Summary
Vaccines are very effective at preventing infectious disease but not all recipients mount a protective immune response to vaccination. Recently, gene expression profiles of peripheral blood mononuclear cell samples in vaccinated individuals have been used to predict the development of protective immunity. However, the magnitude of change in gene expression that separates vaccine responders and non-responders is likely to be small and distributed across networks of genes, making the selection of predictive and biologically relevant genes difficult. Here we apply a new approach to predicting vaccine response based on coordinate up-regulation of sets of biologically informative genes in post vaccination gene expression profiles. We found that enrichment of gene sets related to proliferation and immunoglobulin genes accurately segregated high responders to influenza vaccination from low responders (AUC 0.94) and achieved a prediction accuracy of 88% in an independent clinical trial. Many of the genes in these gene sets would not have been identified using conventional, single-gene level approaches because of their subtle up-regulation in vaccine responders. Our results demonstrate that gene set enrichment method can capture subtle transcriptional changes and may be a generally useful approach for developing and interpreting predictive models of the human immune response.
Keywords: Systems biology, Gene Expression, Vaccine Efficacy, Immune Response, B Cell Proliferation
Introduction
Vaccination is one of the most effective methods of preventing human disease. However many vaccines are not universally protective and even widely used vaccines, such as those against influenza, fail to achieve protective immunity in a significant proportion of vaccinated subjects [1]. Identifying the biological features of the early vaccine response that predict the subsequent development of vaccine immunity is therefore a central goal in human immunology.
New investigative tools such as gene expression profiling have begun to be applied to the problem of predicting vaccine response [2]. Most of these approaches have assayed vaccine-induced changes in gene expression in the peripheral blood mononuclear cell (PBMC) compartment, a bellwether of changes at distant vaccine sites. Two studies have shown that changes in the expression of small numbers of genes in PBMC gene expression profiles a few days after vaccination predict the subsequent magnitude of the immune response measured several weeks later [3, 4]. These studies suggest that gene expression profiles from PBMC samples in vaccinated subjects can provide predictors of the vaccine response. Such approaches would be especially useful both as tools to identify new biological features associated with vaccine response, and as correlates of immunity for the development of new vaccines.
However there are two significant challenges to developing gene expression based predictors of clinical outcome following vaccination. First, the extent of biological change in PBMC caused by direct interaction with the vaccine and PBMC would be expected to be small. While live attenuated vaccines like those developed against yellow fever (YF-17D) are known to replicate systemically and induce readily detectable interferon responses [4-6], non-replicating subunit vaccines such as those against influenza would be expected to have a much smaller effect on the transcriptional profile of PBMC. Thus the selection of individual genes that are strongly associated with response to vaccination can be difficult.
The second challenge is that the biological meaning of gene expression-based predictors is often hard to determine [3, 4]. One reason for this is that the analytic approaches to identify predictive genes are often different from those used to discover biological mechanisms evident in gene expression data. Predictive genes are selected on statistical rather than biological grounds [7] which tends to divorce the identity of the predictive genes from an understanding of their role in vaccine biology [8].
To address these limitations, we applied an approach to developing predictors of vaccine outcome from PBMC gene expression profiles following vaccination that has been used in other domains, e.g., stratifying cancer patients, but is novel to immunology. Rather than building a predictive model based on single differentially expressed genes, we used sets of coordinately regulated, biologically informative gene sets as predictive features in individual samples [9, 10]. As a source of gene sets, we use a compendium of signatures extracted from the published literature and from expert curation [11]. These signatures represent phenotypes of defined cell states and biological perturbations, providing specific biological contexts with which to interpret the predictive models. Moreover, this approach allows changes in networks of genes to be used as predictive features even though the magnitude of change in any individual constituent gene is small [12].
We show that this approach enables the development of gene expression predictors from genes directly related to biological processes that a conventional single-gene level predictor does not identify. We apply this approach to pinpoint the biological hallmarks of response of two different vaccines, and show that signatures consistent with proliferating B cells predict antibody response to influenza vaccination.
Results
YF-17D induces signatures of interferon and inflammatory response
We began by analyzing PBMC microarray data from individuals vaccinated with the yellow fever virus vaccine (YF-17D). YF-17D is a highly potent vaccine that induces a robust interferon gene response in post-vaccination PBMC samples [4-6]. In this small data set, our goal was not to identify predictors of response, but rather to test whether a gene set-based analytic approach could recover known biological features of the effect of YF-17D vaccination such as the interferon response.
To identify sets of genes – rather than individual genes – that were elicited by YF-17D, we used a variant of gene set enrichment analysis (GSEA) [13]. GSEA is an analytic approach that tests for enrichment of an a priori set of genes in a second, rank-ordered list of genes. Such a rank-ordered list of genes is usually created by comparing the average expression values of genes in a group of microarray samples to those in a control group. Enrichment is measured by the degree of over representation of the set of genes of interest at the top (or bottom) of the rank ordered list. Because we wanted to test for enrichment of gene sets in individual samples from vaccinated patients (rather than in a group of samples from vaccinated subjects), we used a single sample version of GSEA (ssGSEA) [14]. In this approach, gene sets are tested for enrichment in the list of genes in a single sample ranked by absolute expression rather than by comparison with another sample.
We analyzed Affymetrix expression profiles of 15 individuals obtained pre-vaccination (Day 0) and seven days following vaccination (Day 7). We used ssGSEA to test each sample for enrichment of signatures in a compendium ∼3,000 gene sets that have been collected by curation of published microarray studies, or are present in pathway databases such as Reactome (described in Methods) [11].
We found that ∼900 gene sets were significantly (FDR < 0.25) enriched in the Day 7 post-vaccine samples (Figure 1A), suggesting marked differences in gene expression profile following vaccination with YF-17D. To identify whether the gene sets represented similar biological processes we tested the gene sets for similarity to each other using two approaches. First, we used the DAVID annotation tool [15] to categorize the genes in each gene set and found that the majority of gene sets were strongly associated with the interferon or inflammatory response (Figure 1A and Supplementary Table 1).
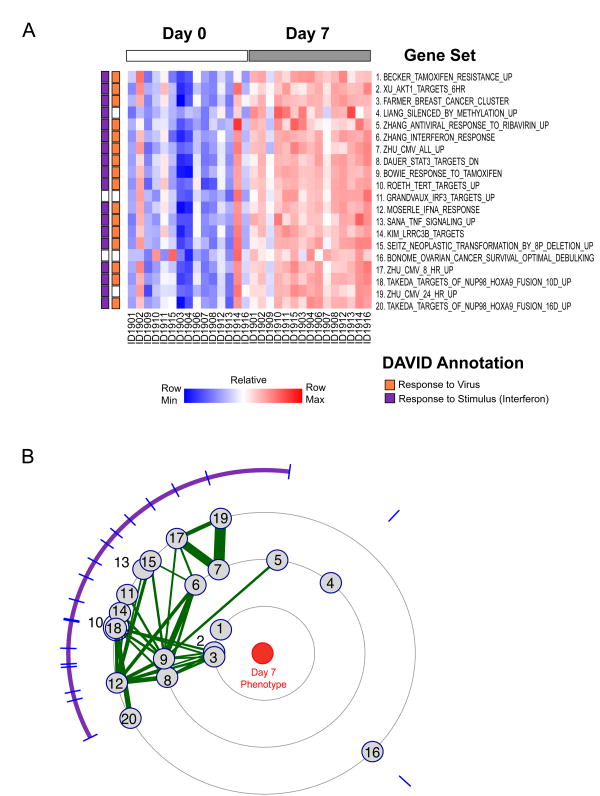
Figure 1. YF-17 vaccination induces upregulation of gene sets related to interferon response.
(A) Heatmap of the top 20 gene sets enriched in Day 7 samples compared to Day 0 samples, with color indicating ssGSEA enrichment scores for each gene set in each sample. Gene sets are ranked by the normalized mutual information score. DAVID annotations of gene sets indicated in the bar on the left; orange indicates a signature enriched for the GO term “Response to Virus”; purple “Response to Stimulus”. (B) Constellation Map of the top scoring 20 gene sets. Purple arc indicates gene-sets with overlapping features. Numbers correspond to gene-sets in (A).
Next, we developed a new visualization and analysis method – a “constellation plot” – to identify the similarity between gene sets whose enrichment correlated with a phenotype of interest (Figure 1B). In this analysis, we project each significantly enriched gene set onto a radial plot. Gene sets that are closer to the center are more enriched in samples of the phenotype of interest (Day 7, post-vaccination). Gene sets that are similar to each other in terms of enrichment patterns will be clustered closely together. To further discern similarities between the gene sets, we connected gene sets with edges whose thickness is proportional to the fraction of genes that they have in common. Groups of gene sets that both show a similar pattern of enrichment in the phenotype of interest and also share genes in common can be easily identified and are indicated by the arc on the perimeter of the radial plot.
Using this method, we found that the majority of the gene sets enriched in Day 7 samples formed a single highly connected cluster, suggesting that the top-scoring gene-sets shared a predominant biological process. (Figure 1B and Supplementary Figure 1). Analysis of the genes common to this cluster of gene sets again showed a striking over representation of interferon response genes consistent with our previous work [4]. Thus the gene sets that are correlated with Day 7 post YF-17D status are associated with a single predominant biological process – interferon response. These findings agree with the up-regulation of individual interferon response genes in response to YF-17D vaccination previously observed [4], and suggest that a gene set-based analytic approach can capture known biological features of the effect of vaccination with a live viral vaccine on PBMC.
Vaccine response to trivalent inactivated influenza vaccine (TIV) is correlated with cell proliferation and immunoglobulin gene signatures
Having validated the analytic approach in samples from subjects vaccinated with YF-17D, we next applied gene set based analysis to a more challenging problem: identifying features that predict the antibody response to the inactivated influenza vaccine.
We analyzed PBMC profiles from individuals vaccinated with the trivalent inactivated influenza vaccine (TIV) that were collected pre-vaccination (Day 0) and 7 days post vaccination [16]. HAI titers for each subject were available pre-vaccination and 28 days post vaccination and were used as the outcome measure of vaccine response. We calculated the magnitude of antibody responses to the vaccine (HAI response) as the maximum difference between the HAI titer at day 28 and the baseline titer (day 0) for any of the three influenza strains contained in the vaccine. We classified the vaccinated subjects as low or high HAI responders based on whether or not a fourfold increase in titer occurred after vaccination. This criterion was based on our prior study [16], and on the US Food and Drug Administration Guidance for Industry document for this field [17]. Using this criterion, 17 vaccinees had a high HAI response and 7 had a low HAI response.
To identify gene sets that correlated with a high HAI response, we compared the PBMC gene expression profile of each individual at Day 7 with the corresponding profile from Day 0, to create a list of genes ranked by their fold changes after vaccination for that subject. We then tested each subject's vaccine response for enrichment of gene sets from the same database collection as used before using ssGSEA and identified the gene sets most differentially enriched in the high responders compared with the low responders. We found 13 gene sets significantly associated with a high HAI response to vaccine (FDR < 0.25) (Figure 2A). The number of gene sets and degree of enrichment of gene sets correlated with TIV antibody response was lower than what we observed in the comparison of pre and post YF-17D vaccination. This suggests that the biological “signal” associated with influenza vaccine response is less pronounced than the effect of vaccination with YF-17D.
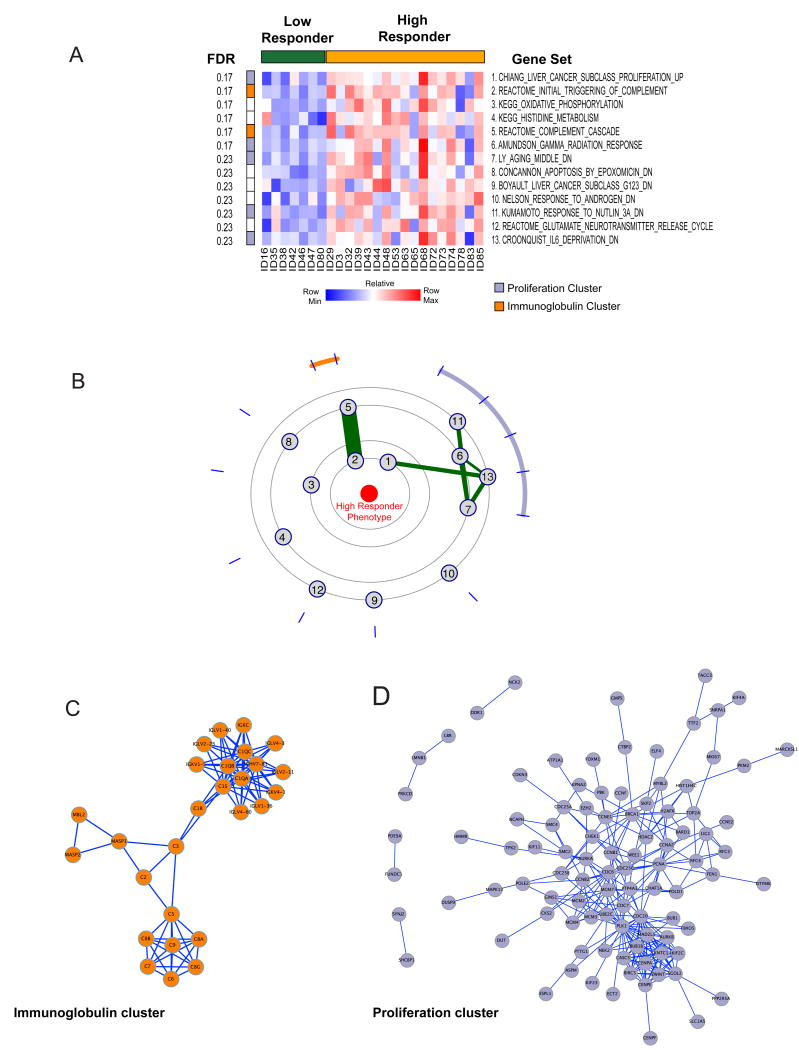
Figure 2. Antibody response to TIV correlates with enrichment of proliferation and immunoglobulin gene sets.
(A) Heatmap of the top 13 gene sets (FDR < 0.25) enriched in high responders (yellow) compared to low responders (green). Gene sets are ranked by the mutual information score. Membership of the clusters detected in the Constellation Map (B) is shown on the left of the heatmap. (B) Constellation Map of the top 13 gene sets. Two connected clusters of gene sets are detected in the constellation map, indicated by orange and lilac arcs. (C and D) Protein-Protein interaction network of two connected clusters in (B). Significant physical connectivity is shown for genes within the antibody cluster (C, orange) and proliferation cluster (D, lilac).
The gene sets that were enriched in responders were from a wide array of studies and sources (Supplementary Table 2) and the genes in most gene sets were non-redundant (Supplementary Figure 2), suggesting that the gene sets represented diverse biologies. However, using a constellation plot we found two distinct but connected clusters of gene sets (Figure 2B). We used DAVID annotation as a tool to provide secondary annotation for the two clusters of genes and found that one cluster (indicated by the orange arc) was strongly enriched for immunoglobulin and complement genes. The second cluster (indicated by the purple arc) was strongly enriched for genes associated with proliferation (Supplementary Table 3). Only a subset of proliferation-related gene sets contained in MSigDB enriched in responders (Supplementary Figure 3) suggesting that the proliferation signature present in vaccine responders is not shared by all tissue types. Alternatively other proliferation-related gene sets in the compendium may also entrain other biologies not present in vaccine responder expression profiles.
We reasoned that if these clusters of highly connected gene sets enriched in samples from vaccine responders represented bona fide biological processes, then the genes shared by each of these clusters should be over-represented for physically interacting genes. To test this, we projected the genes found in the gene set clusters into InWeb [18] a curated protein-protein interaction network (PPI; Figures 2C and D). We found that there was a high degree of physical connectivity between the component genes of the antibody gene cluster (P = 10-3), and between the genes in the proliferation cluster (P = 10-2) (Figure 2C and D). This suggests that the clusters of enriched gene sets found in responders represented coordinated up-regulation of genes in functional networks.
We confirmed these findings using a second, independent source of gene sets, described by Chaussabel et al. [19] and again found that the best-scoring module of genes was related to B cell biology, although individual modules from that collection did not score as highly as those contained in the MSigDB (Supplementary Figure 4).
We compared the performance of gene sets with their constituent genes in profiles from high versus low HAI responders to influenza vaccination. We found that the top-scoring gene sets in TIV responders were more strongly correlated with the high antibody response phenotype than any constituent gene in either gene set (Supplementary Figure 5A). Moreover, although both complement and antibody genes were present in gene sets enriching in responders, the antibody genes were among those most up-regulated (Supplementary Figure 5A and 5B). Thus a gene set-based analytic approach identifies signatures of proliferation and immunoglobulin genes that are strongly correlated with high antibody response.
Immunoglobulin and proliferation gene sets accurately predict vaccine response to TIV
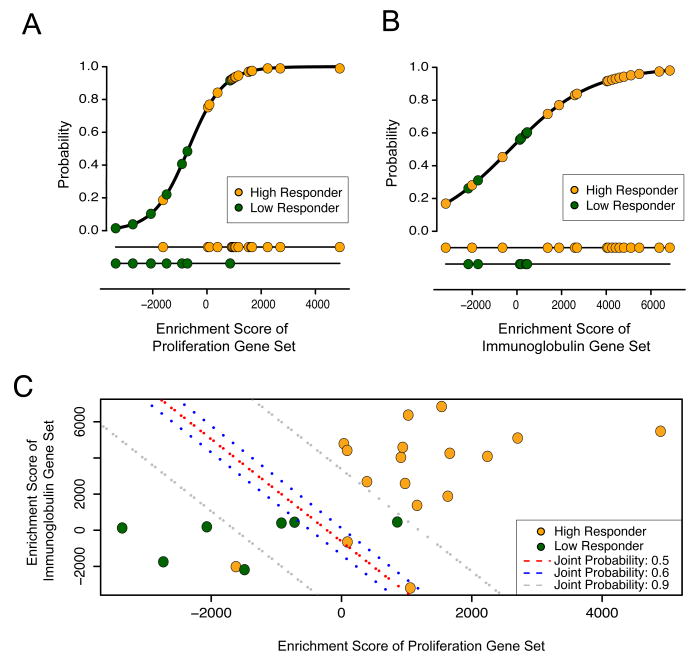
We next sought to determine if enrichment of the immunoglobulin and/or proliferation gene sets could be used as a predictor of vaccine response, using high or low HAI titers as an outcome. To do this, we selected the most differentially enriched gene set from each of the two clusters, and fitted them into logistic regression models. Both models closely fit the data and yielded an AUC of ∼0.9 (Figure 3A and B), suggesting that each independent gene set could provide a strongly predictive model of vaccine response. To integrate both biological processes into a single model, we applied Bayes' rule, and found that the integrated model achieved an AUC of 0.94 (Figure 3C).
Figure 3. Model fit of response to TIV using proliferation and immunoglobulin gene sets.
(A and B) Logistical Regression Model of probability of vaccine response for proliferation (A, CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP) and immunoglobulin (B, REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT) gene set enrichment scores. (C) Combined model using Bayes rule.
To compare our integrated gene set-based model with the single-gene level model previously described for this dataset [16], we tested our model in a validation dataset comprised of PBMC samples from an independent trial of TIV vaccination. We found that our predictive model yielded an accuracy of 88% in the test set, comparable to the performance of the single-gene level predictor [16]. This indicates that gene set-based analysis of expression profiles provide accurate predictors of response to vaccination.
Gene set-based predictors capture subtle alterations in gene expression profile
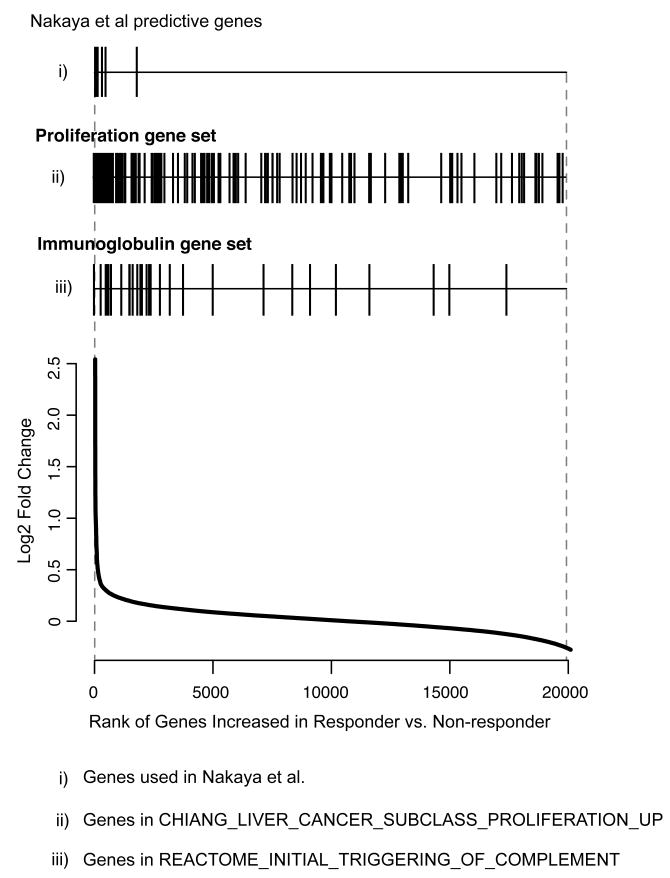
An advantage of a gene set enrichment analysis is that it can capture subtle changes in gene expression distributed across transcriptional networks. We therefore compared the degree of differential expression of genes in the predictive gene sets (proliferation and immunoglobulin gene sets) with that of the genes selected in the single-gene level predictor originally applied to this dataset (Figure 4). Predictive genes selected in the study by Nakaya et al. [16] were all highly differentially expressed in Day 7 PBMC expression profiles from responders compared to non-responders, as expected (mean fold change 3.36). In contrast, the gene sets identified in our analysis included many genes that were much less differentially expressed (mean fold change of proliferation cluster 2.13; mean fold change of immunoglobulin cluster 2.53) (Figure 4).
Figure 4. Predictive gene sets capture genes with subtle changes in expression.
Rank of genes identified in a single-gene predictor of TIV response (i), compared to the rank of genes contained in the proliferation (ii) and immunoglobulin (iii) gene-sets. Each gene indicated by a vertical line and its relative rank on the list of differentially expressed genes comparing TIV responders compared to non-responders indicated by the line graph below.
Although the genes in the B cell and proliferation gene sets were enriched in respsonders, the majority of their constituent genes were not individually identified as significantly up-regulated in TIV responders in the previously published predictor (Figure 4). Indeed analysis of the functional annotations of genes in the previously published single-gene level predictor of influenza vaccine response [16] did not include terms related to B cell biology or proliferation (Supplementary Table 4). Thus a gene-set based approach can identify networks of predictive genes and biologies not otherwise detected by conventional, single-gene level approaches.
The frequency of antibody producing cells correlates with the proliferation and antibody clusters
The simplest explanation for the predictive power of gene sets containing proliferation and antibody genes in individuals with high HAI response to vaccination is that it represents the increased frequency of proliferating B cells in post-vaccination samples. To test this hypothesis, we compared the frequency of antibody-producing B cells in the peripheral blood of vaccinated subjects at Day 7 post vaccination with the enrichment score for the top scoring proliferation and immunoglobulin clusters.
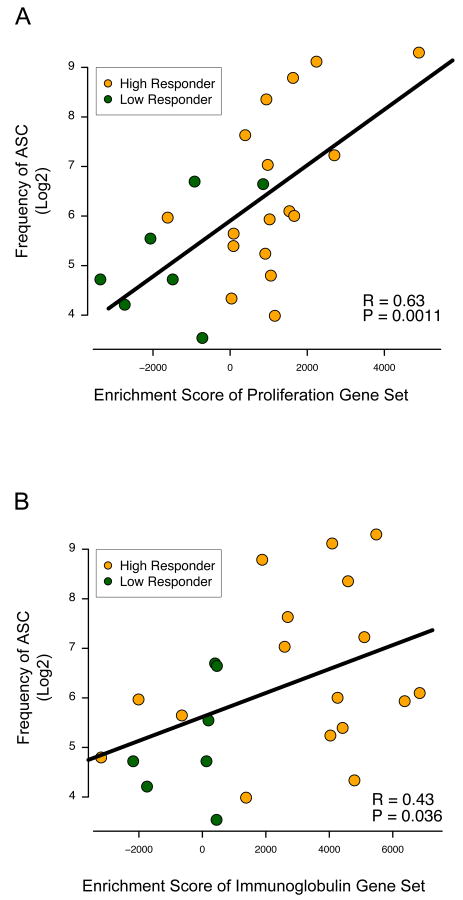
We found that the enrichment score of both gene sets was correlated significantly with the frequency of IgG antibody spot-forming cells (ASC; Figure 5) but not IgM or IgA (data not shown). This is most consistent with the interpretation that enrichment of these gene sets was caused by increased representation of proliferating plasmablasts in PBMC samples from vaccinated subjects with high antibody responses.
Figure 5. Enrichment of proliferation and immunoglobulin gene sets correlate with the frequency of antibody spot forming cells.
(A and B) Enrichment scores of the proliferation gene set (A, CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP) and immunoglobulin gene set (B, REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT and frequency of IgG secreting cells (ASC). Significance is calculated by comparison to the null distribution, calculated by correlations derived from random gene sets.
Discussion
In this study, we applied a gene set enrichment-based approach to developing predictors of vaccine outcome and showed that enrichment of signatures corresponding to proliferating B cells accurately segregate vaccine responders to TIV with an AUC of 0.94 in a training set and an accuracy of 88% in an independent clinical trial. Our approach uses the differential enrichment of sets of biologically related genes rather than single genes as predictive features. This allows subtle biological changes manifest over networks of genes to be captured in a way that conventional gene expression predictors do not because they focus on small numbers of highly differentially expressed genes.
Rapid expansion of plasmablasts following influenza vaccination has been previously observed [20], and it is intuitive that the magnitude of the plasmablast response would correlate with the humoral response to vaccination. However even at their peak, proliferating plasmablasts represent only a tiny fraction of the cells present in the PBMC samples analyzed by microarray in this study. As result, although detailed analysis of gene expression data from influenza vaccinated subjects had revealed that genes related to B cell biology were related to the HAI response, the magnitude of change in these B cell genes was not sufficiently large for them to be incorporated into the previously published gene expression predictor [16]. In contrast, our approach allows subtle changes in sets of coordinately expressed genes related to B cell biology and proliferation to function as a predictive model of vaccine outcome.
Gene set enrichment analysis is ideally suited to identifying small but coordinated changes in gene expression in sets of biologically related genes [13, 21]. It has been used to identify biological processes such as metabolic changes [21] and signaling flux [22] that are evident across networks of genes but subtle at the level of individual gene expression. The ability to build predictive models from small but coordinated changes in transcriptional programs is particularly important for clinical applications such as the detection of a vaccine response in which the transcriptional signal in responders compared to non-responders is small. We therefore anticipate that this approach to gene expression predictor development will be generally useful in clinical situations in which the difference in gene expression between outcome classes is limited. Future studies will be able to use this approach to test whether analogous enrichment of B cell and proliferation signatures are characteristic of vaccine response in different vaccines. Alternatively, analysis of different vaccines and in larger cohorts may be able to identify different gene sets representing other biological processes that underlie vaccine response.
An advantage of gene set-based predictors is that their biological meaning is more transparent. While predictive features based on individual genes may contain important, novel information about the vaccine response, their mechanistic basis is not always obvious without additional experimental inquiry [4, 16]. Instead, we developed our predictive model from a library of well-annotated signatures derived from previously published microarray experiments and expert curation. Together with a novel analysis and visualization method – the constellation plot (Figures 1 and 2) – this allowed the predominant biological themes that correlated with vaccination response to be readily identified. We also anticipate that in addition to vaccine response, this approach may also be useful for identifying subtle features that vary across a group of responders, allowing the heterogeneity that is part of all human studies to be better interrogated. Moreover, the use of gene set-based classifiers may also prove useful in features predictive of adverse effects to vaccines.
A theoretical concern with our method is that the biological processes involved in the vaccine response may not be represented in the compendium of signatures currently used in the analysis. However, our results suggest that at least some of the biological signatures that predict vaccine response – such as proliferation – are already present in the database of signatures used for this study. Moreover, because the method we used can draw on any collection of annotated gene sets, it can easily be extended to additional collections of gene sets. For instance, we and others are developing libraries of modules or gene-sets specifically devoted to cellular states and perturbations in the immune system [19, 23, 24] which should increase the biological resolution of gene set predictors even further.
Finally, knowledge-driven gene expression-based predictors can be translated assays that are simpler and more robust than measurement of transcript abundance for many genes. Gene expression predictors have historically been limited by a lack of reproducibility between experiments [10, 25]. This is thought to be related to the high variance of individual gene measurements commonly seen in datasets of relatively few replicates. This variance results in discordance between lists of predictive genes even in high quality experiments. Using a larger set of genes rather than a small number of genes may provide some degree of robustness lacking in single gene-level predictors. Indeed several platforms have now been developed [26, 27] that allow focused sets of genes to be profiled at high throughput and low cost. Moreover, because gene set-based predictors can identify not just predictive genes but predictive biologies this approach could overcome the limits of predicting clinical responses by measuring gene expression. For instance, our analysis shows that signatures associated with cellular proliferation are predictive of a protective antibody response. It would be relatively easy to translate this to a flow-cytometry based assay of cellular proliferation in PBMC using Ki67 staining, for example, that could rapidly be applied to many samples. In contrast, developing and validating a multi-gene predictive signature of unknown biological significance may prove to be more significantly more complex. Future studies will be required to determine how successfully biologies discovered by gene set-based approaches can be deployed as simpler, more robust diagnostic tools. Gene set-based predictors predicated on biological knowledge may therefore provide a sensitive, relevant and robust analysis of the human immune response.
Methods
Data Preprocessing
We analyzed two existing datasets of gene expression profiles of PBMC from vaccinated subjects: raw Affymetrix array data for subjects vaccinated with YF-17D from Gene Expression Omnibus (GEO) with the accession number GSE13486 [4], and raw Affymetrix array data from subjects vaccinated with influenza TIV with accession number GSE29619 [16]. The Genepattern module “CollapseDataset” was used to extract the expression values of genes from the raw data file and to map Affymetrix probes to gene symbols [28]. Then we applied quantile normalization and a log2 transformation. The final transformed data were used for the single sample GSEA projection (see below). For analysis of data from the influenza vaccinated subjects, gene expression fold change was calculated as the ratio of expression levels from PBMC profiles Day 7 (post-vaccine) / Day 0 (pre-vaccine).
Single Sample Gene Set Enrichment Analysis (ssGSEA)
GSEA yields a quantitative measure of the over representation of a set of genes S (e.g., genes encoding products in a same metabolic pathway) at the top or bottom of a ranked list of genes L. Candidate genes are ranked by their differential expression between two phenotypes. The statistic is a weighted Kolmogorov-Smirnov-like statistic and significance is calculated using an empirical permutation test [13]. Here we applied an extended version of conventional GSEA in order to produce an enrichment score in a single sample as we have previously [14]. Such a score is necessary if one is to make a predictive call on single samples without reference to a larger group of samples. In this approach, the genes are ordered based on either absolute expression (as in the yellow fever vaccine study) or the relative changes with respect to the baseline level (as in the influenza TIV vaccine study).
In this study, we used C2 collection from Molecular Signature Database (MsigDB). The MsigDB is a public available database of annotated gene sets hosted at Broad Institute (http://www.broadinstitute.org/gsea/msigdb/index.jsp) [11]. Currently there are 6 major collections from C1 to C6 while C2 is a special collection of gene sets carefully curated from online pathway databases, publications in PubMed, and knowledge of domain experts. Each of the ∼3000 gene sets in C2 collection is well described in the MsigDB website including the source, annotation as well as other useful information, thus facilitate the interpretation of the biological meaning associated with it.
Gene Set Feature Selection
To detect gene sets whose enrichment scores are highly correlated with phenotypes, we used an Normalized Mutual Information (NMI) score (Eq. [3]) to evaluate the association between phenotypes (Day 7 vs. Day 0 in the yellow fever vaccine study; or high vs. low HAI antibody response in the influenza TIV vaccine study) and gene set enrichment scores.
| [1] |
| [2] |
| [3] |
| [4] |
Using mutual information to detect association is advantageous because it does not require assumptions about the distribution of samples (whereas e.g., t-statistic assumes a normal distribution of samples) and it allows the detection of non-linear associations (whereas Pearson correlation can only detect linear associations). Although the NMI scores do not inherently indicate positive or negative correlation, we used the sign of the Pearson correlation to decide the direction of the association and focus on positive associations in this study (Eq. [4]).
Constellation Plot
The constellation plot is designed to visualize and thus to elucidate groups of gene sets enriched in a phenotype of interest (e.g., vaccine response) that correspond to distinct biological processes. We reasoned that gene sets that i) demonstrate high mutual information with respect to the phenotype; ii) demonstrate high mutual information with respect to each other; and iii) share overlapping member genes would be likely to reflect similar biological processes. We estimated similarities between N gene sets using a NMI score and further transformed it into a dissimilarity score, d = 1 – NMI. Previous studies [29] have proved that this dissimilarity metric has all the properties of a true mathematical distance (metric), allowing us to represent the association of gene sets with a proper distance matrix D.
We visualized this distance matrix D as a radial plot in which the angle between two gene sets represents the distance d between them, and their proximity to the center reflects the their differential enrichment with respect to the phenotype (1 - NMI). The radial plot angular distribution is computed using a circular multidimensional scaling projection, specifically the angular distance matrix Δ is obtained by minimize the objective function (Eq. [5]), where δij is the angular distance between gene sets i and j in the radial plot, while dij is the original distance stored in D.
| [5] |
The radial plot angular distribution is computed using the R package “SMACOF”, version 1.2-1 [30]. Using this plot it is easy to detect clusters of gene sets based on enrichment pattern similarities. To further facilitate the interpretation of clusters, we connected the gene sets by edges whose thickness indicates the Jaccard index between them. The Jaccard index is equal to the number of genes shared by two sets divided by the number of genes in their union and here we used 0.1 as the lower boundary to include an edge [31]. Connected clusters of gene sets can then be extracted and interpreted based on their constituent genes. All the details about the number of genes in each gene set and heat map of Jaccard index of each pair of gene sets are shown in the Supplementary Table 1 and 2 and Supplementary Figure 1 and 2.
Protein-Protein Interaction (PPI) Construction and Module Detection
We constructed the PPI network based on the InWeb database [18]. We identified the modules of the PPI network using the “FastCommunityMH” software package, a simulated annealing algorithm that optimizes the modularity of the network [32]. Here modularity measures the ratio between number of edges within modules and the number of edges between modules. The optimized modularity indicates the best partition of the network that there are many edges within modules and only few between them.
General Linear Model and Bayes' Rule
We first built two logistic regression models using the best scoring gene sets from each of the two identified clusters of differentially enriched gene sets in TIV responders. The outcome of the logistic regression model is the probability that a sample belongs to the high response group given the enrichment score. We further combined the probabilities from these two models using Bayes' rule as follows: for sample x with enrichment scores Ex1 and Ex2 for the gene sets used in the logistic regression model above and with corresponding probability of belonging to the high response group H, P(H | Ex1) and P(H | Ex2), we calculate the likelihood ratio that x belonging to the high response group as shown in Eq. [6]. To validate the combined model, we used a dataset of PBMC gene expression profiles from a second, independent trial to evaluate the predictive accuracy. The second trial (2007-2008 trial) was also used as a validation data set in the study by Nakaya et al.[16] which consisted of 9 subjects vaccinated with TIV in the previous year.
| [6] |
Supplementary Material
Supplementary Figure 1. Jaccard index of highly enriched gene sets in samples 7 days post-vaccination of YF-17D. Heatmap of Jaccard index of top 20 gene sets enriched in the PBMC samples 7 days after vaccination.
Supplementary Figure 2. Jaccard index of highly enriched gene sets in high responders to TIV. Heatmap of Jaccard index of top 13 gene sets enriched in the PBMC samples of high responders comparing to low responders 7 days after vaccination.
Supplementary Figure 3. Mutual information score and FDRs of all the proliferation-related gene sets. All the gene sets that are related to proliferation (based on DAVID annotation) were identified in MsigDB C2 collection. Gene sets are ranked based on their mutual information score with respect to high responders from left to right. A bar graph of 1 – FDR is shown on top of the heatmap of mutual information. Orange bars represent gene sets in the proliferation cluster of constellation map, blue bars represent other gene sets.
Supplementary Figure 4. The best-scoring Chaussabel module of genes is related to B cell biology. Heatmap of the enrichment of Chaussabel modules in high responders (yellow) compared to low responders (green). Modules of genes are ranked by the NMI score and the best scored module (module M1.1) is related to B cell biology. The modules are annotated based on the keyword selection proposed by Chaussabel et al. and the full annotation and interpretation can be found in [19].
Supplementary Figure 5. Proteins encoded by genes in each cluster share a strong physical connectivity. A) Heatmap of the gene sets in the immunoglobulin cluster and their constituent genes. Gene sets and genes are ranked based on the NMI score. B) The protein-protein interaction network of constituent genes. Two modules are detected. The cyan module is composed of antibody genes while the orange module contains the complement component genes. Color bars in (A) represents the module membership of genes and it shows that most of the antibody related genes have higher NMI score.
Supplementary Table 1. Top 20 Gene Sets Enriched in PBMC Samples 7 Days Post-vaccination of YF-17D
Supplementary Table 2. Top 13 Gene Sets Enriched in PBMC Samples from Responders to TIV
Supplementary Table 3. Functional Annotations of Genes in Two Clusters of Gene Sets
Supplementary Table 4. Functional Annotations of Genes in Immunoglobulin Gene Set, Proliferation Gene Set and Nakaya et al. Predictive Genes
Acknowledgments
Supported by R01AI091493 to W.N.H.; U19AI090023 to B.P. and W.N.H, by an Infrastructure and Opportunity Fund Grant from the Human Immune Phenotyping Consortium to W.N.H. and J.M.; and by the Bill and Melinda Gates Foundation OPP50092 to J.M.
References
- 1.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nature Immunology. 2011;131:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley interdisciplinary reviews Systems biology and medicine. 2011 doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature Immunology. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaucher D, Therrien R, Kettaf N, Angermann B, Boucher G, Filali-Mouhim A, Moser J. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. The Journal of Experimental Medicine. 2008 doi: 10.1084/jem.20082292. jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. The Journal of Experimental Medicine. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon R. Roadmap for Developing and Validating Therapeutically Relevant Genomic Classifiers. Journal of Clinical Oncology. 2005;23:7332. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 8.Haining WN, Pulendran B. Identifying gnostic predictors of the vaccine response. Current Opinion in Immunology. 2012:1–5. doi: 10.1016/j.coi.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular Systems Biology. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamayo P, Cho YJ, Tsherniak A, Greulich H, Ambrogio L, Schouten-van Meeteren N, Zhou T. Predicting relapse in patients with medulloblastoma by integrating evidence from clinical and genomic features. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1415–1423. doi: 10.1200/JCO.2010.28.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4:R60. [PubMed] [Google Scholar]
- 16.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.US Department of Health and Human Services Food and Drug Administration Center for Biologics Evaluation and Research. Guidance for Industry: Clinical Data Needed to Support the Licensure of Pandemic Influenza Vaccines. Office of Communication, Training and Manufactures Assistance; Rockville, Maryland: 2007. [Google Scholar]
- 18.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha V, Lindgren C, Eriksson K, Subramanian A, Sihag S, Lehar J, Puigserver P. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 22.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nature medicine. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heng TSP, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D. The Immunological Genome Project: networks of gene expression in immune cells. Nature Immunology. 2008;9:1091. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 24.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T. Cell type-specific gene expression differences in complex tissues. Nature methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nature biotechnology. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haining WN, Angelosanto J, Brosnahan K, Ross K, Hahn C, Russell K, Drury L. High-throughput gene expression profiling of memory differentiation in primary human T cells. BMC immunology. 2008;9:44. doi: 10.1186/1471-2172-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amit I, Regev A, Hacohen N. Strategies to discover regulatory circuits of the mammalian immune system. Nature reviews Immunology. 2011;11:873–880. doi: 10.1038/nri3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nature genetics. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 29.Vinh NX, Epps J, Bailey J. Information Theoretic Measures for Clusterings Comparison: Variants, Properties, Normalization and Correction for Chance. Journal of Machine Learning Research. 2010;11 [Google Scholar]
- 30.Leeuw Jd, Mair P. Multidimensional Scaling Using Majorization: SMACOF in R. Journal of Statistical Software. 2009;31:1–30. [Google Scholar]
- 31.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Physical Review E. 2004;70 doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Jaccard index of highly enriched gene sets in samples 7 days post-vaccination of YF-17D. Heatmap of Jaccard index of top 20 gene sets enriched in the PBMC samples 7 days after vaccination.
Supplementary Figure 2. Jaccard index of highly enriched gene sets in high responders to TIV. Heatmap of Jaccard index of top 13 gene sets enriched in the PBMC samples of high responders comparing to low responders 7 days after vaccination.
Supplementary Figure 3. Mutual information score and FDRs of all the proliferation-related gene sets. All the gene sets that are related to proliferation (based on DAVID annotation) were identified in MsigDB C2 collection. Gene sets are ranked based on their mutual information score with respect to high responders from left to right. A bar graph of 1 – FDR is shown on top of the heatmap of mutual information. Orange bars represent gene sets in the proliferation cluster of constellation map, blue bars represent other gene sets.
Supplementary Figure 4. The best-scoring Chaussabel module of genes is related to B cell biology. Heatmap of the enrichment of Chaussabel modules in high responders (yellow) compared to low responders (green). Modules of genes are ranked by the NMI score and the best scored module (module M1.1) is related to B cell biology. The modules are annotated based on the keyword selection proposed by Chaussabel et al. and the full annotation and interpretation can be found in [19].
Supplementary Figure 5. Proteins encoded by genes in each cluster share a strong physical connectivity. A) Heatmap of the gene sets in the immunoglobulin cluster and their constituent genes. Gene sets and genes are ranked based on the NMI score. B) The protein-protein interaction network of constituent genes. Two modules are detected. The cyan module is composed of antibody genes while the orange module contains the complement component genes. Color bars in (A) represents the module membership of genes and it shows that most of the antibody related genes have higher NMI score.
Supplementary Table 1. Top 20 Gene Sets Enriched in PBMC Samples 7 Days Post-vaccination of YF-17D
Supplementary Table 2. Top 13 Gene Sets Enriched in PBMC Samples from Responders to TIV
Supplementary Table 3. Functional Annotations of Genes in Two Clusters of Gene Sets
Supplementary Table 4. Functional Annotations of Genes in Immunoglobulin Gene Set, Proliferation Gene Set and Nakaya et al. Predictive Genes