Abstract
Although rabbit antibodies are widely used in research, no structures of rabbit antigen-binding fragments (Fab) have been reported. M204 is a rabbit monoclonal antibody that recognizes a generic epitope that is common to prefibrillar amyloid oligomers formed from many different amyloidogenic sequences. Amyloid oligomers are widely suspected to be a primary causative agent of pathogenesis in several age-related neurodegenerative diseases, such as Alzheimer’s disease. The detailed structure of these amyloid oligomers is not known nor is the mechanism for the recognition of the generic epitope by conformation-dependent monoclonal antibodies. As a first approach to understanding the mechanism of conformation-dependent antibody recognition, we have determined the structure of the Fab of M204 at 1.54 Å resolution. The crystal structure reveals structural features unique to rabbit Fabs, and it rules out “steric zipper” formation as a mechanism of antibody-antigen recognition. The details of the first rabbit immunoglobulin Fab structure might also be useful for exploiting the potential of rabbit monoclonal antibodies for the development of humanized rabbit antibodies as therapeutic agents.
Keywords: immunoglobulin, rabbit, IgG, amyloid, oligomer
Introduction
The accumulation of insoluble fibrils in amyloid deposits is a common characteristic of several age-dependent neurodegenerative dementias, such as AD (reviewed in1). However, recent evidence suggests the importance of soluble amyloid oligomers in the pathology of these diseases (reviewed in2). The specific immunoreactivity of conformation-dependent polyclonal antisera indicates that there are at least three general classes of amyloid aggregates that are shared by amyloids of different sequences: fibrils and fibrillar oligomers (FOs), annular protofibrils (APFs), and prefibrillar oligomers (PFOs)1. Many amyloid fibrils are known to be parallel, in-register β-sheet structures, where the peptide strands are oriented perpendicular to the fibril axis1; 3; 4; 5. FOs are defined as oligomers that react with the fibril specific antisera, OC, and appear to represent small pieces of a single fibril protofilament of fibrils that are known to have a parallel, in-register β-sheet organization of the peptide strands6; 7. Although some aggregates with fibrillar morphology are known to be antiparallel β-sheet structures, it is not yet clear whether they are recognized by OC serum. The parallel, in-register fibril structure gives rise to tracts of stacked homogeneous amino acid side chains, known as “spines”5, that run up and down the β-sheets parallel to the fibril axis. These spines mediate the stacking interaction of two β-sheets by interdigitation of the side chain tracts to form what are known as “steric zippers”5. The fibril and FO-specific polyclonal antiserum, OC, specifically recognizes a set of generic epitopes that are specific to amyloid fibrils from many unrelated amyloid-forming sequences6 and the side chain spines may constitute the generic structural features recognized by these antibodies. APFs are believed to be β-barrel structures, because they are specifically recognized by anti-APF antisera that also recognizes β-barrels formed by pore-forming toxins, such as α-hemolysin8. The structures of PFOs is not known, but it is immunologically distinct from that of fibrils and APFs6. Infrared spectroscopy data suggests that PFOs may have an antiparallel β-sheet structure9, but they may also include oligomers that have a “molten” structure characterized by hydrophobic coalescence, analogous to protein micelles10. The PFO-specific polyclonal antiserum, A11, recognizes generic epitopes specific to PFOs formed by several amyloidogenic peptides and proteins11. More recent data from monoclonal antibodies contained in the A11 serum indicate that there are several distinct conformational variants or strains of amyloid beta (Aβ) oligomers within the class of PFOs as exemplified by the rabbit monoclonal antibody, M20412. M204 recognizes high molecular weight (25–250 kDa) PFOs derived from Aβ, α-synuclein, prion (PrP) peptide 106–126, polyglutamine, and calcitonin, but it does not recognize the monomers or amyloid fibrils of these peptides and proteins12. The ambiguity regarding the structure of PFOs impedes an understanding of their relationship to fibrils, the mechanism of their toxicity and the nature of the common, generic epitopes recognized by A11. Using the specificity of antibody-antigen interactions and the crystallizability of antigen-binding fragments (Fabs), our goal is to co-crystallize PFOs with the Fabs of PFO-specific monoclonal antibodies, such as M204. As a first step to achieve this ultimate goal of co-crystallization to solve the PFO structure, we have crystallized the apo form of M204 Fab (204Fab). Here we report the crystal structure of the 204Fab at 1.54 Å resolution. Although hundreds of Fab structures and a crystal structure of rabbit immunoglobulin Fc fragments have been reported13 in the Protein Data Bank (PDB), this is the first rabbit Fab crystal structure that is deposited in the PDB. The structure identifies structural features that are unique to rabbit immunoglobulin G (IgG), in particular an interdomain disulfide bond on the light chain. The structure also rules out a simple mechanism for the recognition of generic epitopes by M204 based on the formation of steric zippers between two β-sheets. The structural insight into the unique features of rabbit Fabs may be useful for facilitating the humanization of rabbit monoclonal antibodies for therapeutic development.
Results
Overall structure of 204Fab
The 204Fab structure was determined to 1.54 Å resolution using x-ray crystallography (PDB ID: 3NL4). The crystal space group of 204Fab was determined to be C2. Table 1 shows the diffraction data collection, data processing, and refinement statistics. The final model of 204Fab was refined to Rwork and Rfree values of 19.9% and 25.8%, respectively. The asymmetric unit is a heterodimer that consists of one H and one L chain. The overall 204Fab crystal structure is shown in Figure 1 (a) and with its complementarity-determining regions (CDRs) in Figure 1 (b). The details of the Fab’s CDRs and other structural features, such as rabbit-unique disulfide bonds, will be discussed in later sections.
Table 1.
Data collection and refinement statistics of the 204Fab crystal
| Data Collection | |
| Beamline | SSRL 9-1 (Menlo Park, CA) |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.979 |
| Spacegroup | C2 |
| Cell dimensions (Å) | a=132.93 b=42.18 c=86.18 |
| Cell angles (°) | α=90.00 β=114.93 γ=90.00 |
| Mosaicity (°) | 1.50 |
| Number of molecules in asymmetric unit | 2 |
| Total number of reflections | 364,971 |
| Number of unique reflections | 56,536 |
| Resolution Range (Å) (highest resolution shell) | 31.01-1.54 (1.60-1.54) |
| Average redundancy | 6.46 (4.67) |
| Completeness (%) | 87.6 (49.7) |
| Rmerge | 0.081 (0.530) |
| Reduced χ2 | 0.96 (1.25) |
| Average I/οI | 10.2 (2.0) |
| Refinement | |
| Resolution (Å) | 31.0-1.54 |
| Atoms | |
| Protein | 3,215 |
| Ions | 5 (sulfate ion) |
| Waters | 610 |
| Mean B Value (Å2) | 18.0 |
| R/Rfreea (%) | 19.9/25.8 |
| RMSD from ideal bond lengths (Å) | 0.012 |
| RMSD from ideal bond angles (°) | 1.52 |
| Ramachandran plot analysis (PROCHECK) | 90.6% most favored regions 8.8% additionally allowed regions 0.3% generously allowed regions 0.3% disallowed regions |
calculated from a random sample containing 5% of the total number of reflections
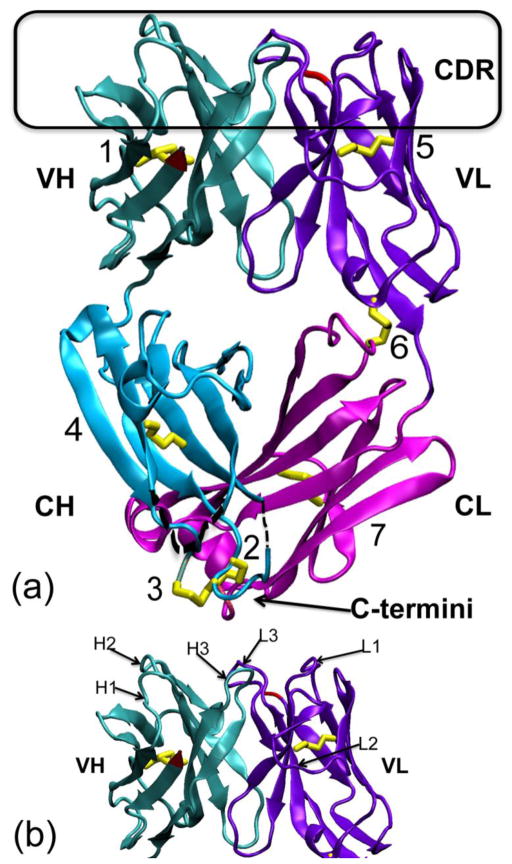
Figure 1. Crystal structure of the M204 antigen-binding fragment (Fab).
(a) The 204Fab structure (PDB ID: 3NL4) in ribbon representation. The four domains of the Fab, the variable (V) and constant (C) domains of the heavy (H) and light (L) chains, are noted as VH (teal), CH (aqua), VL (violet), and CL (magenta), respectively. The complementarity determining region (CDR) loops are boxed in black at the top of the figure. The N-termini of both chains are depicted in red in the VH and VL domain. The C-termini of the both chains are very close to each other and are shown at the bottom of the figure, noted with an arrow. The seven disulfide bonds (1. H-Cys21:H-Cys92, 2. H-Cys129:L-Cys213, 3. H-Cys130:H-Cys215, 4. H-Cys142:H-Cys195, 5. L-Cys23:L-Cys88, 6. L-Cys80:L-Cys172, and 7. L-Cys136:L-Cys195) are highlighted in yellow. Of these, the disulfide bond, rabbit unique disulfide bonds are boxed in orange on the left side and at the bottom of the figure. These disulfide bonds are shown and described in more details in Figure 2. Both alternative conformations of the disulfide bond between H-Cys21 and H-Cys92 are shown in the VH domain. Two short regions not modeled in the CH domain, from H-135 to H-136 (2 residues) and from H-189 to H-191 (3 residues), are denoted with black dotted lines. (b) The figure shows each individual CDR loop with respect to the variable domains of the H and L chains indicated as VH and VL, respectively. CDR-H1, -H2, -H3 and CDR-L1, -L2, -L3 are labeled H1, H2, H3 and L1, L2, L3, respectively.
The four domains of the Fab are noted as: variable domain, heavy chain (VH); constant domain, heavy chain (CH); variable domain, light chain (VL); and constant domain, light chain (CL). Each of these four domains consists of an immunoglobulin fold with a β-sheet sandwich. Additionally, six short helices and a total of seven disulfide bonds were observed in the Fab structure. These disulfide bonds are shown in Figure 1 (a) and described in more detail below. The H chain model consists of residues 1–134, 137–188, and 192–215. The H chain residues 135–136 and 189–191 are not included in the model due to their weak and/or discontinuous electron density. The L chain model consists of residues 1–213.
The results of a Ramachandran plot analysis using the program PROCHECK showed that the fractions of residues found in the allowed, additionally allowed, generously allowed, and disallowed regions are 90.6%, 8.8%, 0.3%, and 0.3%, respectively. Although the PROCHECK analysis indicates that the ϕ and φ angles of L-Ser30 lie in the generously allowed region, the results of the Ramachandran plot analysis of the program Coot suggests that the backbone conformation of this residue lies in the allowed region of the plot. And while the conformation of the peptide backbone for residue L-Thr51 was found in the disallowed region of the Ramachandran plots, the 2Fo-Fc electron density map of the Fab structure supports the model conformation.
Description of the conserved, interdomain, and other unusual disulfide bonds
Four of the seven disulfide bonds are conserved intradomain disulfide bonds that are commonly found in the β-sheet sandwich that makes up the immunoglobulin fold. These intradomain disulfide bonds are found among Fabs across species and will not be discussed further.
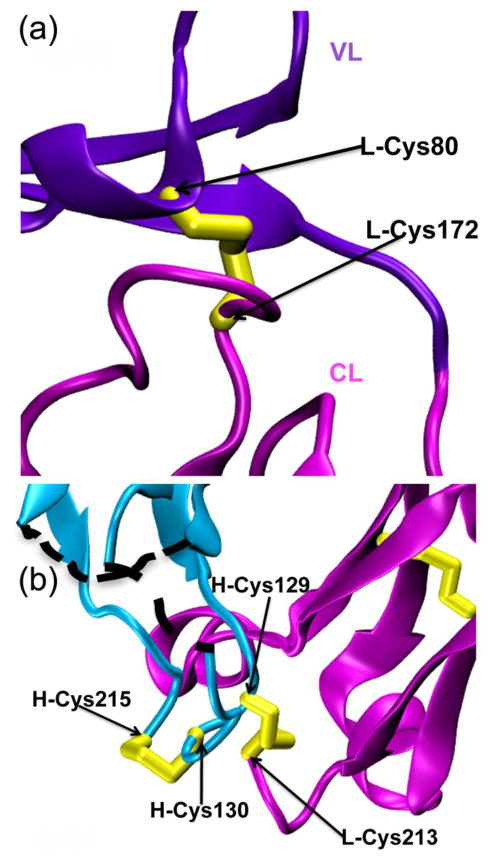
One disulfide bond of particular interest is the interdomain disulfide bond of the 204Fab L chain. This disulfide bond is an unusual feature when compared with the L chains of Fab structures from other species commonly deposited in the PDB, such as human and mouse. The interdomain disulfide bond of 204Fab is found near the interface between the VL and CL domains. The bond is formed between L-Cys80 and L-Cys172 (L-Cys170 by Kabat numbering) and connects the VL and CL domains. Each of the two cysteines is located in a loop (Figure 2 (a)). The disulfide bond has clear electron density in the 2Fo-Fc map. The distance between the two alpha carbons (Cα) of these cysteines is 5.0 Å.
Figure 2. Unique disulfide bonds on the rabbit 204Fab L chain.
(a) A rabbit-unique interdomain disulfide bond (yellow) is observed for the L chain between L-Cys80 and L-Cys172. This bond is found between the VL (violet) and CL (magenta) domains. (b) The connectivity of the disulfide bonds at the two consecutive cysteines on the 204Fab H chain is shown. Two consecutive cysteines are present in the CH domain (aqua) at H-Cys129 and H-Cys130. These residues form two disulfide bonds with the 204Fab H-and L-chain carboxyl terminus. An interchain disulfide bond is formed between H-chain (aqua) H-Cys129 and L chain (magenta) L-Cys213. An intrachain disulfide bond formed between H-Cys130 and H-Cys215. The black-dotted lines show the two disordered sections of the H chain: H-135 and H-136, and H-189 to H-191.
In the CH domain of the 204Fab, there are two consecutive cysteine residues, H-Cys129 and H-Cys130. Each of these residues forms a disulfide bond with the Fab C-terminus (Figure 2 (b)). H-Cys129 forms an interchain disulfide bond with the C-terminal residue of the L chain, L-Cys213. Disulfide bonds similar to this one have been reported for other Fab structures (i.e. PDB ID: 1FLR, between H-Cys133 and L-Cys219) as reviewed in14. The second cysteine, H-Cys130, is not usually observed among human or murine IgGs, but commonly observed in rabbit, ovine, and goat IgGs15. H-Cys130 forms an intrachain disulfide bond with the C-terminal residue of the H chain, H-Cys215. Although the connectivity of these consecutive cysteines on rabbit H-chains remained ambiguous after their identifications by sequencing, the M204 Fab structure shows the exact connectivity of these cysteines in two disulfide bonds in the structure.
Antigen-combining site
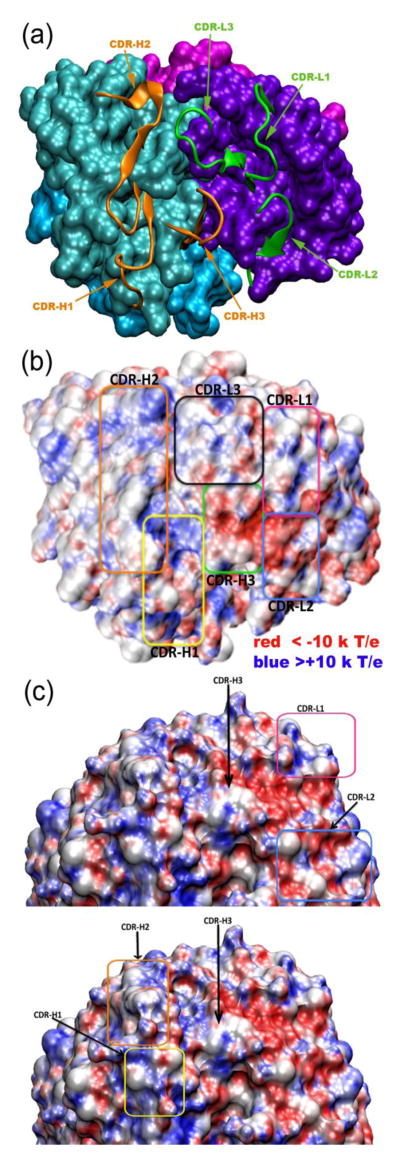
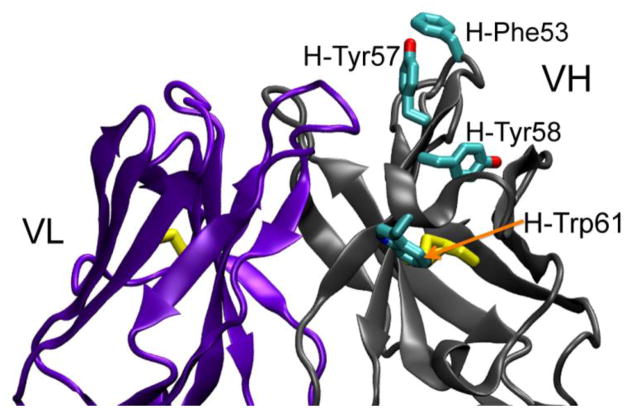
Antigen recognition by immunoglobulins occurs in the variable domain of the H and L chains. In these domains, there are six solvent-exposed loops, which are referred to as hypervariable loops (reviewed in14) due to the high sequence variability that is observed in these loops. These six loops make up the complementarity-determining regions (CDRs) of an antibody, and they play important roles in antigen recognition. There are three loops each on the H and L chains, and they are referred to as CDR-H1, -H2, and -H3 and CDR-L1, -L2, and -L3, respectively. The overall protein-backbone features of these loops of the 204Fab CDRs are shown as a cartoon representation in Figure 1 (b) and Figure 3(a). The 2Fo-Fc electron density map supports the conformations of the six 204Fab CDRs. Canonical classes of the 204Fab CDR loops, with the exception of CDR-H3, were predicted using an antibody canonical loop server available at http://bioinf.org.uk/abs/chothia.html. This server determines loop class based on the length of each CDR and several structurally important amino acid residues (also referred to as structurally determining residues (SDRs)) using two methods called SDR template and Chothia SDR methods. Based on the results of this server, the canonical class for the CDR-H1, -H2, and CDR-L2 were determined to be Class 1 while CDR-L1 was found to be similar to Class 2. However, the results showed some inconsistency in the amino acid sequences between the 204Fab CDR-L2 and those found in the representative Class 2 loops. The class for the CDR-L3 loop was found to be Class 5 in one classification, but it was undetermined using the Chothia classification system. Apparently this server did not predict the class for CDR-H3 due to higher variability in the length and amino acid sequence in the H3 loops compared to those found in other CDR loops. Figure 3(b) shows the surface features and electrostatic potential of the 204Fab CDRs. The figure depicts a groove defined by the H-chain CDRs (at the bottom of the figure). In this concave region, there are approximately ten water molecules. Using the volume of one water molecule (30 Å3) an estimate of the volume of the groove is 300 Å3. The red- and blue-colored regions correspond to the regions with negative and positive electrostatic potential, respectively. Figures 3 (b) and 3 (c) show that positive electrostatic potential lines the groove in the H-CDRs. Adjacent to this groove, H-Phe53 of CRD-H2 protrudes, exposing the hydrophobic side chain to the aqueous environment (Figure 4). In addition to H-Phe53, CDR-H2 is found to be generally rich in aromatic residues (one phenylalanine (H-Phe53), two tyrosines (H-Tyr56 and H-Tyr57), and one tryptophan (H-Trp61)). In the vicinity of the CDR-H3, -L1, and -L2 there are small regions with more negative potential than in the rest of the CDRs (Figure 3 (c)). It is also obvious from the structure of the antigen-combining site that it lacks the repetitive pattern of parallel, in-register or antiparallel β-sheets observed in amyloids5. Therefore, we can rule out steric zipper formation as a mechanism for antigen-antibody recognition for M204, an amyloid-oligomer-specific antibody.
Figure 3. Complementarity-determining regions (CDRs) of 204Fab.
(a) The six CDRs of the 204Fab, H chain (CDR-H1, -H2, and -H3) and L chain (CDR-L1, -L2, and -L3), are shown in ribbon representation in orange and green, respectively. The remainder of the four domains of the Fab (VH, VL, CH, and CL domains) is depicted in surface representation in teal, violet, aqua, and magenta, respectively. (b) Same orientation as (a): the electrostatic potential at the surface of the 204Fab CDRs. CDR-H1, -H2, and -H3 are boxed in yellow, orange, and green, respectively. CDR-L1, -L2, and -L3 are boxed in magenta, blue, and black, respectively. The surface representation of the CDRs with electrostatic potential was generated using the Adaptive Poisson-Boltzmann Solver (APBS) of the program VMD. The red- and blue-colored regions show the regions with negative (red) and positive (blue) electrostatic potential below −10 and above +10 kT/e, respectively. (c) the CDR regions shown in a vertical orientation with respect to the long axis of the Fab molecule. This figure highlights the areas in which positive and negative potential is concentrated with respect to the surrounding CDR loops.
Figure 4. The 204Fab CDR-H2 is rich in aromatic residues.
The main chains of the variable regions of the H and L chain are labeled and shown in ribbon representation in gray (changed from cyan in Figure 1 for color contrast) and violet, respectively. The two variable domains are in an orientation that is rotated 180° clockwise from Figure 1. The side chains of the H-Phe53, H-Tyr57, H-Ty58, and H-Trp61 from the CDR-H2 loop are shown in cyan in licorice representation. The yellow licorice bonds denote the interdomain disulfide bonds in each domain. The red and blue atoms indicate oxygen and nitrogen atoms, respectively.
Discussion
Despite the frequent use of rabbit immunoglobulins in laboratory experiments, there are no structures of rabbit Fabs in the PDB. Here we report a crystal structure of a rabbit Fab, 204Fab, at 1.54 Å resolution. Taken together with the structure of rabbit Fc fragments (PDB ID: 2UVO13), the structural information from 204Fab enhances our knowledge about rabbit IgG structures. In the H chain, residues 135–136 and 189–191 were excluded from the structure due to their ambiguous electron density. However, because these residues are located in the constant domain of the H chain, away from the antigen-combining site or the CDRs, they are unlikely to participate in the conformation-specific antigen recognition of 204Fab.
If rabbit IgGs, such as M204, are to be considered as potential targets for humanization, it is important to examine the consequences of the rabbit IgG unique features as seen in this Fab, such as the interdomain disulfide bond on the rabbit L chains, with regards to retaining antibody-antigen specificity. This bond is unique to rabbit IgGs with κB L chains. The presence of two extra cysteines at residues 80 and 171 has been noted previously for most of rabbit κB L chains when compared to the rabbit κα L chains and human and murine L chains16,17. The variation in the lengths of the CDR loops accounts for the difference in exact numbering of the cysteine residue in the CL domain between their study and the 204Fab structure. The presence of an interdomain disulfide bond for rabbit L chains has been inferred by the Haber group and others by amino acid sequencing combined with proteolytic fragmentations of rabbit immunoglobulins18. Additionally, the distance between the two structurally analogous residues, L-Ala79 and L-Asn172, was shown to be within a disulfide-bond-forming distance of 5.5 Å in the crystal structure of a human λ L chain 19. Because the two analogous residues in the human λ L chain structure shows that these two residues already locate close to each other, the authors predicted that the presence of an interdomain disulfide bond on the rabbit κB L chains would be similar to the other L chain structures without the interdomain disulfide bond19. Yet, the L-Cys80 and L-Cys172 disulfide bond could potentially restrict the flexibility and interdomain rotation at the elbow angle for the rabbit Fabs with κB chain. The elbow angle of a given Fab is an angle that describes the relative orientations between the constant and variable domains of this Fab with respect to the pseudo-two-fold axes that relates VL to VH, and CL to CH (CH1). The elbow angle of the 204Fab L chain was calculated to be 145° by the program RBOW20. The result of the elbow angle calculation of the 204Fab indicates that its elbow angle is smaller when compared the mean (156°) and median (150°) values reported for approximately 300 κ chain structures of different species that are available on the PDB20. The authors from this study note that 2–3° differences in elbow angles should not be considered significant due to a molecular dynamic study of Fab domains showing that their relative positions can fluctuate by this amount. There are also approximately 150 structures that either had their elbow angle equal to or less than 145° in this study, suggesting that the 204Fab elbow angle is not substantially smaller than other κ chains without interdomain disulfide bonds. These results suggest that the presence of an inter-domain disulfide bond does not necessarily lead to less flexibility at its elbow angle. This view is further strengthened by the close alignment (RMSD 1.46 Å) between 204Fab and a human-rabbit chimera Fab structure (PDB ID: 2X7L21) when these two structures were superposed using the secondary structure matching (SSM) calculations available in the program Coot. The distance between the α carbon atoms of the two analogous residues in this human-rabbit chimera Fab structure is very similar (6.12 Å between L-Ala86 and L-Ser177) to the distance measured between the two cysteine residues that form the interdomain disulfide bond. These findings suggest that the overall structure and stability of 204Fab would not be altered via chimerization of M204.
The 204Fab shows additional unusual disulfide bonds when compared to the Fab structures currently found in the PDB. These are the two separate disulfide bonds formed by the consecutive cysteine residues on in the CH domain of rabbit Fabs. The presence of these two consecutive cysteine residues near the residue 130 in the CH (CH1) domain for rabbit IgG has been previously reported by O’Donnell et al.22 The authors identified a proteolytic fragment from the rabbit CH domain containing two consecutive cysteines using diagonal peptide mapping. They also showed that one of the cysteine residues is connected to the terminal cysteine on the L chain, forming an interchain disulfide bond, while the other cysteine residue is bonded to a cysteine in the hinge region adjacent to CH1 domain, the C-terminus of a Fab, forming an intrachain disulfide bond. However, they did not know how exactly which residues are connected via disulfide bonds. The 204Fab structure shows that it is the first of the two cysteines, H-Cys129, that makes an interchain disulfide bond to the terminal cysteine on its L chain, L-Cys213, while the second one, H-Cys130, connects the terminal cysteine on its H chain at the end of the CH1 region, H-Cys215. The presence of two consecutive cysteines that are near these positions has been reported for ovine IgGs (residue 127 and 128 in the Kabat numbering)15. Similar consecutive cysteines have been also reported for the primate ε chains and Xenopus IgY H chains in their CH1 region15. Because the presence of a disulfide bond from the CH1 region to a cysteine in the hinge region is variable among different species, the bond may not have a functional purpose and evolutional importance. However, the extra disulfide bond will likely to contribute to increased stability and rigidity of the antibodies.
The CDRs of 204Fab do not reveal detailed surface characteristics that would readily explain how its antigen might bind to the antibody. This problem is complicated by the possibility that the CDRs may change conformation upon antigen binding. Figure 3(b) shows that there is one concave region surrounded by the CDR-H loops that define a groove. This groove features positive electrostatic potential. Proximal to this groove, in CDR-H3 and -L2, there are small regions of the CDRs that feature negative electrostatic potential. Haupt et al. suggest that the positively-charged electrostatic characteristics of the CDRs of B10, a camelid VH fragment that is specific to amyloid fibrils made of Aβ and other amyloidogenic peptides, may play an important role in recognizing fibrils in a conformation-dependent manner23. The authors showed that mutating the positively-charged amino acid residues to non-charged residues on the B10 CDRs decreased the VH fragment’s binding affinity to amyloid fibrils. Additionally, the binding affinity between B10 and these fibrils decreased when the Aβ and other amyloid fibrils were made from the respective peptides with their negative-charges masked using chemical modifications. Although it is not possible to rule out the contribution of electrostatics to the binding mechanism, the antigen-combining site of M204 contains fewer charged residues than B10. The lack of binding of M204 to amyloid fibrils also argues that its binding mechanism is distinct from that of B10.
If the conformations of the M204 CDRs represent the binding mode of the antibody, one possibility is that the epitope of the antigen displays the charges and shapes that are complementary to the deep groove, as outlined by the CDRs-H1, -H2, and -H3, and has both positive and negative charges located close by. In addition, the surface-exposed and protruding hydrophobic aromatic residue, H-Phe53, could play an important role in mediating hydrophobic interactions between M204 and its antigen. In addition to H-Phe53, CDR-H2 is found to be generally rich in aromatic residues (one phenylalanine, two tyrosines, and one tryptophan). Therefore, there are additional hydrophobic residues that can promote hydrophobic interactions between the antibody and its intended antigens. In a co-crystal structure (PDB ID: 1CU4) of an anti-hamster prion protein Fab called 3F4 with a peptide derived from the prion protein, a tyrosine residue, Tyr-H33 (the notation follows the publication’s notation of the residue), found in this Fab’s CDR-H1 was shown to act as a spindle to its antigen in a loop conformation24. It is possible that the 204Fab’s H-Phe53 may also flip out further from the CDRs to accommodate a turn or loop region of an antigen.
The most interesting and unusual aspect of M204 is that it recognizes an epitope that is generic to PFOs of several distinct amyloidogenic sequences12. One potential recognition mechanism that could account for the generic nature of the epitope shared among PFOs derived from different sequences is the presence of repetitive patterns on the surface of β-sheets derived from tracts of amino acid side chains running perpendicular to the strands. In the case of parallel, in-register β-sheets, identical homogeneous side chain tracts would occur anywhere the same amino acid occurs in the sequence. In the case of antiparallel, alternating register β-sheets, as have been proposed for PFO structures, the tracts would be composed of pairs of alternating amino acids9. These tracts are known to interact by interdigitation of amino acid side chains to form steric zippers during the stacking of β-sheets that occurs during amyloid fibril formation or during crystallization of short β-sheet-forming peptides5. Although this would be a simple and elegant mechanism for antigen-antibody interaction, the structure of the M204 antigen-combining site lacks this type of regular structure, thus ruling out steric zipper formation as a mechanism for antigen binding.
Another possible recognition mechanism is based on antigen-antibody interactions at the edge of a β-sheet on oligomers. Protein-protein interactions by addition of a β-strand of one molecule at the edge of a β-sheet from another protein have been described for small ubiquitin-like modifiers (SUMOs) (reviewed in25). Similar mechanisms involving β-sheets are suggested as the potential mechanism for the recognition of A11-positive, natively-folded heat shock proteins (HSP) by their computational structural similarity search results among these proteins26. The authors of this study proposed that the solvent-exposed β-strand at the edge of a β-sheet among the A11-positive, natively-folded proteins play a key role in the immunorecognition by A11 antibody. Strand-β-sheet-edge interactions have been proposed as the mechanism for the specific recognition of Aβ fibrils and oligomers by designed “gammabodies” made by grafting small, 6–7 amino acid amyloidogenic peptide segments of the Aβ sequence into the CDRs of a VH scaffold 27. These gammabodies recognize Aβ fibrils and oligomers in a sequence-specific fashion presumably by the homologous interaction of the Aβ sequence of the CDR with the same sequence at the edge of the β-sheet of the Aβ fibrils or oligomers. This homologous recognition mechanism does occur in M204 because the CDRs have no significant sequence identity to Aβ. A recent publication by Ekiert et al. reveals how a Fab can recognize β-sheet edges with varying amino acid sequence28. In this case, the sequence-independent binding is mediated by the hydrogen bonding interactions between the edge of the antigen’s β-sheet main chain and the Fab’s CDRs. Perhaps, PFO-specific monoclonal antibodies recognize various PFOs composed of different amyloidogenic peptides and proteins in a similar manner. One or more of these potential recognition mechanisms may be involved in the antibody-PFO generic epitope recognition by M204, but determining which mechanism is correct will require the structural determination of Fab-PFO co-crystals.
Materials and Methods
Production of the Fab fragments and crystallization
M204, a rabbit IgG, was expressed in human embryonic kidney 293 (293HEK) cells by Epitomics, Inc. (Burlingame, CA). Fab fragments of M204 (204Fab) were generated using immobilized papain resin (Pierce) by an overnight cleavage with shaking at 37 °C according to the manufacturer’s instructions. The Fc-containing fragments and intact IgG were removed from the papain-digested M204 samples using a Protein-A column (Pierce). The flow-through fractions from the column were dialyzed against a pH 7.5 buffer containing 20 mM HEPES and 50 mM NaCl and concentrated to 4 mg/mL. Crystallization of 204Fab was achieved using the hanging-drop, vapor-diffusion method with 1 mL well solution. Each crystallization drop consisted of 0.75 μL of the protein and 0.75 μL of the well solution. The QIAGEN PEG Suite, solution 80 (0.2 M magnesium sulfate, 20% (w/w) PEG 3350), was used to grow the crystal at 4 °C for the collection of the diffraction data.
X-ray diffraction, data processing, model building, and refinement
The 204Fab crystal was diffracted to the 1.54 Å resolution at the Beamline 9-1 at the Stanford Synchrotron Radiation Lightsource (SSRL). One crystal was used to collect 360 frames with 1° rotation per frame at 100 K. 10% glycerol was used as the cryoprotectant for this crystal. The indexing, integrating, and scaling and averaging of the collected diffraction data was performed with the program d*TREK29 (see Table 1 for the data collection and processing, and refinement statistics). For the determination of the phases of the 204Fab structure, molecular replacement was performed using the program Phaser30 in the CCP4 suite31. The search model for the heavy (H) and light (L) chains were a mouse Fab H chain (52% sequence identity, PDB ID: 1QKZ), and a rat Fab L chain (5% sequence identity, PDB ID: 1ZAN), respectively. One copy of each search model was found per an asymmetric unit. The Rwork value of the resultant 204Fab model before any refinement cycles with a 2 Å cutoff using with the program Refmac32 was 49.9%. The program Coot33 was used for model rebuilding. The programs Refmac and CNS34; 35 were used for model refinement. Water molecules were placed into unoccupied electron density that was located between 2.2–3.4 Å away from protein molecules using the programs Coot and CNS. The structural validation was conducted using the program PROCHECK36. The figures for this publication were prepared with the program VMD37 and the Adaptive Poisson-Boltzmann Solver (APBS)38 plug-in.
Accession numbers
The 204Fab structure factors and model have been deposited with the PDB under PDB ID 3NL4. The PDB IDs of other structures cited in this article are: 2UVO, 1FLR, 2XL4, 1CU4, 1QKZ, and 1ZAN.
Highlights.
First rabbit Fab crystal structure
First structure of a conformation-dependent, amyloid oligomer specific antibody
Structure shows disulfide bonds unique to rabbit IgG
The data rule out steric zipper formation as the Fab antigen-binding mechanism
Acknowledgments
This work was supported by NIH AG 033069, the Cure Alzheimer’s Fund, the Larry L. Hillblom Foundation, and the UC Irvine Center for Biomembrane Systems.
Abbreviations
- Fab
Antigen-binding fragment
- PDB
Protein Data Bank
- AD
Alzheimer’s disease
- M204
monoclonal antibody 204
- PFOs
prefibrillar oligomers
- Aβ
amyloid beta peptide
- FOs
fibrillar oligomers
- APFs
annular protofibrils
- IgG
immunoglobulin G
- HEK293 cells
human embryonic kidney 293
- VH
variable domain, heavy chain
- CH
constant domain, heavy chain
- VL
variable domain, light chain
- CL
constant domain, light chain
- CDRs
complementarity-determining regions
- RMSD
root-mean-square deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283:29639–43. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–12. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 3.Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys. 2008;41:265–97. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 4.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 5.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–7. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 6.Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, Rasool S, Gurlo T, Butler P, Glabe CG. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, Glabe C. Fibrillar Oligomers Nucleate the Oligomerization of Monomeric Amyloid {beta} but Do Not Seed Fibril Formation. J Biol Chem. 2010;285:6071–9. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kayed R, Pensalfini A, Margol L, Sokolov Y, Sarsoza F, Head E, Hall J, Glabe C. Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J Biol Chem. 2009;284:4230–7. doi: 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrene YF, Narayanaswami V, Goormaghtigh E, Ruysschaert JM, Raussens V. Antiparallel beta-sheet: a signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421:415–23. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 10.Cheon M, Chang I, Mohanty S, Luheshi LM, Dobson CM, Vendruscolo M, Favrin G. Structural reorganisation and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. PLoS Comput Biol. 2007;3:1727–38. doi: 10.1371/journal.pcbi.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–9. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 12.Kayed R, Canto I, Breydo L, Rasool S, Lukacsovich T, Wu J, Albay R, 3rd, Pensalfini A, Yeung S, Head E, Marsh JL, Glabe C. Conformation dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar abeta oligomers. Mol Neurodegener. 2010;5:57. doi: 10.1186/1750-1326-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girardi E, Holdom MD, Davies AM, Sutton BJ, Beavil AJ. The crystal structure of rabbit IgG-Fc. Biochem J. 2009;417:77–83. doi: 10.1042/BJ20081355. [DOI] [PubMed] [Google Scholar]
- 14.Nezlin RS. The immunoglobulins: structure and function. Academic Press; San Diego: 1998. [Google Scholar]
- 15.Clarkson CA, Beale D, Coadwell JW, Symons DB. Sequence of ovine Ig gamma 2 constant region heavy chain cDNA and molecular modelling of ruminant IgG isotypes. Mol Immunol. 1993;30:1195–204. doi: 10.1016/0161-5890(93)90138-2. [DOI] [PubMed] [Google Scholar]
- 16.Rejnek J, Appella E, Mage RG, Reisfeld RA. Subtypes of rabbit kappa light polypeptide chains associated with the beta locus. Biochemistry. 1969;8:2712–8. doi: 10.1021/bi00835a004. [DOI] [PubMed] [Google Scholar]
- 17.Popkov M, Mage RG, Alexander CB, Thundivalappil S, Barbas CF, 3rd, Rader C. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: the impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol. 2003;325:325–35. doi: 10.1016/s0022-2836(02)01232-9. [DOI] [PubMed] [Google Scholar]
- 18.Strosberg AD, Margolies MN, Haber E. The interdomain disulfide bond of a homogeneous rabbit pneumococcal antibody light chain. J Immunol. 1975;115:1422–4. [PubMed] [Google Scholar]
- 19.Poljak RJ, Amzel LM, Avey HP, Chen BL, Phizackerley RP, Saul F. Three-dimensional structure of the Fab’ fragment of a human immunoglobulin at 2,8-A resolution. Proc Natl Acad Sci U S A. 1973;70:3305–10. doi: 10.1073/pnas.70.12.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–74. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 21.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A. 2010;107:5810–4. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donnell IJ, Frangione B, Porter RR. The disulphide bonds of the heavy chain of rabbit immunoglobulin G. Biochem J. 1970;116:261–8. doi: 10.1042/bj1160261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haupt C, Morgado I, Kumar ST, Parthier C, Bereza M, Hortschansky P, Stubbs MT, Horn U, Fandrich M. Amyloid fibril recognition with the conformational B10 antibody fragment depends on electrostatic interactions. J Mol Biol. 2011;405:341–8. doi: 10.1016/j.jmb.2010.10.059. [DOI] [PubMed] [Google Scholar]
- 24.Kanyo ZF, Pan KM, Williamson RA, Burton DR, Prusiner SB, Fletterick RJ, Cohen FE. Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change. J Mol Biol. 1999;293:855–63. doi: 10.1006/jmbi.1999.3193. [DOI] [PubMed] [Google Scholar]
- 25.Remaut H, Waksman G. Protein-protein interaction through beta-strand addition. Trends Biochem Sci. 2006;31:436–44. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Yoshiike Y, Minai R, Matsuo Y, Chen YR, Kimura T, Takashima A. Amyloid oligomer conformation in a group of natively folded proteins. PLoS One. 2008;3:e3235. doi: 10.1371/journal.pone.0003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perchiacca JM, Ladiwala AR, Bhattacharya M, Tessier PM. Structure-based design of conformation- and sequence-specific antibodies against amyloid beta. Proc Natl Acad Sci U S A. 2012;109:84–9. doi: 10.1073/pnas.1111232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. A Highly Conserved Neutralizing Epitope on Group 2 Influenza A Viruses. Science. 2011 doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–33. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]