factor expressed to a significant extent during exponential growth (92). Thus, toxin expression is suppressed by TcdC during early and exponential growth of C. difficile through the protein's inhibitory effects on TcdR. The variations in TcdC as they relate to hypervirulence are discussed in subsequent sections of this review.
TcdB (∼270 kDa) is encoded on the PaLoc and is a major virulence factor in all pathogenic C. difficile strains. TcdB functions as an intracellular bacterial toxin and translocates into the cytosol of cells, where it modifies small GTPases through glycosylation. The details of TcdB's role in pathogenesis and its mechanisms of action are described further in the following sections.
TcdA (∼308 kDa) is closely related to TcdB and is also encoded on the PaLoc of C. difficile. It has been suggested that TcdA and TcdB arose from a single gene duplication, given their close proximity on the genome and their overall sequence similarity. Several studies indicate that TcdA is a more potent enterotoxin than TcdB (10, 93), while TcdB has a broader tropism and is more cytotoxic (94, 95). Disease-causing strains of C. difficile lacking TcdA production (TcdA− TcdB+) have been identified but are not common (96–98). Confounding the matter further are reports that TcdA− TcdB+ and TcdA+ TcdB− mutants of C. difficile were found to cause disease in hamsters (99, 100), though both single mutants were attenuated compared to the wild type. In contrast, an earlier study found that only TcdB is essential for virulence, while TcdA is dispensable (101). To date, these conflicting findings have not been explained or resolved. Nonetheless, both toxins are lethal in animal challenge models, and antibodies against TcdA and TcdB can protect against disease (102). The fact remains that despite extensive study for more than 3 decades, the general roles of TcdA and TcdB in disease and their contributions to the overall virulence of C. difficile are not well understood.
The temporal course of expression for genes encoded by the PaLoc has been examined in one study, which found that most of the genes, including toxin genes, are not expressed until later stages of growth (92). Only TcdC is expressed during early growth, which corresponds to the factor's role in suppressing transcription until TcdR is expressed near stationary phase. Using a variety of methods, two studies have shown that transcripts of tcdB and tcdA arise from both upstream readthrough and the immediate uncoded 5′ regions of these genetic elements (92, 103). The extent to which the polycistronic messages contribute to toxin expression and whether these arise from more than two genes have not been clearly established; however, earlier work by Hammond et al. detected an ∼17.5-kb transcript which could encompass a large portion of the PaLoc (104). Interestingly, analysis of transcription at later stages of growth indicated that the genes for both toxins are transcribed from their respective upstream promoters and are not part of a larger transcript, suggesting a growth-related effect on the type of messages present at different stages of growth (103).
Toxin expression is subject to catabolite repression involving a variety of carbon sources (103). Levels of biotin and amino acids can also influence expression of TcdA and TcdB (105, 106). Although the mechanisms by which these factors influence toxin expression differ, in general each appears to involve increased toxin expression as the availability of these factors declines toward the stationary phase of growth. CodY, a common transcriptional regulator in Gram-positive organisms, regulates the expression of TcdA and TcdB in response to changes in the levels of GTP and branched-chain amino acids (BCAAs) (107). In this system, CodY functions as a negative regulator of toxin expression by promoting transcription of tcdR. Under nutrient-rich conditions, GTP and BCAAs increase the affinity of CodY for the tcdR promoter, thereby blocking the expression of TcdR. As a result, TcdR levels remain low during exponential growth, and toxin expression from the polycistronic transcript and from the TcdR-dependent promoters of tcdA and tcdB is minimal. As nutrient levels decline, CodY's affinity for the TcdR promoter declines, and tcdA and tcdB transcription is derepressed. Toxin expression is also regulated by the carbon catabolite repression system, involving the catabolite control protein (CcpA) (108), similar to the case in other Gram-positive bacteria. However, in C. difficile, the CcpA-related expression of TcdA and TcdB is independent of Hpr but can be influenced directly by levels of fructose-1,6-bisphosphate. The understanding of systems related to toxin expression in C. difficile is improving; for example, toxin expression has also been linked to the onset of sporulation, with a dependence on SigH (109). In total, these findings support the notion that the metabolic and growth state of C. difficile largely determines when the organism induces expression of the toxin. How this relates to regulation of toxin expression in vivo has not been determined.
Toxinotypes
Despite the fact that TcdA and TcdB are important for C. difficile virulence, many strains exhibit variations in the PaLoc. These PaLoc variants, referred to as toxinotypes, include variants with intact genes but sequence changes, forms with truncated tcdA, variants of tcdB, and forms with tcdC encoding mutations and deletions. Thus, it is notable that this region, which is very critical to C. difficile virulence, is so malleable in its sequence. Toxinotypes and the method of toxinotyping have been described and summarized in several excellent reviews (110–112) and, as such, are only briefly summarized here. There are at least 31 different toxinotypes, which vary relative to the reference toxinotype 0. In humans, toxinotype 0 is by the far the most common historical isolate, but toxinotype III is more often associated with epidemic strains and strains that cause more serious disease. Four toxinotypes (VIII, X, XVI, and XVII) are TcdA− TcdB+ strains of different varieties. The relevance of different toxinotypes to clinical illness remains unclear, and other than toxinotype III (IIIb) association with the NAP1/027/BI epidemic strains, there is little evidence that toxinotype itself influences the outcome of disease or supports outbreaks. However, toxinotypes may provide a valuable tool for tracking particular strains of C. difficile.
Mechanism of Action of TcdA and TcdB
TcdA and TcdB are both intracellular bacterial toxins that glucosylate small GTPases within intoxicated cells (113, 114). These relatively large (250 to 308 kDa) toxins are members of the large clostridial cytotoxin family which, along with toxins from Clostridium sordellii and Clostridium novyi, are able to hydrolyze UDP-glucose and transfer the liberated sugar moiety to a reactive threonine in the GTP-binding domain of Rho, Rac, Cdc42, and particular isoforms of these small GTPases. Glucosylation prevents the GTPase from forming a magnesium-dependent association with the gamma phosphate of GTP, thereby rendering these regulatory proteins inactive (115). Rho, Rac, and Cdc42 are essentially locked in the off position following glucosylation.
Four functional domains have been characterized in TcdA and TcdB. Both toxins contain an amino-terminal enzymatic domain of approximately 550 residues which engages UDP-glucose, hydrolyzes the molecule, and transfers glucose to the target substrate (116). Several critical residues have been characterized in this domain, including a DXD motif at residues 286 to 288 in TcdB, which coordinates UDP-glucose binding along with Mn2+ (117), and W102, which stabilizes the DXD motif's interaction with UDP-Glc by aromatic stacking (118). Proximal to the enzymatic region is a cysteine protease domain (CPD), which carries out autoproteolytic processing of the toxin following binding of inositol hexakisphosphate, found abundantly in the cytosol of eukaryotic cells (119–121). This autoproteolytic processing releases the enzymatic domain during receptor-mediated endocytosis, and it appears that this amino-terminal portion of the toxin is the only region delivered into the cytosol (122). The CPD includes a catalytic dyad (H653/C698) important for intramolecular cleavage of substrate proximal to the amino terminus and D597, a residue important for binding inositol hexakisphosphate (121). The third domain, found near the middle of TcdB, is a hydrophobic patch of residues associated with membrane insertion, and this domain most likely fulfills the requirement for penetrating the membrane during translocation of the enzymatic region into the cell. Two residues, E970 and E976, have been found to be essential for membrane insertion (123). Finally, the carboxy-terminal region encodes combined repetitive oligopeptide repeats (CROPs), which are known to bind trisaccharide glycans in vitro (124, 125). The CROP domain exhibits a beta-solenoid-like structure similar to that found in choline-binding domains of cell wall-associated proteins such as LytA (124, 125). Important to the area of vaccine development, the CROP domain is highly antigenic, and antibodies toward this region of the protein can neutralize the toxin in vitro (78).
Although similar in sequence, domain organization, and mechanism of action, TcdA and TcdB are different in some notable ways. First, the CROP region is more extensive in TcdA, with as many as 38 repeats, although this number can vary among different toxinotypes. TcdB's CROP region is smaller than that of TcdA and has fewer repeats (as few as 16) within this domain (126). Structural studies indicate that the CROP domain of TcdA has at least 7 glycan-binding sites, and by extrapolation based on primary sequence, TcdB maintains only 4 such binding sites (126). It is not known whether this difference in avidity accounts for variations in tropism between the two toxins; however, TcdB and TcdA differ in their cell targeting. In this regard, TcdA is well known to function as an enterotoxin, but it intoxicates only a narrow range of cell types at doses similar to those of TcdB (84, 127–130). On the other hand, TcdB targets a broad range of cells and is considered the major cytotoxin produced by C. difficile (131–133). Based on more recent studies, it appears that TcdB is also enterotoxic (134). Indeed, TcdB+ TcdA− strains of C. difficile can cause disease similarly to TcdB+ TcdA+ strains, suggesting that TcdB can function as an enterotoxin or at least substitute as the enterotoxin in the absence of TcdA (135). More direct evidence from recent studies indicates that TcdB can function as an enterotoxin in mice implanted with human colonic tissue (136). Thus, it appears that TcdB along with TcdA can target and damage human intestinal tissue.
TcdA and TcdB also exhibit slight differences in their substrate specificities. In addition to Rho, Rac, and Cdc42, TcdA appears to target a broader range of small GTPases than TcdB, including Rap2. Until recently, the structural reasons for this difference in substrate targeting were not known. A study by Pruitt and colleagues found that the overall structure of the glucosyltransferase domain (GTD) in TcdA is similar to that in TcdB; however, the GTD of TcdA has a larger net negative charge of surface-exposed residues than that in TcdB (137). Moreover, the UDP-glucose-binding pocket of TcdA is positively charged, but the corresponding region in TcdB exhibits a large net negative charge. How these charge differences influence substrate specificity is not known.
In addition to TcdA and TcdB, C. difficile possesses virulence factors that are encoded outside the PaLoc and that vary between historical and hypervirulent strains. These factors, which are involved in many processes, including motility, antigenicity, and antibiotic resistance, are discussed below and summarized in Fig. 3.
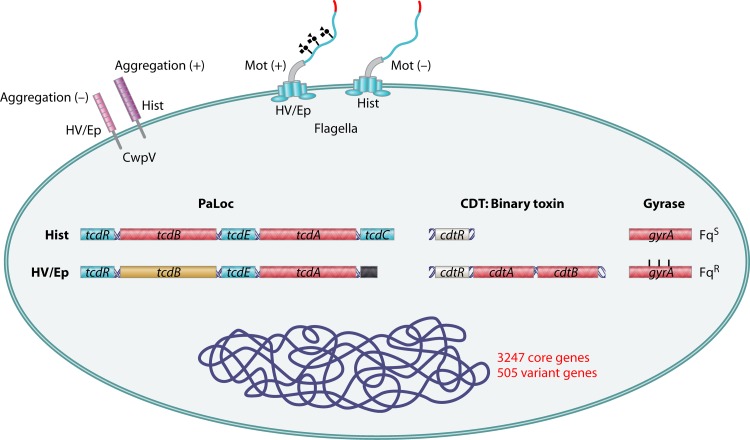
Fig 3.
Comparison of virulence-associated factors between historical and hypervirulent C. difficile strains. A cartoon of C. difficile summarizing the differences between historical (Hist) and hypervirulent/epidemic (HV/Ep) strains is shown. Historical strains contain full-length tcdC but lack the binary toxin. Additionally, they are generally sensitive to fluoroquinolones, have a positive aggregation phenotype, and are nonmotile. Hypervirulent strains lack a full-length tcdC gene and have the opposite phenotype of historical strains for the above virulence factors.
Binary Toxin
Specific strains of C. difficile produce a binary toxin, CDT, that ADP-ribosylates G actin in target cells (138). The toxin, a member of the iota family of binary clostridial toxins, consists of two distinct protein components, CDTa and CDTb, whose genes are located separately from the PaLoc (139, 140). CDT is similar to other clostridial binary toxins which ADP-ribosylate actin, including Clostridium perfringens iota toxin, Clostridium botulinum C2 toxin, and the Clostridium spiroforme toxin CST. Only a small percentage of C. difficile strains of all known ribotypes produce CDT (141); however, 027 epidemic hypervirulent ribotypes are among these strains (142), and this has heightened interest in the possible contribution this toxin makes to virulence. The genotypes related to CDT-producing and non-CDT-producing strains are particularly interesting. Several characterized strains carry the fully intact LytR-like regulator (CdtR) gene, which is proximal to the cdtA and cdtB genes, making up the entire binary toxin locus (143, 144). Despite the presence of an intact CdtR-encoding gene, many strains contain only remnants of cdtA and cdtB (145). Other strains are completely devoid of the entire locus, containing a 68-bp insert at this site (143). Thus, similar to the PaLoc encoding TcdA, TcdB, and their regulators of expression, the Cdt locus varies extensively between strains.
A model of CDT's mechanism of cellular intoxication has begun to emerge. CDTb, the cell-binding and entry portion of the binary toxin, binds to lipolysis-stimulated lipoprotein receptor (LSR) to localize at the cell surface, where it is thought to then bind to and mediate the delivery of CDTa into the cell (146). CDTb-mediated translocation of CDTa into the cell requires the chaperone proteins Hsp90 and cyclophilin A (147). Within the cytosol, CDTa-mediated ADP-ribosylation of G actin leads to the formation of protrusion bodies composed of microtubules that extend from the cell to contact C. difficile, thereby increasing colonization efficiency (148). Recently, Barth and colleagues identified CD44 as being important for intoxication by toxins in the iota clostridial binary toxin family (149). Cells deficient in CD44 or treated with anti-CD44 antibody are protected from intoxication. More importantly, CD44 knockout mice are less susceptible to killing by iota toxin administered intraperitoneally. The importance of this finding remains to be seen, as a prominent role for CDT in pathogenesis has largely been dismissed because many disease-causing strains do not produce this toxin. However, recent studies may start to change this perspective. In one study, the 30-day fatality rates of CDI patients infected with a binary or nonbinary toxin-producing strain of C. difficile were determined. Patients infected with a CDT-positive strain were more likely to succumb to CDI than patients infected with a CDT-negative strain (150). Moreover, CDT has been detected in the stools of CDI patients, which indicates that the toxin is produced within the intestinal tract during disease (142).
Cell Wall-Associated Proteins
Early studies found that C. difficile produces a pair of proteins that can spontaneously assemble into an organized structure surrounding the organism (151, 152). These were found to be S-layer proteins (Slp) that distributed around C. difficile. Unlike many other S layers, composed of only one protein or two proteins encoded by different genes, the S layer of C. difficile arises from the processing of a single Slp protein (153). This processing, via the cell wall protease Cwp84, results in a high-molecular-weight S-layer protein (HMW Slp) and a low-molecular-weight S-layer protein (LMW Slp) (154, 155). Following processing, the two proteins assemble into a paracrystalline array (156). There is evidence to suggest that LMW Slp can mediate adherence of C. difficile in the intestinal tract (157). Moreover, patients mount antibody responses to the S-layer proteins, indicating their expression in vivo (158).
In addition to variability in the sequences of the PaLoc and toxin genes, studies have shown that cell wall proteins, particularly CwpV, vary among different strains (159). CwpV causes bacterial aggregation in all strains tested but exhibits very different antigenic characteristics among various isolates of C. difficile. Structurally, CwpV consists of a membrane-anchoring amino-terminal domain, a putative flexible linker, and a carboxy-terminal antigenic region with a variable number of 120-amino-acid repeats (160). Expression of CwpV is phase variable and regulated by an inverted repeat region downstream of the promoter and proximal to the start codon of cwpV (160). RecV mediates the recombination and inversion of the DNA switch, which either supports formation of a stem-loop to block transcription or removes the stem-loop to allow expression (160). Thus, individual bacteria may have a CwpV-on or CwpV-off status, depending on the RecV-directed orientation of the regulatory sequence. When CwpV is expressed, it represents the majority of cell wall proteins in C. difficile. The amino-terminal region of CwpV is found throughout all strains of C. difficile examined, but substantial variability exists in the repeat region found at the carboxy-terminal portion of the protein (159). In particular, the carboxy-terminal region of CwpV from the hypervirulent strain (NAP1/027/BI) contains only eight repeats which are 79 amino acids long, which is much different from the nine 120-amino-acid repeats found in the historical, nonhypervirulent 630 strain of C. difficile (159). This sequence and repeat variation leads to distinct antigenic characteristics, and antisera to the carboxy-terminal regions of the different strains do not cross-react (159). Thus, CwpV represents an antigenic protein which is expressed in a phase-variable manner and has antigenic characteristics that are substantially different between historical and hypervirulent strains of C. difficile. The overall importance of CwpV in virulence and as a target of neutralizing antibodies is not yet known; however, it is reasonable to speculate that host responses against CwpV from historical C. difficile would not detect or protect against CwpV from hypervirulent strains of C. difficile.
Flagella and Motility
Most strains of C. difficile produce flagella and are motile in semisolid agar assays. The extent to which motility contributes to virulence is not entirely known, but the flagella may be important for interaction with the mucosa and may enhance colonization. Structural components of the flagella and proteins involved in posttranslational modification of flagella are encoded within three adjacent gene clusters on the chromosome of C. difficile. The genes, grouped into the F1, F2, and F3 regions, are found throughout different C. difficile strains, but with some notable variations. Genome analyses have found that C. difficile 630 retains similar structural genes (e.g., fliC and flgB) but differs from other hypervirulent strains, which contain up to 7 other open reading frames (ORFs) located between the F1 and F3 regions (161). Based on comparison analysis, these genes appear to be involved in glycosylation precursor synthesis and direct glycosylation. The glycosyltransferases and related proteins encoded at the 5′ region of the F2 loci are involved in posttranslational modification of flagella. Genetic disruption of these genes leads to a loss in motility and a reduction in the posttranslational modification of the flagella (162). Glycosylation of FliC (flagellin) improves flagellar stability in C. difficile and appears to be necessary for motility (162). Given the notable differences in glycosylation-related genes in various strains of C. difficile, it is not surprising that the glycosylation patterns vary among serotypes (162).
Recent studies have found a connection between toxin expression and the flagellum regulon (163). The hierarchy of the regulator system appears to be complicated. FliA, a sigma-28-related transcriptional regulator that supports late-stage flagellar gene expression, influences transcription of genes in the PaLoc. In FliA-deficient strains, transcription of tcdA is reduced almost 100-fold, but tcdB transcription is unaffected. Further adding to the complexity is the finding that disruption of several other F3 genes, irrespective of the type of encoded protein, results in a similar transcriptional profile in the PaLoc. In contrast to the reduction in tcdA transcription observed in F3 mutants, loss of FliC results in increased levels of toxin accumulating in culture supernatants of this mutant. Thus, there appear to be multiple flagellum-related effects on toxin levels in C. difficile, some of which may be related to secretion and not necessarily transcription of the toxin.
HISTORY AND EMERGENCE OF NAP1/027/BI STRAINS
Concerns about CDI heightened when several hospitals noted unexpected increases in the severity of illness in patients with CDI. For example, the University of Pittsburgh noted a significant increase in the number of CDI patients requiring a colectomy in the year 2000 (14). Similar reports came from North American hospitals over the next 2 years and led to the discovery that the NAP1 strain of C. difficile contributed to this change in morbidity and mortality. Two very prominent outbreaks, one in Quebec, Canada (15, 164), and one at the Stoke Mandeville Hospital in the United Kingdom, further focused attention on the NAP1 strain (165). In both locations, there was a concerning increase in the severity of disease. The Quebec outbreak peaked in 2003, with a short-term mortality rate reaching 13.8% at the Sherbrooke Hospital (15). This compared to a mortality rate of 4.7% about 10 years earlier. The outbreak at the Stoke Mandeville Hospital was perhaps even more severe, with over 300 CDI cases and 38 deaths in a 2-year period. Between 2000 and 2008, the rate of hospital discharges with any diagnosis code for C. difficile roughly doubled, from 3.82 to 8.75 per 1,000 discharges (19, 166), in the United States. The incidence of CDI in the U.S. population aged 65 years or greater is approximately 5 times the rate for all ages (228 versus 61 cases per 100,000 persons) (L. C. McDonald, S. Banerjee, and D. B. Jernigan, presented at the 14th Annual Scientific Meeting of the Society for Healthcare Epidemiology of America, Philadelphia, PA, 2004), indicating that the elderly are the most at risk for this disease. However, it should be noted that several reports have demonstrated the changing epidemiology of CDI. In at least one large study, the incidence of community-acquired CDI (CA-CDI) in patients with no traditional risk factor was equal to the CDI incidence in at-risk populations (167). This observation is backed up by reports from others (168, 169), as well as by data from the U.S. Centers for Disease Control and Prevention (170), and suggests that the incidence of CA-CDI is on the rise.
The characterization of NAP1/BI/027 strains as hypervirulent or highly pathogenic is not without controversy. Clearly, the NAP1/BI/027 ribotype became the predominant strain in many geographical locations during the past decade, and this has been well documented in several large-scale epidemiological studies, such as the one performed by the National Ribotype-Based Surveillance Scheme in England (171). Results from this surveillance program demonstrate a correlation between the frequency of NAP1/BI/027 infections and increased mortality. In the years when NAP1/BI/027 strains were reported more frequently, the mortality rate was higher. Years with lower mortality rates also had a reduced frequency of NAP1/BI/027 infections. For example, this network reported that 55% of CDI cases were caused by NAP1/BI/027 infections in 2007 to 2008 and 21% were caused by NAP1/BI/027 in 2009 to 2010. The mortality rate among CDI cases from this surveillance network was 29% in 2007 to 2008 and 14% in 2009 to 2010. This correlates with hospital-specific reports dating back to 2000, where many of the outbreaks were found to have been caused by a NAP1/BI/027 strain and an unexpectedly high mortality rate alerted clinicians to a possible change in CDI epidemiology. In addition, a recent study at the University of Pittsburgh found that patients initially infected by NAP1/BI/027 strains were more likely to relapse than patients infected with other strains of C. difficile (172). Thus, the clinical and surveillance evidence from the past decade shows that an increased frequency of 027 ribotypes correlates with increased mortality. But does this mean that death is more likely in a patient infected with a NAP1/BI/027 strain? Some studies suggest that this may not always be the case. A recent study by Walk and colleagues found that while the NAP1/BI/027 strain is associated with severe disease, it is not the sole predictor of disease severity (173). Other factors, such as white blood cell count, may be better clinical indicators of the severity of disease. Several other studies also bring into question the relevance of using strain type to predict the severity of disease (174–176). A more conservative idea may be that differences in NAP1/BI/027 virulence, along with other factors, allowed this strain to penetrate into the population and rapidly replace previously more common strains of C. difficile.
Factors Accounting for Epidemics and Hypervirulence of NAP1/027/BI Strains
The reasons for hypervirulence and epidemics associated with NAP1/027/BI strains have not been easy to resolve. As mentioned above, there are significant differences in the genomes of historical and hypervirulent strains of C. difficile, and which of these differences matter in regard to changes in disease severity is difficult to know. In early studies, it was proposed that NAP1 strains produced higher levels of toxin than historical strains (177); however, this observation has not been made consistently among these isolates. The idea is appealing because NAP1 strains are known to carry a mutation in tcdC, which has been thought to render it inactive. As such, it seems reasonable that loss of this repressor of toxin production could lead to higher levels of toxin. As TcdC is known to negatively regulate toxin expression and tcdC from hypervirulent strains is known to carry multiple mutations that prevent proper expression of a functional TcdC protein, it is thought that this may account for increased virulence, perhaps through increased production of TcdA and TcdB. The specific mutations were initially characterized as a 39-bp deletion or 18-bp deletion most likely arising from recombination within a DNA repeat region (178). The deletions are in frame and result in loss of either 6 or 13 residues, starting at A114 and continuing through E119 or E126, respectively. The gene encoding the A114-E126 deletion mutant of TcdC also contains a DNA nonsense mutation (C to T) at nucleotide 184 which leads to a premature stop codon and a form of TcdC of only 61 residues. A more expansive analysis of isolates from 199 C. difficile-associated disease (CDAD) cases identified different truncating mutations resulting in TcdC proteins of either 61, 63, 65, or 85 amino acids (179). Thus, the two general mutations are one in which a premature stop codon leads to production of a severely truncated TcdC protein and one in which internal deletions reduce the size of the protein by 6 residues. Other strains of C. difficile known to produce variant forms of TcdB (e.g., strain 8864) have also been found to produce a truncated form of TcdC of only 22 amino acids (180).
Two general observations have suggested that mutated tcdC cannot entirely account for the differences in virulence among emerging and historical strains. First, other strains, such as the well-studied VPI 10463 strain, produce large amounts of toxin yet possess an unmutated, intact tcdC gene (181, 182). Second, tcdC mutants have been identified in strains that do no not appear to produce larger-than-normal (relative to historical strains such as 630) amounts of toxin (183).
Direct experimental assessments have not resolved the conflicting issues regarding TcdC's role in hypervirulence. Studies involving full-length TcdC expressed from a plasmid conjugated into a TcdC-negative strain of C. difficile found that intact functional TcdC can cause the organism to produce smaller-than-normal amounts of toxin (184). Moreover, this tcdC+ strain was found to be attenuated in virulence relative to the parent strain. The extent of virulence in the complemented tcdC+ strain was almost identical to that of C. difficile strain 630, which endogenously expresses normal TcdC, further indicating that the lesion in tcdC accounted for the increased virulence in the hypervirulent strain. In contrast to these findings, other work has found that allelic exchange using intact tcdC to replace the mutant form of tcdC does not alter levels of toxin production in a hypervirulent strain (185). Similarly, allelic replacement of an unmutated tcdC gene with a tcdC gene carrying the mutation did not cause the 630 strain to express larger amounts of the toxin (186).
An additional consideration is the fact that NAP1 strains produce the binary toxin, CDT, and that this could enhance colonization. Indeed, in one mouse study, C. difficile strains that produced CDT were found to have significantly improved colonization and to increase intestinal damage (148). Thus, while it is unlikely to be as critical to the virulence of all pathogenic C. difficile strains, CDT may be an important accessory factor that improves colonization by epidemic strains known to cause more severe disease. However, arguing against CDT as the sole contributor to increased virulence is the fact that CDT is produced by other nonhypervirulent strains of C. difficile. Thus, the presence of CDT does not solely account for the increase in virulence, although this could be among the contributing factors to this effect.
It has also been suggested that NAP1 strains sporulate more efficiently than historical strains (187); however, a more recent analysis of over 40 C. difficile NAP1 strains found substantial variations in the sporulation efficiencies of these isolates, some of which sporulate no better than historical strains (188). In mice colonized with NAP1/BI/027, the “supershedder” phenotype has been observed in response to antibiotic treatment (189), and this phenotype prolongs transmission to naive mice (190). However, the role of sporulation in these studies is not known, as the transmission by “supershedders” is likely due to more prolific disease.
Another phenomenon that has been observed in both the human population and animal models is the decreased diversity of gut flora in individuals with CDI (190–193). This decrease in diversity appears to coincide with antibiotic use, which C. difficile exploits, rather than by C. difficile directly altering the host flora. Thus, the extent to which differences in the microbiota affect the host's interactions with NAP1 strains and the overall virulence of these strains is not fully known but is likely to be an important area of study in the future.
Toxin Variation and Hypervirulence
Recent work from Lanis et al. examined the significance of sequence variation in TcdB between historical and hypervirulent strains of C. difficile (194, 195). Wren and colleagues noted that TcdB from NAP1/027/BI strains was more cytotoxic against certain cell lines than TcdB from historical strains, suggesting that the increased toxicity of TcdB could contribute to hypervirulence (161). Lanis et al. then compared the sequences of the PaLoc between a historical strain and a hypervirulent strain (NAP1/027/BI) and noted that the TcdB gene sequence varied substantially, while other genes within the locus remained virtually identical (194). Further analysis found that NAP1/027/BI TcdB causes more extensive tissue damage in a zebrafish model. Studies of the mechanism of these differences in TcdB found that NAP1/027/BI TcdB is capable of entering cells more quickly and translocating to the cytosol at an earlier step in endocytosis than the historical form of the toxin. Both forms of the toxin appear to be equally efficient at modifying substrate and engaging target cells, further restricting the difference to a step in translocation.
Inositol hexakisphosphate-induced activation of autoprocessing in TcdB is thought to be an important step in cellular intoxication. Analysis of autoprocessing found that hypervirulent TcdB is more efficient at autoprocessing and that this is due to the toxin's ability to more effectively engage the intramolecular substrate (195). It appears that hypervirulent TcdB exists in a structure more favorable to intramolecular self-cleavage and that this allows this form of the toxin to enter cells more efficiently.
Genomic Differences in Hypervirulent and Historical C. difficile Strains
Sequence, multilocus sequence typing (MLST), and microarray analyses have led to the classification of C. difficile strains into five distinct clades. An initial microarray-based analysis using a C. difficile strain 630 target sequence found a substantial variation among 75 isolates, with hypervirulent strains, livestock strains, toxin-negative strains, and historical strains forming distinct clades.
Stabler and colleagues (161) examined and compared the genomes of a historical nonepidemic and nonhypervirulent C. difficile strain (strain 630), a nonepidemic 027 strain (CD196), and a recently emerged epidemic strain (R20291). Several notable differences were identified between the genomes of these three strains. The strains share a core of 3,247 genes, and 505 potential genes are unique to the 630 strain and absent in both 027 strains of C. difficile. Both 027 genomes carry 234 genes unique to their genomes that are absent in the historical 630 strain. In addition to the known differences in tcdB and tcdC and the presence of the binary toxin-encoding genes, the R20291 strain exhibits gyrA mutations that may account for its unique resistance to fluoroquinolones. The strains also differ in the flagellar genes, with a difference in the replacement of phosphotransferase genes found in the 630 strain with glycosyltransferase genes in 027 ribotypes. The variations in flagellar and glycosyltransferase genes may explain the observed differences in motility and autoagglutination between the strains. The 027 ribotype strains both exhibit motility, while the 012 ribotype (strain 630) does not. In addition, the recently emerged 027 ribotype appears to autoagglutinate more efficiently than the historical 027 strain or the 630 strain. Interestingly, an intact agr locus is present in the 027 strains, yet specific agr genes are missing in the 630 strain. Agr is a known growth stage regulator of virulence factor production in other Gram-positive organisms, so its presence in 027 strains could provide a mechanism of virulence regulation unique to these strains and absent in other nonhypervirulent strains.
Strains of the 027 ribotype appear to be clonal, arising from earlier nonepidemic strains such as CD196. CD196 was first described in the mid-1980s, and R20291 is a much more recent isolate. The two strains have many genes in common, but notable changes have been detected in the sequences of these two isolates. For example, R20291 has acquired a phage island that provides a new response regulator and a toxin-antitoxin system to the organism. Collectively, the genomic data indicate that several new genes were recently acquired by the epidemic strain of C. difficile. Which of these genes may contribute to the differences in virulence between hypervirulent 027 strains and older strains of this ribotype is not known; however, the data suggest that the organism continues to acquire new genes during its evolution as a pathogen.
A detailed analysis of the phylogenetic lineages of C. difficile indicates that the species diverged early from other clostridial strains and that lineages were established several million years ago (196). Several factors appear to influence development of the lineages. In particular, there is a remarkable amount of genetic exchange (traced by shared single-nucleotide polymorphisms [SNPs]) among the different strains. Very large portions of genomic DNA appear to have been exchanged across strains. Several of these are associated with mobile elements, but many of these appear to be substantial portions of the genome that have moved, through an unknown mechanism, into various strains. In particular, evolution of hypervirulent strains seems to have accelerated from strains around the year 2000, corresponding to the emergence of epidemic C. difficile. These studies reveal C. difficile to have a highly fluid genome that is rapidly changing in the human population, driven by a high rate of genetic exchange. Thus, the emergence of hypervirulent epidemic strains may be supported in part by the ability of the organism to efficiently alter genomic sequences by acquiring new or altered genes through genetic recombination.
These genetic variations among strains of C. difficile could influence disease severity and relapse by preventing cross-neutralization. Studies on relapse and recurrence have been very informative in this regard. As we discussed above, relapse in CDI is a common and serious clinical problem. Curiously, almost 50% of patients relapse with a different strain of C. difficile (46, 47). Given the fact that immune responses to TcdA and TcdB are correlated with protection against CDI and reduce the likelihood of relapse, it is reasonable to speculate that new strains may be resistant to host immunity through variability in the antigenic makeup of the toxins. We have reported that TcdB proteins from NAP1 and 630 strains differ in sequence and toxic activity (194). Thus, it is worth considering the possibility that host immune responses may be effective against only one form of the toxin and that the host remains susceptible to infection by new strains expressing a different form of TcdB. Along these lines, although in need of more investigation, the lack of appropriate cross-neutralizing immune responses could be one explanation for entry of new strains into the human population.
Additional Emerging Hypervirulent Strains of C. difficile
To date, at least 375 different ribotypes of C. difficile have been identified, and the number continues to increase with every new epidemiological study. The vast majority of these strains have the potential to cause disease, with the exception of rare isolates lacking toxin-encoding genes. Strains of C. difficile fall into five clades based on sequence, especially MLST, analysis (197). The most conservative estimates suggest that the clades originated at least 1 million years ago (196). These clades of C. difficile seem to have evolved virulence independently, although there are some examples to suggest the acquisition of toxic elements via horizontal transfer. The findings on strain diversity emphasize not only the need to better understand the current hypervirulent strains, such as NAP1, but also the need to discover the underlying reasons for the emergence of new strains that could represent the next wave of epidemics causing severe disease. There are several notable points that should be considered. First, given the exceptional variance in sequences, especially in genes such as tcdB, the lack of immune cross-protection could be an important factor in enriching for new strains. Indeed, without appropriate cross-protection, the human population may simply remain susceptible to new strains, and once immunity increases against a new strain, a different one may replace it. This concept is especially important as vaccines and therapeutic antibodies are considered for possible prevention or treatment of CDI. These treatments will need to be broadly protective to prevent selection of resistant strains. Second, more work is needed to discover the environmental source of C. difficile. Recent work has found C. difficile, particularly ribotype 078 of clade 5, to be a common contaminant of food sources (198, 199). Because C. difficile 078 has also been associated with infection of livestock, animals may provide the conduit for strains moving into the human population.
Finally, the fundamental bacteriology of C. difficile remains poorly understood, and little is known about the factors that contribute to mutations in virulence-associated genes. With only a few exceptions, most toxins produced by bacterial pathogens are very stable in their sequences, yet the analysis of variant toxinotypes in C. difficile suggests that this is not the case with this pathogen. It will be interesting to discover the fundamental reasons for such a high mutation rate in tcdB. Relative to many other human bacterial pathogens, C. difficile has been recognized as a human pathogen for only a short period, and the emergence of hypervirulent strains may be just a small glimpse of what may continue to emerge as new strains enter the human population. It is reasonable to predict that new strains will continue to emerge and that some of these could exhibit virulence more severe than what has been found with NAP1.
Despite the conflicting evidence regarding the increased virulence of C. difficile NAP1/027/BI and its association, or lack of association, with more severe disease, it is clear that this strain has emerged from the hundreds of different strains of C. difficile to become one of the most common causes of CDI. Arguably the most important lesson could be that strains of C. difficile emerge in the population, cause large outbreaks and epidemics, and replace otherwise common strains in a hospital setting. Thus, the careful documentation of the increase in NAP1/BI/027 cases over the past decade and understanding how these cases differ from others could help to inform us about the potential for the next wave of C. difficile infections. Much like the annual changes in influenza virus, more longitudinal studies could reveal the ebb and flow of the many strains involved in CDI within the population. Indeed, current studies indicate that changes may already be taking place. The number of NAP1/BI/027 cases is starting to wane, and new strains, such as the 078 ribotype, are starting to emerge (200). A study from the Netherlands found that the number of CDI cases caused by C. difficile 078 ribotypes increased from 3% to 13% in just 3 years (201). Interestingly, 078 strains have been found most commonly in food sources, and the careful assessment of changes in human 078 strains may provide the first opportunity to follow the epidemiology of emerging C. difficile strains while knowing the environmental source of the pathogen.
FINAL COMMENTS
Compared to many prominent human infectious diseases, CDI is relatively new and appears to be a man-made illness. Much of our understanding of CDI is suggestive. Clearly, many variants of C. difficile exist in the environment and are able to move into the human population under the right conditions. Yet the major environmental sources of C. difficile and how events in the environment select for virulence are not known. There is evidence that the emergence of NAP1/BI/027 during the past decade has started to subside, but this change could simply select for another strain, and the wave of disease during the past decade could represent a more common occurrence in the future. Indeed, if increased virulence is a major factor in selecting for new strains in the human population, then the encounter with NAP1/BI/027 may be just the beginning of a long-term health care problem. Moreover, it is reasonable to expect that each new strain could be more virulent than the previous strain. As such, perhaps more important than designing novel treatments against hypervirulent strains will be the thorough study and understanding of why and how the NAP1/BI/027 strains emerged to begin with. In doing so, we can better predict the next epidemics and develop more sophisticated surveillance and treatment systems to deal with the next outbreak of CDI.
ACKNOWLEDGMENTS
Support for preparation of this review was provided by Public Health Service grant R01HL084489.
We thank Aleze Krumholz for technical assistance and editing during the preparation of this review.
REFERENCES
- 1.Blair JE, Hallman FA. 1935. Streptococcal agglutinins and antistreptolysins in rheumatoid (atrophic) arthritis. J. Clin. Invest. 14:505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder ML. 1937. Further studies on Bacillus difficilis (Hall and O'Toole). J. Infect. Dis. 60:223–231 [Google Scholar]
- 3.McBee RH. 1960. Intestinal flora of some Antarctic birds and mammals. J. Bacteriol. 79:311–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LD, King EO. 1962. Occurrence of Clostridium difficile in infections of man. J. Bacteriol. 84:65–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George RH, Symonds JM, Dimock F, Brown JD, Arabi Y, Shinagawa N, Keighley MR, Alexander-Williams J, Burdon DW. 1978. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br. Med. J. i:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson HE, Parry JV, Price AB, Davies DR, Dolby J, Tyrrell DA. 1977. Undescribed toxin in pseudomembranous colitis. Br. Med. J. i:1246–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson HE, Price AB. 1977. Pseudomembranous colitis: presence of clostridial toxin. Lancet ii:1312–1314 [DOI] [PubMed] [Google Scholar]
- 8.Chang TW, Bartlett JG, Gorbach SL, Onderdonk AB. 1978. Clindamycin-induced enterocolitis in hamsters as a model of pseudomembranous colitis in patients. Infect. Immun. 20:526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan NM, Pellett S, Wilkins TD. 1982. Purification and characterization of toxins A and B of Clostridium difficile. Infect. Immun. 35:1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyerly DM, Lockwood DE, Richardson SH, Wilkins TD. 1982. Biological activities of toxins A and B of Clostridium difficile. Infect. Immun. 35:1147–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corthier G, Muller MC, Wilkins TD, Lyerly D, L'Haridon R. 1991. Protection against experimental pseudomembranous colitis in gnotobiotic mice by use of monoclonal antibodies against Clostridium difficile toxin A. Infect. Immun. 59:1192–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyerly DM, Phelps CJ, Toth J, Wilkins TD. 1986. Characterization of toxins A and B of Clostridium difficile with monoclonal antibodies. Infect. Immun. 54:70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pepin J, Saheb N, Coulombe MA, Alary ME, Corriveau MP, Authier S, Leblanc M, Rivard G, Bettez M, Primeau V, Nguyen M, Jacob CE, Lanthier L. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254–1260 [DOI] [PubMed] [Google Scholar]
- 14.Muto CA, Pokrywka M, Shutt K, Mendelsohn AB, Nouri K, Posey K, Roberts T, Croyle K, Krystofiak S, Patel-Brown S, Pasculle AW, Paterson DL, Saul M, Harrison LH. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect. Control Hosp. Epidemiol. 26:273–280 [DOI] [PubMed] [Google Scholar]
- 15.Pepin J, Valiquette L, Alary ME, Villemure P, Pelletier A, Forget K, Pepin K, Chouinard D. 2004. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 17.Miller BA, Chen LF, Sexton DJ, Anderson DJ. 2011. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect. Control Hosp. Epidemiol. 32:387–390 [DOI] [PubMed] [Google Scholar]
- 18.Murphy SL, Xu J, Kochanek MA. 2012. Deaths: preliminary data for 2010. National vital statistics reports, vol 60, no 4 National Center for Health Statistics, Hyattsville, MD [Google Scholar]
- 19.Lucado J, Gould C, Elixhauser A. 2012. Clostridium difficile infections (CDI) in hospital stays, 2009. HCUP statistical brief 124. Agency for Healthcare Research and Quality, Rockville, MD: [PubMed] [Google Scholar]
- 20.Karas JA, Enoch DA, Aliyu SH. 2010. A review of mortality due to Clostridium difficile infection. J. Infect. 61:1–8 [DOI] [PubMed] [Google Scholar]
- 21.Bignardi GE. 1998. Risk factors for Clostridium difficile infection. J. Hosp. Infect. 40:1–15 [DOI] [PubMed] [Google Scholar]
- 22.Welfare MR, Lalayiannis LC, Martin KE, Corbett S, Marshall B, Sarma JB. 2011. Co-morbidities as predictors of mortality in Clostridium difficile infection and derivation of the ARC predictive score. J. Hosp. Infect. 79:359–363 [DOI] [PubMed] [Google Scholar]
- 23.Johnson S, Gerding DN. 1998. Clostridium difficile-associated diarrhea. Clin. Infect. Dis. 26:1027–1034 [DOI] [PubMed] [Google Scholar]
- 24.Jarchum I, Liu M, Lipuma L, Pamer EG. 2011. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect. Immun. 79:1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen A. 2012. Clostridium difficile toxins: mediators of inflammation. J. Innate Immun. 4:149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libby DB, Bearman G. 2009. Bacteremia due to Clostridium difficile—review of the literature. Int. J. Infect. Dis. 13:e305–e309 [DOI] [PubMed] [Google Scholar]
- 28.Rubin MS, Bodenstein LE, Kent KC. 1995. Severe Clostridium difficile colitis. Dis. Colon Rectum 38:350–354 [DOI] [PubMed] [Google Scholar]
- 29.Greenstein AJ, Byrn JC, Zhang LP, Swedish KA, Jahn AE, Divino CM. 2008. Risk factors for the development of fulminant Clostridium difficile colitis. Surgery 143:623–629 [DOI] [PubMed] [Google Scholar]
- 30.Jacob SS, Sebastian JC, Hiorns D, Jacob S, Mukerjee PK. 2004. Clostridium difficile and acute respiratory distress syndrome. Heart Lung 33:265–268 [DOI] [PubMed] [Google Scholar]
- 31.Jacobs A, Barnard K, Fishel R, Gradon JD. 2001. Extracolonic manifestations of Clostridium difficile infections. Presentation of 2 cases and review of the literature. Medicine 80:88–101 [DOI] [PubMed] [Google Scholar]
- 32.Dobson G, Hickey C, Trinder J. 2003. Clostridium difficile colitis causing toxic megacolon, severe sepsis and multiple organ dysfunction syndrome. Intensive Care Med. 29:1030. [DOI] [PubMed] [Google Scholar]
- 33.Johnson S, Kent SA, O'Leary KJ, Merrigan MM, Sambol SP, Peterson LR, Gerding DN. 2001. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann. Intern. Med. 135:434–438 [DOI] [PubMed] [Google Scholar]
- 34.Cunney RJ, Magee C, McNamara E, Smyth EG, Walshe J. 1998. Clostridium difficile colitis associated with chronic renal failure. Nephrol. Dial. Transplant. 13:2842–2846 [DOI] [PubMed] [Google Scholar]
- 35.Parikh VA, Edlund JW. 1996. Fatal Clostridium difficile enteritis after total abdominal colectomy. J. Clin. Gastroenterol. 22:329. [DOI] [PubMed] [Google Scholar]
- 36.Hawker PC, Hine KR, Burdon DW, Thompson H, Keighley MR. 1981. Fatal pseudomembranous colitis despite eradication of Clostridium difficile. Br. Med. J. (Clin. Res. Ed.) 282:109–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele J, Chen K, Sun X, Zhang Y, Wang H, Tzipori S, Feng H. 2012. Systemic dissemination of Clostridium difficile toxins A and B is associated with severe, fatal disease in animal models. J. Infect. Dis. 205:384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qualman SJ, Petric M, Karmali MA, Smith CR, Hamilton SR. 1990. Clostridium difficile invasion and toxin circulation in fatal pediatric pseudomembranous colitis. Am. J. Clin. Pathol. 94:410–416 [DOI] [PubMed] [Google Scholar]
- 39.Hamm EE, Voth DE, Ballard JD. 2006. Identification of Clostridium difficile toxin B cardiotoxicity using a zebrafish embryo model of intoxication. Proc. Natl. Acad. Sci. U. S. A. 103:14176–14181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartlett JG, Tedesco FJ, Shull S, Lowe B, Chang T. 1980. Symptomatic relapse after oral vancomycin therapy of antibiotic-associated pseudomembranous colitis. Gastroenterology 78:431–434 [PubMed] [Google Scholar]
- 41.George WL, Volpicelli NA, Stiner DB, Richman DD, Liechty EJ, Mok HY, Rolfe RD, Finegold SM. 1979. Relapse of pseudomembranous colitis after vancomycin therapy. N. Engl. J. Med. 301:414–415 [DOI] [PubMed] [Google Scholar]
- 42.McFarland LV, Elmer GW, Surawicz CM. 2002. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am. J. Gastroenterol. 97:1769–1775 [DOI] [PubMed] [Google Scholar]
- 43.Johnson S. 2009. Recurrent Clostridium difficile infection: causality and therapeutic approaches. Int. J. Antimicrob. Agents 33(Suppl 1):S33–S36 [DOI] [PubMed] [Google Scholar]
- 44.Johnson S. 2009. Recurrent Clostridium difficile infection: a review of risk factors, treatments, and outcomes. J. Infect. 58:403–410 [DOI] [PubMed] [Google Scholar]
- 45.O'Neill GL, Beaman MH, Riley TV. 1991. Relapse versus reinfection with Clostridium difficile. Epidemiol. Infect. 107:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang-Feldman Y, Mayo S, Silva J, Jr, Cohen SH. 2003. Molecular analysis of Clostridium difficile strains isolated from 18 cases of recurrent Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 41:3413–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbut F, Richard A, Hamadi K, Chomette V, Burghoffer B, Petit JC. 2000. Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2386–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson S, Adelmann A, Clabots CR, Peterson LR, Gerding DN. 1989. Recurrences of Clostridium difficile diarrhea not caused by the original infecting organism. J. Infect. Dis. 159:340–343 [DOI] [PubMed] [Google Scholar]
- 49.Kyne L, Warny M, Qamar A, Kelly CP. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet 357:189–193 [DOI] [PubMed] [Google Scholar]
- 50.Kyne L, Warny M, Qamar A, Kelly CP. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 342:390–397 [DOI] [PubMed] [Google Scholar]
- 51.Warny M, Vaerman JP, Avesani V, Delmee M. 1994. Human antibody response to Clostridium difficile toxin A in relation to clinical course of infection. Infect. Immun. 62:384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH, Society for Healthcare Epidemiology of America, Infectious Diseases Society of America 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455 [DOI] [PubMed] [Google Scholar]
- 53.Chand MA, Fleming MJ, Wellsteed S, Kelsey MC. 2011. Impact of changes in Clostridium difficile diagnostic testing on detection of C. difficile infection and all England mandatory surveillance data. J. Hosp. Infect. 79:8–12 [DOI] [PubMed] [Google Scholar]
- 54.Bamber AI, Fitzsimmons K, Cunniffe JG, Beasor CC, Mackintosh CA, Hobbs G. 2012. Diagnosis of Clostridium difficile-associated disease: examination of multiple algorithms using toxin EIA, glutamate dehydrogenase EIA and loop-mediated isothermal amplification. Br. J. Biomed. Sci. 69:112–118 [PubMed] [Google Scholar]
- 55.Longtin Y, Trottier S, Brochu G, Paquet-Bolduc B, Garenc C, Loungnarath V, Beaulieu C, Goulet D, Longtin J. 2013. Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin. Infect. Dis. 56:67–73 [DOI] [PubMed] [Google Scholar]
- 56.Ylisiurua P, Koskela M, Vainio O, Tuokko H. 2013. Comparison of antigen and two molecular methods for the detection of Clostridium difficile toxins. Scand. J. Infect. Dis. 45:19–25 [DOI] [PubMed] [Google Scholar]
- 57.Garimella PS, Agarwal R, Katz A. 2012. The utility of repeat enzyme immunoassay testing for the diagnosis of Clostridium difficile infection: a systematic review of the literature. J. Postgrad. Med. 58:194–198 [DOI] [PubMed] [Google Scholar]
- 58.Shakir FA, Thompson D, Marlar R, Ali T. 2012. A novel use of rectal swab to test for Clostridium difficile infection by real-time PCR. Am. J. Gastroenterol. 107:1444–1445 [DOI] [PubMed] [Google Scholar]
- 59.Williamson DA, Basu I, Freeman J, Swager T, Roberts SA. 2013. Improved detection of toxigenic Clostridium difficile using the Cepheid Xpert C difficile assay and impact on C difficile infection rates in a tertiary hospital: a double-edged sword. Am. J. Infect. Control 41:270–272 [DOI] [PubMed] [Google Scholar]
- 60.Nistico JA, Hage JE, Schoch PE, Cunha BA. 2013. Unnecessary repeat Clostridium difficile PCR testing in hospitalized adults with C. difficile-negative diarrhea. Eur. J. Clin. Microbiol. Infect. Dis. 32:97–99 [DOI] [PubMed] [Google Scholar]
- 61.Cooper PB, Heuer AJ, Warren CA. 2013. Electronic screening of patients for predisposition to Clostridium difficile infection in a community hospital. Am. J. Infect. Control 41:232–235 [DOI] [PubMed] [Google Scholar]
- 62.Hensgens MP, Goorhuis A, Dekkers OM, Kuijper EJ. 2012. Time interval of increased risk for Clostridium difficile infection after exposure to antibiotics. J. Antimicrob. Chemother. 67:742–748 [DOI] [PubMed] [Google Scholar]
- 63.Goorhuis A, Debast SB, Dutilh JC, van Kinschot CM, Harmanus C, Cannegieter SC, Hagen EC, Kuijper EJ. 2011. Type-specific risk factors and outcome in an outbreak with 2 different Clostridium difficile types simultaneously in 1 hospital. Clin. Infect. Dis. 53:860–869 [DOI] [PubMed] [Google Scholar]
- 64.Vardakas KZ, Konstantelias AA, Loizidis G, Rafailidis PI, Falagas ME. 2012. Risk factors for development of Clostridium difficile infection due to BI/NAP1/027 strain: a meta-analysis. Int. J. Infect. Dis. 16:e768–e773 [DOI] [PubMed] [Google Scholar]
- 65.Dubberke ER, Reske KA, Yan Y, Olsen MA, McDonald LC, Fraser VJ. 2007. Clostridium difficile-associated disease in a setting of endemicity: identification of novel risk factors. Clin. Infect. Dis. 45:1543–1549 [DOI] [PubMed] [Google Scholar]
- 66.Cornely OA, Miller MA, Louie TJ, Crook DW, Gorbach SL. 2012. Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin. Infect. Dis. 55(Suppl 2):S154–S161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pepin J, Routhier S, Gagnon S, Brazeau I. 2006. Management and outcomes of a first recurrence of Clostridium difficile-associated disease in Quebec, Canada. Clin. Infect. Dis. 42:758–764 [DOI] [PubMed] [Google Scholar]
- 68.Wenisch JM, Schmid D, Kuo HW, Allerberger F, Michl V, Tesik P, Tucek G, Laferl H, Wenisch C. 2012. Prospective observational study comparing three different treatment regimes in patients with Clostridium difficile infection. Antimicrob. Agents Chemother. 56:1974–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drekonja DM, Butler M, Macdonald R, Bliss D, Filice GA, Rector TS, Wilt TJ. 2011. Comparative effectiveness of Clostridium difficile treatments: a systematic review. Ann. Intern. Med. 155:839–847 [DOI] [PubMed] [Google Scholar]
- 70.Crook DW, Walker AS, Kean Y, Weiss K, Cornely OA, Miller MA, Esposito R, Louie TJ, Stoesser NE, Young BC, Angus BJ, Gorbach SL, Peto TE, Study 003/004 Teams 2012. Fidaxomicin versus vancomycin for Clostridium difficile infection: meta-analysis of pivotal randomized controlled trials. Clin. Infect. Dis. 55(Suppl 2):S93–S103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Louie TJ, Emery J, Krulicki W, Byrne B, Mah M. 2009. OPT-80 eliminates Clostridium difficile and is sparing of Bacteroides species during treatment of C. difficile infection. Antimicrob. Agents Chemother. 53:261–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Louie TJ, Cannon K, Byrne B, Emery J, Ward L, Eyben M, Krulicki W. 2012. Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin. Infect. Dis. 55(Suppl 2):S132–S142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garborg K, Waagsbo B, Stallemo A, Matre J, Sundoy A. 2010. Results of faecal donor instillation therapy for recurrent Clostridium difficile-associated diarrhoea. Scand. J. Infect. Dis. 42:857–861 [DOI] [PubMed] [Google Scholar]
- 74.Rubin TA, Gessert CE, Aas J, Bakken JS. 2013. Fecal microbiome transplantation for recurrent Clostridium difficile infection: report on a case series. Anaerobe 19:22–26 [DOI] [PubMed] [Google Scholar]
- 75.Louie T, Louie M, Krulicki W, Byrne B, Ward L. 2008. Home-base fecal flora infusion to arrest multiply-recurrent Clostridium difficile infection (CDI), abstr K4201 Abstr. 48th Annu. ICAAC/IDSA 46th Annu. Meet [Google Scholar]
- 76.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, Stollman N, Rohlke F, Surawicz C. 2012. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 107:1079–1087 [DOI] [PubMed] [Google Scholar]
- 77.US FDA 2013. US NIH Clinical Trial Database. http://clinicaltrials.gov/ct2/results?term=%22fecal%22+AND+%22difficile%22&Search=Search Accessed 11 July 2013
- 78.Babcock GJ, Broering TJ, Hernandez HJ, Mandell RB, Donahue K, Boatright N, Stack AM, Lowy I, Graziano R, Molrine D, Ambrosino DM, Thomas WD., Jr 2006. Human monoclonal antibodies directed against toxins A and B prevent Clostridium difficile-induced mortality in hamsters. Infect. Immun. 74:6339–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giannasca PJ, Warny M. 2004. Active and passive immunization against Clostridium difficile diarrhea and colitis. Vaccine 22:848–856 [DOI] [PubMed] [Google Scholar]
- 80.Lowy I, Molrine DC, Leav BA, Blair BM, Baxter R, Gerding DN, Nichol G, Thomas WD, Jr, Leney M, Sloan S, Hay CA, Ambrosino DM. 2010. Treatment with monoclonal antibodies against Clostridium difficile toxins. N. Engl. J. Med. 362:197–205 [DOI] [PubMed] [Google Scholar]
- 81.O'Horo J, Safdar N. 2009. The role of immunoglobulin for the treatment of Clostridium difficile infection: a systematic review. Int. J. Infect. Dis. 13:663–667 [DOI] [PubMed] [Google Scholar]
- 82.Steele J, Mukherjee J, Parry N, Tzipori S. 2013. Antibody against TcdB, but not TcdA, prevents development of gastrointestinal and systemic Clostridium difficile disease. J. Infect. Dis. 207:323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marozsan AJ, Ma D, Nagashima KA, Kennedy BJ, Kang YK, Arrigale RR, Donovan GP, Magargal WW, Maddon PJ, Olson WC. 2012. Protection against Clostridium difficile infection with broadly neutralizing antitoxin monoclonal antibodies. J. Infect. Dis. 206:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitchell TJ, Ketley JM, Haslam SC, Stephen J, Burdon DW, Candy DC, Daniel R. 1986. Effect of toxin A and B of Clostridium difficile on rabbit ileum and colon. Gut 27:78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hammond GA, Johnson JL. 1995. The toxigenic element of Clostridium difficile strain VPI 10463. Microb. Pathog. 19:203–213 [DOI] [PubMed] [Google Scholar]
- 86.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. U. S. A. 98:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dupuy B, Matamouros S. 2006. Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors. Res. Microbiol. 157:201–205 [DOI] [PubMed] [Google Scholar]
- 88.Dupuy B, Raffestin S, Matamouros S, Mani N, Popoff MR, Sonenshein AL. 2006. Regulation of toxin and bacteriocin gene expression in Clostridium by interchangeable RNA polymerase sigma factors. Mol. Microbiol. 60:1044–1057 [DOI] [PubMed] [Google Scholar]
- 89.Olling A, Seehase S, Minton NP, Tatge H, Schroter S, Kohlscheen S, Pich A, Just I, Gerhard R. 2012. Release of TcdA and TcdB from Clostridium difficile CDI 630 is not affected by functional inactivation of the tcdE gene. Microb. Pathog. 52:92–100 [DOI] [PubMed] [Google Scholar]
- 90.Govind R, Dupuy B. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 8:e1002727. 10.1371/journal.ppat.1002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matamouros S, England P, Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 64:1274–1288 [DOI] [PubMed] [Google Scholar]
- 92.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244:735–742 [DOI] [PubMed] [Google Scholar]
- 93.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. 1985. Effects of Clostridium difficile toxins given intragastrically to animals. Infect. Immun. 47:349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Donta ST, Sullivan N, Wilkins TD. 1982. Differential effects of Clostridium difficile toxins on tissue-cultured cells. J. Clin. Microbiol. 15:1157–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chaves-Olarte E, Weidmann M, Eichel-Streiber C, Thelestam M. 1997. Toxins A and B from Clostridium difficile differ with respect to enzymatic potencies, cellular substrate specificities, and surface binding to cultured cells. J. Clin. Invest. 100:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elliott B, Squire MM, Thean S, Chang BJ, Brazier JS, Rupnik M, Riley TV. 2011. New types of toxin A-negative, toxin B-positive strains among clinical isolates of Clostridium difficile in Australia. J. Med. Microbiol. 60:1108–1111 [DOI] [PubMed] [Google Scholar]
- 97.Kim H, Riley TV, Kim M, Kim CK, Yong D, Lee K, Chong Y, Park JW. 2008. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J. Clin. Microbiol. 46:1116–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Komatsu M, Kato H, Aihara M, Shimakawa K, Iwasaki M, Nagasaka Y, Fukuda S, Matsuo S, Arakawa Y, Watanabe M, Iwatani Y. 2003. High frequency of antibiotic-associated diarrhea due to toxin A-negative, toxin B-positive Clostridium difficile in a hospital in Japan and risk factors for infection. Eur. J. Clin. Microbiol. Infect. Dis. 22:525–529 [DOI] [PubMed] [Google Scholar]
- 99.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 100.Kuehne SA, Cartman ST, Minton NP. 2011. Both, toxin A and toxin B, are important in Clostridium difficile infection. Gut Microbes 2:252–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Davies NL, Compson JE, Mackenzie B, O'Dowd VL, Oxbrow AK, Heads JT, Turner A, Sarkar K, Dugdale SL, Jairaj M, Christodoulou L, Knight DE, Cross AS, Herve KJ, Tyson KL, Hailu H, Doyle CB, Ellis M, Kriek M, Cox M, Page MJ, Moore AR, Lightwood DJ, Humphreys DP. 2013. A mixture of functionally oligoclonal humanized monoclonal antibodies that neutralize Clostridium difficile TcdA and TcdB with high levels of in vitro potency shows in vivo protection in a hamster infection model. Clin. Vaccine Immunol. 20:377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27:107–120 [DOI] [PubMed] [Google Scholar]
- 104.Hammond GA, Lyerly DM, Johnson JL. 1997. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb. Pathog. 22:143–154 [DOI] [PubMed] [Google Scholar]
- 105.Yamakawa K, Karasawa T, Ikoma S, Nakamura S. 1996. Enhancement of Clostridium difficile toxin production in biotin-limited conditions. J. Med. Microbiol. 44:111–114 [DOI] [PubMed] [Google Scholar]
- 106.Karlsson S, Burman LG, Akerlund T. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145:1683–1693 [DOI] [PubMed] [Google Scholar]
- 107.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206–219 [DOI] [PubMed] [Google Scholar]
- 108.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79:882–899 [DOI] [PubMed] [Google Scholar]
- 109.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 193:3186–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rupnik M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 32:541–555 [DOI] [PubMed] [Google Scholar]
- 111.Drudy D, Fanning S, Kyne L. 2007. Toxin A-negative, toxin B-positive Clostridium difficile. Int. J. Infect. Dis. 11:5–10 [DOI] [PubMed] [Google Scholar]
- 112.Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, Aktories K. 1995. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 270:13932–13936 [DOI] [PubMed] [Google Scholar]
- 114.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500–503 [DOI] [PubMed] [Google Scholar]
- 115.Sehr P, Joseph G, Genth H, Just I, Pick E, Aktories K. 1998. Glucosylation and ADP ribosylation of rho proteins: effects on nucleotide binding, GTPase activity, and effector coupling. Biochemistry 37:5296–5304 [DOI] [PubMed] [Google Scholar]
- 116.Hofmann F, Busch C, Prepens U, Just I, Aktories K. 1997. Localization of the glucosyltransferase activity of Clostridium difficile toxin B to the N-terminal part of the holotoxin. J. Biol. Chem. 272:11074–11078 [DOI] [PubMed] [Google Scholar]
- 117.Busch C, Hofmann F, Selzer J, Munro S, Jeckel D, Aktories K. 1998. A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem. 273:19566–19572 [DOI] [PubMed] [Google Scholar]
- 118.Busch C, Hofmann F, Gerhard R, Aktories K. 2000. Involvement of a conserved tryptophan residue in the UDP-glucose binding of large clostridial cytotoxin glycosyltransferases. J. Biol. Chem. 275:13228–13234 [DOI] [PubMed] [Google Scholar]
- 119.Reineke J, Tenzer S, Rupnik M, Koschinski A, Hasselmayer O, Schrattenholz A, Schild H, von Eichel-Streiber C. 2007. Autocatalytic cleavage of Clostridium difficile toxin B. Nature 446:415–419 [DOI] [PubMed] [Google Scholar]
- 120.Rupnik M, Pabst S, Rupnik M, von Eichel-Streiber C, Urlaub H, Soling HD. 2005. Characterization of the cleavage site and function of resulting cleavage fragments after limited proteolysis of Clostridium difficile toxin B (TcdB) by host cells. Microbiology 151:199–208 [DOI] [PubMed] [Google Scholar]
- 121.Pruitt RN, Chagot B, Cover M, Chazin WJ, Spiller B, Lacy DB. 2009. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. J. Biol. Chem. 284:21934–21940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. 2007. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 282:25314–25321 [DOI] [PubMed] [Google Scholar]
- 123.Genisyuerek S, Papatheodorou P, Guttenberg G, Schubert R, Benz R, Aktories K. 2011. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol. Microbiol. 79:1643–1654 [DOI] [PubMed] [Google Scholar]
- 124.Greco A, Ho JG, Lin SJ, Palcic MM, Rupnik M, Ng KK. 2006. Carbohydrate recognition by Clostridium difficile toxin A. Nat. Struct. Mol. Biol. 13:460–461 [DOI] [PubMed] [Google Scholar]
- 125.Ho JG, Greco A, Rupnik M, Ng KK. 2005. Crystal structure of receptor-binding C-terminal repeats from Clostridium difficile toxin A. Proc. Natl. Acad. Sci. U. S. A. 102:18373–18378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dingle T, Wee S, Mulvey GL, Greco A, Kitova EN, Sun J, Lin S, Klassen JS, Palcic MM, Ng KK, Armstrong GD. 2008. Functional properties of the carboxy-terminal host cell-binding domains of the two toxins, TcdA and TcdB, expressed by Clostridium difficile. Glycobiology 18:698–706 [DOI] [PubMed] [Google Scholar]
- 127.Aronson B, Granstrom M, Mollby R, Nord CE. 1982. Toxin A (enterotoxin) from Clostridium difficile in antibiotic-associated colitis. Lancet ii:1279. [DOI] [PubMed] [Google Scholar]
- 128.Chang TW. 1983. Interaction between Clostridium difficile toxin A and mammalian cells. Acta Pharmacol. Sin. 4:210–214 [PubMed] [Google Scholar]
- 129.Gianfrilli P, Luzzi I, Pantosti A, Occhionero M, Gentile G, Panichi G. 1984. Cytotoxin and enterotoxin production by Clostridium difficile. Microbiologica 7:375–379 [PubMed] [Google Scholar]
- 130.Stephen J, Redmond SC, Mitchell TJ, Ketley J, Candy DC, Burdon DW, Daniel R. 1984. Clostridium difficile enterotoxin (toxin A): new results. Biochem. Soc. Trans. 12:194–195 [DOI] [PubMed] [Google Scholar]
- 131.Wedel N, Toselli P, Pothoulakis C, Faris B, Oliver P, Franzblau C, LaMont T. 1983. Ultrastructural effects of Clostridium difficile toxin B on smooth muscle cells and fibroblasts. Exp. Cell Res. 148:413–422 [DOI] [PubMed] [Google Scholar]
- 132.Pothoulakis C, Triadafilopoulos G, Clark M, Franzblau C, LaMont JT. 1986. Clostridium difficile cytotoxin inhibits protein synthesis in fibroblasts and intestinal mucosa. Gastroenterology 91:1147–1153 [DOI] [PubMed] [Google Scholar]
- 133.Meador J, 3rd, Tweten RK. 1988. Purification and characterization of toxin B from Clostridium difficile. Infect. Immun. 56:1708–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Limaye AP, Turgeon DK, Cookson BT, Fritsche TR. 2000. Pseudomembranous colitis caused by a toxin A(−) B(+) strain of Clostridium difficile. J. Clin. Microbiol. 38:1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wilcox MH, Fawley WN. 2001. Virulence of Clostridium difficile toxin A negative strains. J. Hosp. Infect. 48:81. [DOI] [PubMed] [Google Scholar]
- 136.Savidge TC, Pan WH, Newman P, O'Brien M, Anton PM, Pothoulakis C. 2003. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125:413–420 [DOI] [PubMed] [Google Scholar]
- 137.Pruitt RN, Chumbler NM, Rutherford SA, Farrow MA, Friedman DB, Spiller B, Lacy DB. 2012. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J. Biol. Chem. 287:8013–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Popoff MR, Boquet P. 1988. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem. Biophys. Res. Commun. 152:1361–1368 [DOI] [PubMed] [Google Scholar]
- 139.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect. Immun. 65:1402–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chang SY, Song KP. 2001. ADP-ribosylating binary toxin genes of Clostridium difficile strain CCUG 20309. DNA Seq. 12:115–120 [DOI] [PubMed] [Google Scholar]
- 141.Martin H, Willey B, Low DE, Staempfli HR, McGeer A, Boerlin P, Mulvey M, Weese JS. 2008. Characterization of Clostridium difficile strains isolated from patients in Ontario, Canada, from 2004 to 2006. J. Clin. Microbiol. 46:2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Carman RJ, Stevens AL, Lyerly MW, Hiltonsmith MF, Stiles BG, Wilkins TD. 2011. Clostridium difficile binary toxin (CDT) and diarrhea. Anaerobe 17:161–165 [DOI] [PubMed] [Google Scholar]
- 143.Carter GP, Lyras D, Allen DL, Mackin KE, Howarth PM, O'Connor JR, Rood JI. 2007. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J. Bacteriol. 189:7290–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bouvet PJ, Popoff MR. 2008. Genetic relatedness of Clostridium difficile isolates from various origins determined by triple-locus sequence analysis based on toxin regulatory genes tcdC, tcdR, and cdtR. J. Clin. Microbiol. 46:3703–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 146.Papatheodorou P, Carette JE, Bell GW, Schwan C, Guttenberg G, Brummelkamp TR, Aktories K. 2011. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc. Natl. Acad. Sci. U. S. A. 108:16422–16427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kaiser E, Kroll C, Ernst K, Schwan C, Popoff M, Fischer G, Buchner J, Aktories K, Barth H. 2011. Membrane translocation of binary actin-ADP-ribosylating toxins from Clostridium difficile and Clostridium perfringens is facilitated by cyclophilin A and Hsp90. Infect. Immun. 79:3913–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schwan C, Stecher B, Tzivelekidis T, van Ham M, Rohde M, Hardt WD, Wehland J, Aktories K. 2009. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 5:e1000626. 10.1371/journal.ppat.1000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wigelsworth DJ, Ruthel G, Schnell L, Herrlich P, Blonder J, Veenstra TD, Carman RJ, Wilkins TD, Van Nhieu GT, Pauillac S, Gibert M, Sauvonnet N, Stiles BG, Popoff MR, Barth H. 2012. CD44 promotes intoxication by the clostridial iota-family toxins. PLoS One 7:e51356. 10.1371/journal.pone.0051356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bacci S, Molbak K, Kjeldsen MK, Olsen KE. 2011. Binary toxin and death after Clostridium difficile infection. Emerg. Infect. Dis. 17:976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Takeoka A, Takumi K, Koga T, Kawata T. 1991. Purification and characterization of S layer proteins from Clostridium difficile GAI 0714. J. Gen. Microbiol. 137:261–267 [DOI] [PubMed] [Google Scholar]
- 152.Takumi K, Endo Y, Koga T, Oka T, Natori Y. 1992. In vitro self-assembly of the S layer subunits from Clostridium difficile GAI 0714 into tetragonal arrays. Tokushima J. Exp. Med. 39:95–100 [PubMed] [Google Scholar]
- 153.Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, Dell A, Dougan G, Fairweather N. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187–1199 [DOI] [PubMed] [Google Scholar]
- 154.Kirby JM, Ahern H, Roberts AK, Kumar V, Freeman Z, Acharya KR, Shone CC. 2009. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284:34666–34673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dang TH, de la Riva L, Fagan RP, Storck EM, Heal WP, Janoir C, Fairweather NF, Tate EW. 2010. Chemical probes of surface layer biogenesis in Clostridium difficile. ACS Chem. Biol. 5:279–285 [DOI] [PubMed] [Google Scholar]
- 156.Cerquetti M, Molinari A, Sebastianelli A, Diociaiuti M, Petruzzelli R, Capo C, Mastrantonio P. 2000. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb. Pathog. 28:363–372 [DOI] [PubMed] [Google Scholar]
- 157.Calabi E, Calabi F, Phillips AD, Fairweather NF. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Spigaglia P, Galeotti CL, Barbanti F, Scarselli M, Van Broeck J, Mastrantonio P. 2011. The LMW surface-layer proteins of Clostridium difficile PCR ribotypes 027 and 001 share common immunogenic properties. J. Med. Microbiol. 60:1168–1173 [DOI] [PubMed] [Google Scholar]
- 159.Reynolds CB, Emerson JE, de la Riva L, Fagan RP, Fairweather NF. 2011. The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 7:e1002024. 10.1371/journal.ppat.1002024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Emerson JE, Reynolds CB, Fagan RP, Shaw HA, Goulding D, Fairweather NF. 2009. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74:541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Twine SM, Reid CW, Aubry A, McMullin DR, Fulton KM, Austin J, Logan SM. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Aubry A, Hussack G, Chen W, KuoLee R, Twine SM, Fulton KM, Foote S, Carrillo CD, Tanha J, Logan SM. 2012. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect. Immun. 80:3521–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pepin J, Valiquette L, Cossette B. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 173:1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.While A. 2006. Lessons from Stoke Mandeville. Br. J. Community Nurs. 11:406. [DOI] [PubMed] [Google Scholar]
- 166.HCUPnet 30 July 2013, accession date. Healthcare Cost and Utilization Project (HCUP). 2000, 2008. Agency for Healthcare Research and Quality, Rockville, MD. http://hcupnet.ahrq.gov/HCUPnet.jsp [PubMed]
- 167.Kuntz JL, Chrischilles EA, Pendergast JF, Herwaldt LA, Polgreen PM. 2011. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case-control study. BMC Infect. Dis. 11:194. 10.1186/1471-2334-11-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kutty PK, Woods CW, Sena AC, Benoit SR, Naggie S, Frederick J, Evans S, Engel J, McDonald LC. 2010. Risk factors for and estimated incidence of community-associated Clostridium difficile infection, North Carolina, U.S.A. Emerg. Infect. Dis. 16:197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Naggie S, Frederick J, Pien BC, Miller BA, Provenzale DT, Goldberg KC, Woods CW. 2010. Community-associated Clostridium difficile infection: experience of a Veteran Affairs medical center in southeastern U.S.A. Infection 38:297–300 [DOI] [PubMed] [Google Scholar]
- 170.Centers for Disease Control and Prevention 2005. Severe Clostridium difficile-associated disease in populations previously at low risk—four states, 2005. MMWR Morb. Mortal. Wkly. Rep. 54:1201–1205 [PubMed] [Google Scholar]
- 171.Wilcox MH, Shetty N, Fawley WN, Shemko M, Coen P, Birtles A, Cairns M, Curran MD, Dodgson KJ, Green SM, Hardy KJ, Hawkey PM, Magee JG, Sails AD, Wren MW. 2012. Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin. Infect. Dis. 55:1056–1063 [DOI] [PubMed] [Google Scholar]
- 172.Marsh JW, Arora R, Schlackman JL, Shutt KA, Curry SR, Harrison LH. 2012. Association of relapse of Clostridium difficile disease with BI/NAP1/027. J. Clin. Microbiol. 50:4078–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, Almassalha LM, Ewing SA, Ring C, Galecki AT, Rogers MA, Washer L, Newton DW, Malani PN, Young VB, Aronoff DM. 2012. Clostridium difficile ribotype does not predict severe infection. Clin. Infect. Dis. 55:1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Morgan OW, Rodrigues B, Elston T, Verlander NQ, Brown DF, Brazier J, Reacher M. 2008. Clinical severity of Clostridium difficile PCR ribotype 027: a case-case study. PLoS One 3:e1812. 10.1371/journal.pone.0001812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cloud J, Noddin L, Pressman A, Hu M, Kelly C. 2009. Clostridium difficile strain NAP-1 is not associated with severe disease in a nonepidemic setting. Clin. Gastroenterol. Hepatol. 7:868–873 [DOI] [PubMed] [Google Scholar]
- 176.Sirard S, Valiquette L, Fortier LC. 2011. Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes. J. Clin. Microbiol. 49:4040–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 178.Spigaglia P, Mastrantonio P. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45:215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Soehn F, Wagenknecht-Wiesner A, Leukel P, Kohl M, Weidmann M, von Eichel-Streiber C, Braun V. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864—implications for transcription, expression and enzymatic activity of toxins A and B. Mol. Gen. Genet. 258:222–232 [DOI] [PubMed] [Google Scholar]
- 181.Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T. 2003. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect. Immun. 71:1784–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Akerlund T, Persson I, Unemo M, Noren T, Svenungsson B, Wullt M, Burman LG. 2008. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J. Clin. Microbiol. 46:1530–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Murray R, Boyd D, Levett PN, Mulvey MR, Alfa MJ. 2009. Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of toxin B or toxin A. BMC Infect. Dis. 9:103. 10.1186/1471-2334-9-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, Buckley AM, Antunes A, Kotsanas D, Jenkin GA, Dupuy B, Rood JI, Lyras D. 2011. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog. 7:e1002317. 10.1371/journal.ppat.1002317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl. Environ. Microbiol. 78:4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bakker D, Smits WK, Kuijper EJ, Corver J. 2012. TcdC does not significantly repress toxin expression in Clostridium difficile 630DeltaErm. PLoS One 7:e43247. 10.1371/journal.pone.0043247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192:4904–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Burns DA, Heeg D, Cartman ST, Minton NP. 2011. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS One 6:e24894. 10.1371/journal.pone.0024894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8:e1002995. 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197:435–438 [DOI] [PubMed] [Google Scholar]
- 192.Rea MC, O'Sullivan O, Shanahan F, O'Toole PW, Stanton C, Ross RP, Hill C. 2012. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J. Clin. Microbiol. 50:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Skraban J, Dzeroski S, Zenko B, Mongus D, Gangl S, Rupnik M. 2013. Gut microbiota patterns associated with colonization of different Clostridium difficile ribotypes. PLoS One 8:e58005. 10.1371/journal.pone.0058005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lanis JM, Barua S, Ballard JD. 2010. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 6:e1001061. 10.1371/journal.ppat.1001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lanis JM, Hightower LD, Shen A, Ballard JD. 2012. TcdB from hypervirulent Clostridium difficile exhibits increased efficiency of autoprocessing. Mol. Microbiol. 84:66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Debast SB, van Leengoed LA, Goorhuis A, Harmanus C, Kuijper EJ, Bergwerff AA. 2009. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbiol. 11:505–511 [DOI] [PubMed] [Google Scholar]
- 199.Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. 2009. Clostridium difficile in retail meat products, U.S.A., 2007. Emerg. Infect. Dis. 15:819–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Patterson L, Wilcox MH, Fawley WN, Verlander NQ, Geoghegan L, Patel BC, Wyatt T, Smyth B. 2012. Morbidity and mortality associated with Clostridium difficile ribotype 078: a case-case study. J. Hosp. Infect. 82:125–128 [DOI] [PubMed] [Google Scholar]
- 201.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, Bergwerff AA, Dekker FW, Kuijper EJ. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47:1162–1170 [DOI] [PubMed] [Google Scholar]