Abstract
The Gram-positive soil bacterium Bacillus subtilis is able to choose between motile and sessile lifestyles. The sessile way of life, also referred to as biofilm, depends on the formation of an extracellular polysaccharide matrix and some extracellular proteins. Moreover, a significant proportion of cells in a biofilm form spores. The first two genes of the 15-gene operon for extracellular polysaccharide synthesis, epsA and epsB, encode a putative transmembrane modulator protein and a putative protein tyrosine kinase, respectively, with similarity to the TkmA/PtkA modulator/kinase couple. Here we show that the putative kinase EpsB is required for the formation of structured biofilms. However, an epsB mutant is still able to form biofilms. As shown previously, a ptkA mutant is also partially defective in biofilm formation, but this defect is related to spore formation in the biofilm. The absence of both kinases resulted in a complete loss of biofilm formation. Thus, EpsB and PtkA fulfil complementary functions in biofilm formation. The activity of bacterial protein tyrosine kinases depends on their interaction with modulator proteins. Our results demonstrate the specific interaction between the putative kinase EpsB and its modulator protein EpsA and suggest that EpsB activity is stimulated by its modulator EpsA.
Introduction
Like many other bacteria, the Gram-positive model organism Bacillus subtilis is able to form structured biofilms in order to attach to surfaces and to withstand harsh environmental conditions. Biofilm formation in Bacillus subtilis is usually observed as the formation of either structured complex colonies on solid surfaces or thick floating pellicles in liquid media (Branda et al., 2001). Bacillus subtilis cells in the biofilm form an extracellular matrix composed of polysaccharides, amyloid-like fibres made up of the TasA protein, and a water-repellent surface coat formed by the bacterial hydrophobin BslA (see Vlamakis et al., 2013 for a review). Interestingly, biofilm formation is strongly impaired in the domesticated laboratory strain of Bacillus subtilis, 168. This strain forms only poorly structured colonies and thinner and non-structured yet stable pellicles. Such a loss of phenotype, which is important in natural environments but not in the laboratory, is commonly observed in bacteria. Reduced biofilm formation in the Bacillus subtilis laboratory strain results from the presence of point mutations in the sfp, swrAA, degQ and epsC genes and from the absence of a plasmid that harbours the rapP gene encoding a response regulator aspartate phosphatase. Introduction of wild-type copies of these genes restores biofilm formation in the domesticated laboratory strain (McLoon et al., 2011).
Expression of the epsA-O operon encoding the enzymes for extracellular polysaccharide synthesis and of the tapA–sipW–tasA operon encoding the amyloid-like protein and the factors for its export and assembly is under tight control by the RemA transcription activator and the SinR repressor protein (Chu et al., 2006; Winkelman et al., 2013). The DNA-binding and thus repressing activity of SinR is controlled by regulatory protein–protein interactions with one of three antagonist proteins: SinI, SlrA and SlrR (Chai et al., 2009, 2010; Lewis et al., 1998; Newman et al., 2013). The interaction between SinR and SlrR seems to be of key importance as the complex not only prevents the repression of biofilm gene expression but also acts as a transcriptional repressor for motility and autolysis genes. This mechanism ensures that a single cell of Bacillus subtilis has to choose to express either biofilm or motility genes but not both sets (Chai et al., 2010). The choice of cell fate is also affected by the recently discovered YmdB protein, which is essential for the expression of biofilm genes (Diethmaier et al., 2011, 2014).
While the functions of the proteins of the tapA–sipW–tasA operon have been studied in detail, far less is known about the individual functions of the proteins encoded by the 15-gene epsA-O operon. For the EpsE protein, it was shown that it is required for extracellular polysaccharide synthesis, but that it also inhibits motility by separating the cytoplasmic FliG motor from the MotA–MotB stator (Blair et al., 2008; Guttenplan et al., 2010). The proteins encoded by the two promoter-proximal genes of the eps operon, EpsA and EpsB, are similar to bacterial protein tyrosine kinases (BY-kinases). Interestingly, these BY-kinases are present as single multidomain proteins in Gram-negative bacteria, whereas the kinase and its dedicated transmembrane modulator are separated proteins in Gram-positive bacteria (Grangeasse et al., 2012).
Bacillus subtilis encodes two potential pairs of BY-kinases (PtkA and EpsB) and their modulators (TkmA and EpsA, respectively). While the activity of PtkA/TkmA has been intensively studied, nothing is known about the function(s) of EpsB and its modulator EpsA. For PtkA, it has been established that the kinase phosphorylates and thereby activates the UDP-glucose dehydrogenases Ugd and TuaD as well as the single-stranded DNA-binding proteins SsbA and SsbB (Mijakovic et al., 2003; Petranovic et al., 2007). In addition, PtkA-dependent phosphorylation decreases the DNA-binding activity of the transcription repressor FatR (Derouiche et al., 2013). Moreover, PtkA-dependent protein phosphorylation was proposed to affect protein localization, as observed for the glycolytic enzyme enolase (Jers et al., 2010). Interestingly, the BY-kinase PtkA is attached to its membrane modulator during vegetative growth whereas it is associated with its phosphorylated cytosolic target proteins in the stationary phase. Thus, the localization of PtkA is highly dynamic (Jers et al., 2010).
The clustering of BY-kinases with genes required for extracellular polysaccharide matrix and/or capsule production is well established in a variety of bacteria such as Escherichia coli, Staphylococcus aureus and Streptococcus pneumoniae (Bechet et al., 2010; Morona et al., 2003; Soulat et al., 2007; Wugeditsch et al., 2001). Although the precise function of the BY-kinase in extracellular polysaccharide synthesis remains elusive, it is generally assumed that autophosphorylation of the BY-kinases is required for the export of polysaccharides (Bechet et al., 2010; Whitfield, 2006).
Recently, we have shown that protein tyrosine phosphorylation is also implicated in biofilm formation in Bacillus subtilis (Kiley & Stanley-Wall, 2010). In the absence of the BY-kinase PtkA, the complex colonies are strongly wrinkled, but lack the rough outer region of the colony where usually the fruiting bodies are formed. This phenotype was attributed to the kinase activity of PtkA that is required for efficient sporulation under conditions of biofilm formation. In this study, we have investigated the potential role of the putative BY-kinase EpsB and its transmembrane modulator protein EpsA. Based on the similarity of EpsB and PtkA and their presumptive modulators, we have also investigated whether these proteins act in concert to control biofilm formation. Our results indicate that EpsB and PtkA fulfil distinct but additive functions in biofilm formation and that they require their cognate modulators for this.
Methods
Bacterial strains and growth conditions.
All Bacillus subtilis strains used in this work are derived from the laboratory wild-type strain 168 or the non-domesticated strain NCIB3610. Mutations were transferred to the NCIB3610 background using SPP1-mediated generalized transduction (Yasbin & Young, 1974). All strains are listed in Table 1. E. coli XL1-Blue (Stratagene) was used for plasmid constructions and transformation using standard techniques (Sambrook et al., 1989).
Table 1. Bacillus subtilis strains used in this study.
| Strain | Genotype | Source/construction†‡ |
| 168 | trpC2 | Laboratory collection |
| 8G5 sinR : : tet | trpC2 tyr-1 his ade met rib ura nic sinR : : tet | Oscar Kuipers, Groningen, the Netherlands |
| AM373 | sfp+ ermC epsC+ swrA+ degQ+ amyE : : (P-rapP phrP cat) (yvzG/yvyD spc) | McLoon et al. (2011) |
| GP736 | trpC2 ΔsinR : : tet | 8G5 sinR : : tet → 168 |
| GP1517 | trpC2 ΔepsA : : aphA3 | this work |
| GP1518 | trpC2 ΔepsB : : aphA3 | this work |
| GP1519 | trpC2 ΔepsAB : : aphA3 | this work |
| GP1526 | trpC2 epsA-3x FLAG spc | pGP2127 → 168 |
| GP1528 | trpC2 sfp+ ermC epsC+ swrA+ degQ+ amyE : : (P-rapP phrP cat)(yvzG/yvyD Ωspc) ΔepsAB : : aphA3 | GP1519 → AM373 |
| GP1535 | trpC2 sfp+ ermC epsC+ swrA+ degQ+ amyE : : (P-rapP phrP cat)(yvzG/yvyD Ωspc) ΔepsB : : aphA3 | GP1518 → AM373 |
| GP1540 | trpC2 sfp+ ermC epsC+ swrA+ degQ+ amyE : : (P-rapP phrP cat)(yvzG/yvyD Ωspc) ΔepsA : : aphA3 | GP1517 → AM373 |
| GP1542 | trpC2 xkdE : : Pxyl-epsB ermC | pGP2129 → 168 |
| GP1566 | trpC2 ΔtkmA : : spc | this work |
| GP1567 | trpC2 ΔepsA : : aphA3 ΔtkmA : : spc | GP1566 → GP1517 |
| GP1568 | trpC2 ΔepsA : : aphA3 ΔtkmA : : spc xkdE : : Pxyl-epsA ermC | pGP2147 → GP1567 |
| GP1575 | ΔepsB : : aphA3 | GP1535 → NCIB3610* |
| GP1577 | ΔptkA ΔepsB : : aphA3 | GP1535 → NRS2544* |
| GP1589 | trpC2 sinR : : tet epsA-3x FLAG spc | GP736 → GP1526 |
| GP1600 | ΔepsA : : aphA3 | GP1540 → NCIB3610* |
| GP1602 | tkmA : : spc | GP1566 → NCIB3610* |
| GP1611 | ΔepsA : : aphA3 tkmA : : spc | GP1566 → GP1600* |
| GP1622 | ΔsinR-tasA : : cat | GP1672 → NCIB3610* |
| GP1623 | ΔepsB : : aphA3 ΔsinR-tasA : : cat | GP1672 → GP1575* |
| GP1624 | ΔptkA ΔsinR-tasA : : cat | GP1672 → NRS2544* |
| GP1625 | ΔepsB : : aphA3 ΔptkA ΔsinR-tasA : : cat | GP1672 → GP1577* |
| GP1626 | ΔepsA : : aphA3 ΔsinR-tasA : : cat | GP1672 → GP1600* |
| GP1627 | ΔtkmA : : spc ΔsinR-tasA : : cat | GP1672 → GP1602* |
| GP1628 | ΔepsA : : aphA3 ΔtkmA : : spc ΔsinR-tasA : : cat | GP1672 → GP1611* |
| GP1629 | ΔepsA-O : : tet ΔsinR-tasA : : cat | GP1672 → NRS2450* |
| GP1634 | ΔepsB : : aphA3 ΔptkA xkdE : : Pxyl-epsB ermC | GP1542 → GP1577* |
| GP1636 | ΔepsA : : aphA3 ΔtkmA : : spc xkdE : : Pxyl-epsA ermC | GP1568 → GP1611* |
| GP1637 | ΔepsAB : : aphA3 | GP1528 → NCIB3610* |
| GP1672 | trpC2 ΔsinR-tasA : : cat | this work |
| NRS2450 | ΔepsA-O : : tet | Ostrowski et al. (2011) |
| NRS2499 | epsB (D81A+D83A) | pNW329 → NCIB3610* |
| NRS2544 | ΔptkA | Kiley & Stanley-Wall (2010) |
Arrows indicate construction by transformation.
Asterisks indicate construction by SPP1 phage transduction. BSGC represents the Bacillus genetic stock centre. Antibiotic resistance cassettes are: aphA3, kanamycin; cat, chloramphenicol; tet, tetracycline; spc, spectinomycin; ermC, erythromycin.
Luria–Bertani (LB) broth was used to grow E. coli and Bacillus subtilis. When required, media were supplemented with the following antibiotics – for E. coli: ampicillin (100 µg ml−1); and for Bacillus subtilis: spectinomycin (150 µg ml−1), kanamycin (10 µg ml−1), chloramphenicol (5 µg ml−1), erythromycin plus lincomycin (2 and 25 µg ml−1, respectively) and tetracycline (12 µg ml−1).
Bacillus subtilis was grown in C minimal medium supplemented with glucose, succinate, glutamate and auxotrophic requirements (at 50 mg l−1) (Commichau et al., 2007). SP (sporulation) and MSgg plates were prepared by the addition of 17 g Bacto agar l−1 (Difco) to SP (8 g nutrient broth, 1 mM MgSO4 and 13 mM KCl l−1, supplemented after sterilization with 2.5 µM FeSO4, 500 µM CaCl2 and 10 µM MnCl2) or 15 g Bacto agar l−1 (Difco) to MSgg medium (Branda et al., 2001).
Assays of complex colony and pellicle formation.
For colony architecture analysis, bacteria were precultured in LB to an OD600 of 0.6–0.8. Then, 10 µl of this cell suspension was spotted onto minimal MSgg medium (Branda et al., 2001) containing 1.5 % agar and incubated at 22 °C for 3–5 days or at 37 °C for 48 h as indicated. To study the formation of pellicles, 8 µl of the above-mentioned preculture was used to inoculate 8 ml liquid MSgg medium and incubated at room temperature for 3–4 days.
DNA manipulation and transformation.
Plasmid DNA extraction was performed using standard procedures (Sambrook et al., 1989). Restriction enzymes, T4 DNA ligase and DNA polymerases were used as recommended by the manufacturers. DNA fragments were purified from agarose gels using the QIAquick PCR purification kit (Qiagen). Phusion DNA polymerase was used for the PCR as recommended by the manufacturer. Primer sequences are detailed in Table S1 (available in the online Supplementary Material). DNA sequences were determined using the dideoxy chain-termination method (Sambrook et al., 1989). All plasmid inserts derived from PCR products were verified by DNA sequencing. Standard procedures were used to transform E. coli (Sambrook et al., 1989) and transformants were selected on LB plates containing ampicillin (100 µg ml−1) or kanamycin (50 µg ml−1).
Chromosomal DNA of Bacillus subtilis was isolated using the DNeasy Tissue kit (Qiagen) according to the supplier’s protocol. Bacillus subtilis was transformed with plasmid or chromosomal DNA according to the two-step protocol (Kunst & Rapoport, 1995). Transformants were selected on either LB or SP plates containing chloramphenicol (5 µg ml−1), kanamycin (10 µg ml−1), spectinomycin (150 µg ml−1), erythromycin plus lincomycin (2 and 25 µg ml−1, respectively) or tetracycline (12 µg ml−1).
Regulated expression of genes.
To allow the controlled expression of genes, we placed the gene behind a xylose-regulated promoter and integrated this cassette into the xkdE gene in the Bacillus subtilis chromosome. First, the origin of replication and the bla resistance gene were amplified from plasmid pUC19 (Sambrook et al., 1989) using the primer pair KG50/KG51 (for primer sequences see Table S1). This fragment was ligated in a three-arm ligation to the flanking regions of the Bacillus subtilis xkdE gene (amplified with KG70/KG71 and KG72/KG73). The resulting plasmid was pGP883. Next, the Bacillus subtilis xylA promoter as well as the ermC erythromycin resistance gene from pDG647 (Guérout-Fleury et al., 1995) were amplified (KG69/KG59 and KG48/KG49, respectively) and cloned between the BamHI and SmaI sites of pGP883 in a three-arm ligation. With the oligonucleotide KG59, the translation initiation signals of the strongly expressed gapA gene were attached downstream of the xylA promoter. The resulting plasmid was pGP885. Then, the DNA region corresponding to the N-terminal fragment of the improved yellow fluorescent protein from plasmid pIYFP (Veening et al., 2004) was amplified using the primer pair KG74/KG75. With KG74, a multiple cloning region encompassing XbaI, BamHI, KpnI and EcoRI sites was added. The fragment was cloned between the BamHI and SalI sites of pGP885 to give pGP886. A map of the vector pGP886 is available at http://subtiwiki.uni-goettingen.de/wiki/index.php/PGP886 (Michna et al., 2014).
Construction of deletion and complementation strains.
Deletion of the epsA, epsB and tkmA genes as well as of the epsAB and sinR–tasA chromosomal regions was achieved by transformation with PCR products constructed using oligonucleotides (see Table S1) to amplify DNA fragments flanking the target genes and intervening antibiotic resistance cassettes (Guérout-Fleury et al., 1995), as described by Wach (1996). To avoid polar effects on the expression of downstream genes of the eps and the tkmA–ptkA–ptpZ–ugd operons, we used resistance cassettes lacking a transcription terminator. Moreover, expression of the downstream epsC gene was verified in both the epsA and epsB mutants by quantitative real time reverse-transcription (qRT) RT-PCR. While the epsC gene was expressed as in the wild-type in the epsA mutant, the expression was slightly increased in the epsB mutant (data not shown), indicating that the downstream genes of the eps operon were expressed in both mutants.
To allow ectopic expression of epsA and epsB, we constructed the Bacillus subtilis strains GP1568 and GP1542, respectively. In these strains, the genes of interest are expressed in the xkdE locus under the control of the inducible xylose operon promoter PXyl. For this purpose, we used the integrative complementation vector pGP886. The complementing plasmids were constructed as follows. The coding sequences of the epsA and epsB genes were amplified (for oligonucleotides, see Table S1), digested with XbaI and KpnI and cloned into pGP886 linearized with the same enzymes. The resulting plasmids pGP2147 and pGP2129 were linearized with ScaI and NotI, respectively, and used to transform competent cells of Bacillus subtilis 168.
Construction of strains with chromosomal nucleotide substitutions.
Plasmid pNW323 was used for the construction of an in-frame, markerless point mutation of epsB at the ‘DxD’ motif. Primers NSW209 and NSW210 were used to amplify epsB, which was cloned into pCR2.1 (Invitrogen). Primers NSW213 and NSW214 were used to introduce point mutations into the plasmid. Following PCR, template DNA was digested using DpnI. With the remaining DNA E. coli was transformed and colonies were selected based on ampicillin resistance. The resulting plasmid, pNW325, was sequenced to ensure that only the desired mutations were introduced. The region of DNA carrying epsB (D81A–D83A) was cloned into pMAD (Arnaud et al., 2004) using the restriction sites engineered into primers NSW209 and NSW210. To introduce the mutations into NCIB3610, pNW329 was first transformed into Bacillus subtilis strain 168 and phage transduction was utilized to transfer the plasmid to Bacillus subtilis NCIB3610 (Kearns & Losick, 2003). NRS2499 was constructed by integration and curing of pNW329 in NCIB3610 (Arnaud et al., 2004).
Construction of ΔepsA, ΔepsB and ΔepsAB mutant strains with an intact epsC gene.
In the laboratory strain, Bacillus subtilis 168, the epsC gene carries a point mutation and encodes an inactive protein (McLoon et al., 2011). Any genetic transfer of constructed epsA and epsB alleles is likely to co-transfer this epsC mutation. Therefore, we constructed strains GP1540, GP1535 and GP1528 that carry a single deletion of epsA and epsB or a simultaneous deletion of epsAB and the epsC wild-type (epsC+) allele, respectively. For this, we transformed the epsC+ strain AM373 with chromosomal DNA of the epsA mutant GP1517, the epsB mutant GP1518 and the epsAB mutant GP1519. For the resulting ΔepsA, ΔepsB and epsAB transformants the chromosomal epsC allele of ten of the clones was analysed by sequencing, and in each case a single epsC+ transformant was re-isolated and termed GP1540, GP1535 and GP1528.
Precipitation and staining of exopolysaccharides (EPSs).
To analyse the formation of extracellular polysaccharides, precipitation and staining of polymers present in the culture supernatant were performed as described by Guttenplan et al. (2010). Briefly, we used the sinR tasA mutant to facilitate release of extracellular polysaccharides from the cell. The polysaccharides were precipitated using 75 % ethanol, separated from protein in the sample by SDS-PAGE and stained using the Stains-all dye (Applichem). In addition, polysaccharides in the supernatant were precipitated in 24-well plates by ethanol. For better visualization glycerol was added.
Real-time qRT-PCR.
For RNA isolation, cells were grown in CSE minimal medium containing 0.5 % (w/v) glucose (CSE-glucose) to an OD600 of 0.5–0.8 and harvested. Preparation of total RNA was carried out as described previously (Ludwig et al., 2002). cDNAs were synthesized using the One-Step RT-PCR kit (Bio-Rad). qRT-PCR was carried out on an iCycler instrument (Bio-Rad) following the manufacturer’s recommended protocol by using the primers indicated in Table S1. The rpsE and rpsJ genes encoding constitutively expressed ribosomal proteins were used as internal controls. Data analysis and the calculation of expression ratios as fold changes were performed as described by Diethmaier et al. (2011). qRT-PCR experiments were performed in duplicate.
Spore quantification.
For the quantification of spores within biofilms we used the procedure described by Vlamakis et al. (2008). Briefly, complex colonies were collected, disrupted, sonicated and then plated in serial dilutions. To kill any surviving vegetative cells, aliquots were heated to 80 °C and plated again.
In vivo detection of protein–protein interactions.
To address the possible interaction between EpsA and EpsB, we used Bacillus subtilis GP1589/pGP2126, which encodes EpsA carrying a C-terminal FLAG-tag and EpsB with an N-terminal Strep-tag, to facilitate the purification and detection of the protein. Strain GP1526 was constructed as follows: The 3′ end of the epsA gene was amplified using the primer pair JG107/JG111, digested with BamHI/PstI and cloned into pGP1331 (Lehnik-Habrink et al., 2010). The resulting plasmid pGP2127 was integrated into Bacillus subtilis 168, giving rise to strain GP1526. To ensure a high expression of the FLAG-tagged EpsA protein we deleted the sinR gene by transforming strain GP1526 with chromosomal DNA of strain GP736, resulting in strain GP1589. Plasmid pGP2126 was constructed by cloning the epsB gene between the BamHI and SalI sites of the expression vector pGP382 (Herzberg et al., 2007).
The isolation of protein complexes from Bacillus subtilis cells was performed using SPINE technology (Herzberg et al., 2007). Briefly, growing cultures of Bacillus subtilis were treated with formaldehyde (0.6 % w/v, 20 min) to facilitate cross-linking of interacting proteins (Herzberg et al., 2007). The Strep-tagged proteins and their potential interaction partners were then purified from crude extracts using a Streptactin column (IBA) and desthiobiotin as the eluent. Interacting proteins were identified by Western blot analysis.
Western blotting.
For Western blot analysis, proteins were separated by 12 % SDS-PAGE and transferred onto PVDF membranes (Bio-Rad) by electroblotting. Rabbit anti-FLAG (Sigma-Aldrich; 1 : 10 000) polyclonal antibodies served as primary antibodies. The antibodies were visualized by using anti-rabbit immunoglobulin alkaline phosphatase secondary antibodies (Promega) and the CDP-Star detection system (Roche Diagnostics), as described previously (Commichau et al., 2007).
Bacterial two-hybrid assay.
Primary protein–protein interactions were studied by bacterial two-hybrid (B2H) analysis. The B2H system is based on the interaction-mediated reconstruction of adenylate cyclase (CyaA) activity from Bordetella pertussis in E. coli (Karimova et al., 1998). Briefly, proteins suspected to interact physically were fused with separated domains of the adenylate cyclase as described previously (Lehnik-Habrink et al., 2010). DNA fragments corresponding to the epsA and epsB genes were obtained by PCR (for primers, see Table S1). The PCR products were digested with KpnI and XbaI and cloned into the vectors of the two-hybrid system that had been linearized with the same enzymes. The resulting plasmids (see Table S2) were used for co-transformations of E. coli BTH101, and the protein–protein interactions were then analysed by plating the cells on LB plates containing ampicillin (100 µg ml−1), kanamycin (50 µg ml−1), X-Gal (80 µg ml−1) and IPTG (0.5 mM), respectively. The plates were incubated for a maximum of 48 h at 30 °C.
Results
In vivo interaction between EpsA and EpsB
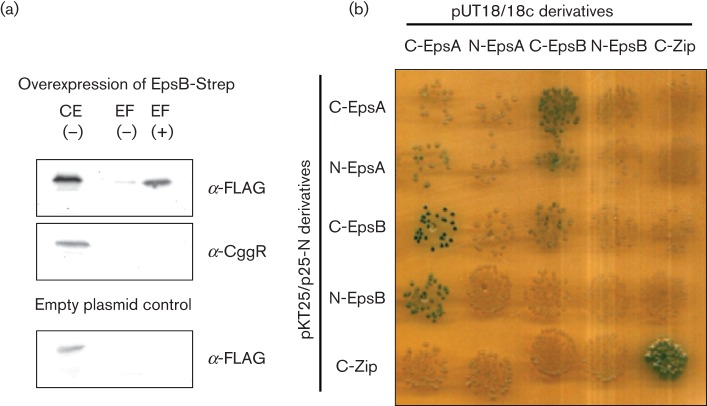
The EpsA and EpsB proteins are similar to the TkmA transmembrane kinase modulator and the PtkA protein tyrosine kinase of Bacillus subtilis, respectively. It is well established that the modulator proteins stimulate the activity of their cognate protein kinases by protein–protein interaction. To address whether this is also the case for EpsA and EpsB, we analysed the potential interaction between these two proteins. For this, we used strain GP1589 carrying plasmid pGP2126. In this strain, EpsB is fused to an N-terminal Strep-tag and EpsA carries a C-terminal FLAG-tag. Additionally, the gene for the anti-activator (and master regulator of biofilm formation) SinR was deleted to ensure a high expression level of EpsA. If the two proteins interacted with each other, one would expect that the FLAG-tag epitope bound to EpsA is detectable in the elution fractions containing Strep-EpsB. As EpsA contains two transmembrane domains and is likely to be a membrane protein, the possible interaction between EpsA and EpsB was fixed using formaldehyde as cross-linker. Strep-EpsB with its bound interaction partners was purified by its binding to Streptactin columns. Both the cell extract and the elution fractions were analysed by SDS-PAGE and subjected to Western blot analysis. As shown in Fig. 1(a), EpsA-FLAG was present in the crude extract. Importantly, EpsA-FLAG co-eluted with EpsB in a protein preparation obtained with cross-linking. To ascertain the specificity of the binding of EpsA to EpsB, we tested whether CggR, a cytoplasmic transcription factor, was also co-purified with EpsB. As shown in Fig. 1(a), CggR was expressed under the tested conditions. However, CggR did not co-elute with EpsB. Additionally, we used an empty vector control to ensure that EpsA-FLAG does not bind to the Streptactin columns unspecifically. In this case we also could not detect EpsA-FLAG in the elution fractions. Therefore, the elution of EpsA is caused by a specific interaction with EpsB.
Fig. 1.
The tyrosine kinase EpsB and its cognate modulator protein EpsA interact physically. (a) EpsA co-purifies with EpsB. The EpsB-Strep fusion protein was expressed from plasmid pGP2126 under the control of a strong constitutive promoter in Bacillus subtilis GP1589 that expresses EpsA-FLAG under the control of its native promoter. To ensure high expression of EpsA-FLAG, the gene for the anti-activator SinR was deleted in this strain. Cells were grown in CSE-glucose medium until the late exponential growth phase. EpsB-Strep was purified in the absence (−) or presence (+) of the cross-linker formaldehyde. To detect the co-purified EpsA-FLAG protein, the elution fractions were heated to reverse the cross-linking and applied to a 12 % SDS polyacrylamide gel. After electrophoresis and blotting onto a PVDF membrane, EpsA was detected via its FLAG-tag. To test that the binding of EpsA to EpsB is not unspecific we tried to detect the CggR protein in the same elution fractions with a specific antibody. Additionally, we used an empty vector control to test unspecific binding of EpsA to the Streptactin column. CE, crude extract; EF, elution fraction. (b) EpsA and EpsB interact in the B2H system. The genes encoding EpsA and EpsB were cloned in the low-copy plasmids p25N and pKT25 and the high-copy plasmids pUT18 and pUT18c. These plasmids allow the expression of the genes of interest fused to the N or C terminus of the T18 or T25 domains of the Bordetella pertussis adenylate cyclase, respectively. The E. coli transformants harbouring both vectors were incubated for 48 h at 30 °C. Degradation of X-Gal and the resulting blue colour of the cells indicate interaction due to the presence of a functional adenylate cyclase.
The results presented above demonstrate an interaction between the membrane protein EpsA and the putative protein kinase EpsB. However, they do not allow us to conclude whether this interaction is direct or indirect. To address this question, we studied the interaction using the B2H system. As shown in Fig. 1(b), EpsA and EpsB clearly interacted with each other in this heterologous E. coli system whereas neither of the two proteins exhibited an interaction with the control protein (leucine zipper of yeast Gcn4p). Thus, EpsA and EpsB are capable of interacting specifically and directly with each other.
The role of tyrosine protein kinases in complex colony and pellicle formation
It is well established that BY-kinases are implicated in extracellular polysaccharide synthesis in many species (Grangeasse et al., 2012). The location of the epsA and epsB genes in the eps operon encoding the functions for extracellular polysaccharide formation in Bacillus subtilis biofilms was highly suggestive of a role for these proteins in biofilm formation. To address the role of the putative BY-kinase EpsB in biofilm formation, we deleted the epsB gene in the undomesticated NCIB3610 wild-type strain. The resulting mutant was GP1575 (see Table 1). In agreement with previous reports (Branda et al., 2001), the wild-type strain formed well-structured colonies (Fig. 2a) and thick and wrinkled pellicles (Fig. 2b). The isogenic epsB mutant GP1575 also formed structured colonies; however, the wrinkles resulting from EPS accumulation were completely lost (Fig. 2a). Similarly, the pellicle formed by the epsB mutant strain was less structured than observed for the wild-type (see Fig. 2b). The epsB deletion did not, however, have an impact as significant as the deletion of the entire epsA-O operon, thus suggesting that EPS biosynthesis was reduced but not completely lost in the epsB mutant. To rule out the possibility that the replacement of the epsB gene by an aphA3 resistance cassette might have a polar effect on the expression of the downstream genes of the eps operon we used a resistance cassette that lacked a transcription terminator downstream of the aphA3 gene. Moreover, we compared the expression of the downstream epsC gene in the wild-type strain 168 and the epsB mutant GP1518 by qRT-PCR. The epsC expression was not affected by the epsB deletion.
Fig. 2.
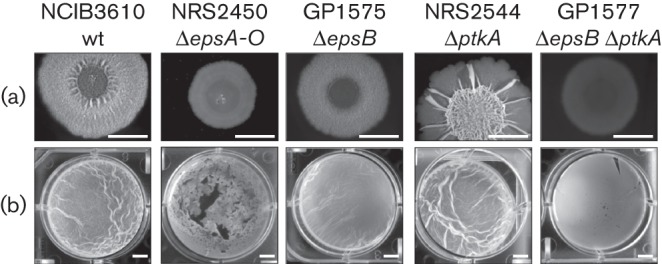
The deletion of tyrosine kinase genes in the wild-type strain NCIB3610 leads to less structured colonies and pellicles. (a) Complex colony formation. Cells were grown on MSgg agar plates solidified with 1.5 % agar for 1 day at 30 °C and for 1–2 days at room temperature prior to photography. Bars, 5 mm. (b) Pellicle formation. Cells were grown in MSgg medium for 2–3 days at room temperature prior to photography. Bars, 5 mm.
To support our hypothesis that the transmembrane modulator EpsA is required for the function of the cognate EpsB protein, we deleted the epsA gene in the undomesticated NCIB3610 wild-type strain. The resulting mutant was GP1600 (see Table 1). As shown for the epsB mutant, the epsA mutant strain formed structured colonies but the wrinkles were lost (Fig. 3a). Also, the pellicle of the epsA mutant strain looked similar to pellicles formed by the epsB mutant (Fig. 3b). The observation that an epsA mutant has the same phenotype as an epsB mutant suggests that EpsA and EpsB act in one pathway of biofilm formation and supports the idea that the EpsA modulator is required for the function of the EpsB protein. To further study the idea that EpsA and EpsB act in one pathway of biofilm formation, the epsAB mutant GP1637 (see Table 1) was constructed and the biofilm phenotype was analysed. As expected, the epsAB double mutant showed the same phenotype as epsA and epsB single mutants, supporting the initial idea.
Fig. 3.
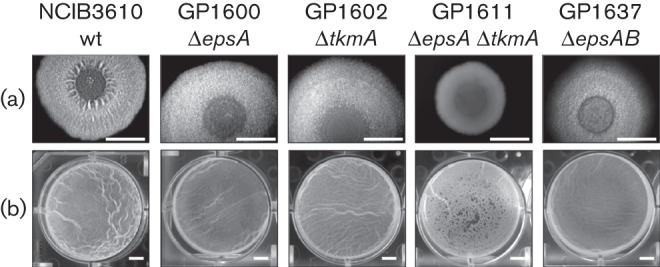
The deletion of tyrosine kinase modulator genes in the wild-type strain NCIB3610 leads to less structured colonies and pellicles. (a) Complex colony formation. Cells were grown on MSgg agar plates solidified with 1.5 % agar for 1 day at 30 °C and for 1–2 days at room temperature prior to photography. Bars, 5 mm. (b) Pellicle formation. Cells were grown in MSgg medium for 2–3 days at room temperature prior to photography. Bars, 5 mm.
As EpsB and PtkA are the only BY-kinases in Bacillus subtilis. we addressed the question of whether both proteins have complementary function for biofilm formation. Therefore, we compared the effect of the inactivation of epsB with that of a ptkA mutation and studied the phenotype resulting from a loss of both BY-kinases. As observed previously (Kiley & Stanley-Wall, 2010), the ptkA mutant NRS2544 formed structured and strongly wrinkled colonies; however, these colonies lacked a rough outer region that is usually the area of sporulation and fruiting body formation (see Fig. 2a). The pellicles formed by the ptkA mutant were similar to those of the isogenic wild-type strain (see Fig. 2b). The most severe phenotype was observed for the epsB ptkA double mutant GP1577. As shown in Fig. 2(a), this strain was unable to form structured colonies, and was thus very similar to epsA-O or ymdB mutants (see Fig. 2; Diethmaier et al., 2011).
Additionally, we performed a complementation assay with the epsB ptkA mutant strains that had a functional copy of the epsB gene under the control of a xylose-induced promoter in the non-essential xkdE locus (GP1634). Expression of the ectopic epsB gene upon addition of xylose to the biofilm medium restored the formation of a thick and structured pellicle (compare Figs S1 and S2). In conclusion, our data support the idea of a role for the BY-kinases in biofilm formation.
Interestingly, the simultaneous deletion of the genes coding for the two kinase modulators EpsA and TkmA in the NCIB3610 strain (GP1611) leads to the same phenotype as the deletion of both BY-kinases (Fig. 3). As shown for the epsB ptkA mutant, the ectopic expression of epsA in the epsA tkmA mutant (GP1636) also restored the formation of a thick and wrinkled pellicle (compare Figs S3 and S4). Again, this supports the idea that the two modulator proteins, EpsA and TkmA are required for the function of their cognate BY-kinases, EpsB and PtkA, respectively.
Together, the results reported above (i) clearly demonstrate the implication of EpsB and PtkA in biofilm formation and (ii) suggest distinct roles for the two enzymes in this process. As reported previously, PtkA seems to be required for biofilm-associated sporulation whereas EpsB is mainly responsible for EPS biosynthesis.
Mutation of the active centre of EpsB does not impact sporulation
Biofilm formation by Bacillus subtilis involves cell fate differentiation and culminates in the formation of environmentally resistant spores (Lopez et al., 2009; Vlamakis et al., 2008). The formation of spores during biofilm formation is dependent on the synthesis of the extracellular matrix. The regulator Spo0A is required for both sporulation and biofilm formation (Lopez et al., 2009). Spo0A is activated by phosphorylation upon detection of environmental stimuli that are perceived by sensor kinases. KinD links spore formation to extracellular matrix production (Aguilar et al., 2010). To establish whether the BY-kinase EpsB has an impact on cell differentiation, we compared the level of sporulation of wild-type and mutant strains after 72 h of incubation under biofilm formation conditions (Vlamakis et al., 2008). In contrast to the epsA-O deletion strain NRS2450 that was used as a control (mean±SEM 2.8±1 % sporulation), 95±0.1 % of the epsB mutant strain NRS2499 had sporulated; this level that was not statistically significantly different from the wild-type parental strain NCIB3610 (102±17 %). These findings support the idea that the impact of EpsB on biofilm formation is downstream from the checkpoint triggering sporulation during biofilm formation, which is controlled by KinD (Aguilar et al., 2010).
Tyrosine kinases influence extracellular polysaccharide production
EPSs are a major component of the biofilm matrix (Branda et al., 2001). The proteins for the synthesis and export of the EPS are encoded within the epsA-O operon. As shown in Fig. 2, deletion of the putative BY-kinase EpsB leads to a loss of the wrinkled colony and pellicle structure, which is usually considered as a loss of EPS production. The location of the epsB gene in the epsA-O operon supports this idea. To test our assumption that EpsB is involved in the production of EPSs, we used a strain with deletions of the sinR and tasA genes to enhance the production and release of EPSs from the cells. The cells were cultivated and the EPSs within the supernatant of the culture medium were precipitated by ethanol. Surprisingly, no major effect of the epsB deletion on the amount of EPSs was observed (Fig. 4). This suggests that either the putative kinase activity of EpsB is dispensable for EPS production or EpsB can be functionally replaced by the second BY-kinase, PtkA.
Fig. 4.
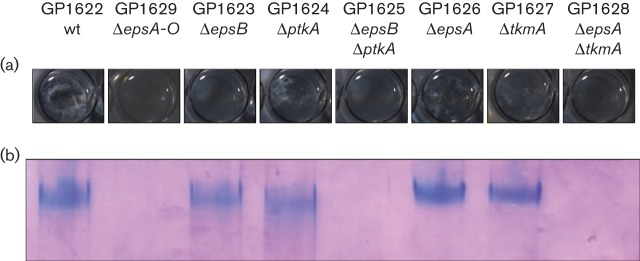
The deletion of tyrosine kinases affects EPS production. All strains contain a ΔsinR-tasA deletion to facilitate the release of EPSs into the culture medium. (a) Ethanol-precipitated supernatant from the indicated strains in the chambers of a 24-well plate. (b) Ethanol precipitates resolved in the stacker of an SDS-PAGE gel stained with Stains-all dye.
Therefore, the implication of the second BY-kinase PtkA in EPS production was addressed. In agreement with the wrinkled colony and pellicle structure of the ptkA mutant, similar amounts of EPSs as in the wild-type were observed. Thus, PtkA is not directly involved in the regulation of EPSs by the proteins encoded within the epsA-O operon. Interestingly, no EPS was detectable in the epsB ptkA double mutant. This observation is in perfect agreement with the complete lack of biofilm formation in the epsB ptkA double mutant and suggests that PtkA also affects EPS production, at least in the absence of EpsB.
Furthermore, we monitored EPS production of the epsA and tkmA modulator single mutants and the epsA tkmA double mutant. As shown for the respective BY-kinase mutants, the modulator single mutants are able to produce EPS, whereas the epsA tkmA mutant shows no EPS production. This supports the requirement of EpsB and PtkA for their modulator proteins.
Discussion
We demonstrate here that the two protein tyrosine kinases of Bacillus subtilis, EpsB and PtkA, participate in biofilm formation. Inactivation of both kinases results in a complete loss of biofilm formation whereas the single mutants exhibit rather mild defects. Analysis of complex colony and pellicle formation also demonstrated that the two enzymes have distinct and complementary functions: PtkA is required to facilitate sporulation in the ‘fruiting body’-like structures in the outer region of the complex colonies (Kiley & Stanley-Wall, 2010). In contrast, EpsB is not involved in controlling sporulation. Instead, EpsB seems to be required for extracellular polysaccharide production. Together, both extracellular matrix production and sporulation seem to be important for the successful formation of a resistant biofilm.
Interestingly, we observed that EPS production is still active in the epsB mutant, whereas no EPS was formed in the epsB ptkA double mutant. Thus, it seems that PtkA can compensate for the loss of EpsB in the activation of extracellular polysaccharide synthesis. Such partial take-over of a function by a paralogous protein is not unprecedented: in Bacillus subtilis the HPr protein of the phosphotransferase system is involved in the phosphorylation and uptake of sugars. In addition, HPr phosphorylated at a specific serine residue acts as co-factor for the transcription factor CcpA, thus controlling carbon catabolite repression. In the absence of HPr, the paralogous protein Crh can partially replace HPr and bind to CcpA, although with significantly lower affinity. As observed here with EpsB and PtkA, this take-over of a function seems to be limited to situations in which the protein normally in charge of the function is absent whereas the paralogues normally have different functions.
It is well established that the BY-kinases require activation by transmembrane modules. These modules are either a domain of a larger kinase protein (as in the enzymes of Gram-negative bacteria) or separate proteins that are encoded by genes upstream of their cognate kinase genes (in Firmicutes) (Grangeasse et al., 2012). In Bacillus subtilis, PtkA is activated by TkmA (Mijakovic et al., 2003). Similar to the arrangement of the tkmA and ptkA genes, the epsB kinase-encoding gene is located downstream of the epsA gene coding for the modulator protein. Here, we have shown that the two proteins EpsA and EpsB do indeed physically interact in vivo, and that this interaction is direct (see Fig. 1). Moreover, our results demonstrate that the epsA mutant has phenotypes with respect to complex colony and pellicle formation identical to that of the epsB mutant. The genetic clustering, the physical interaction of the two proteins and the identity of mutant phenotypes indicate that EpsA is required for the presumptive protein kinase activity of EpsB.
For several bacterial tyrosine kinases it has been demonstrated that they regulate the activity, the localization or the interaction properties of proteins by the phosphorylation of their targets. This suggests that EpsB may also act by directly phosphorylating other proteins. This prompted us to perform an intensive search for phosphorylation targets of the BY-kinase EpsB; unfortunately, no phosphorylation targets could be identified.
To study the role of EpsA and EpsB in more detail, we will attempt to identify interaction partners of the two proteins and will analyse potential EpsB-dependent phosphorylation of these proteins.
Acknowledgements
We thank Kerstin Kruse, Sumana Sharma, Katrin Bäsell, Dörte Becher, Julia Motz, Jan Oberdiek, Chuan Qin and David Pawlowicz for their help with some experiments and Julia Busse for technical assistance. We are grateful to R. Losick for the gift of strain AM373. This work was supported by grants of the DFG (SFB860) to J. S, N. S.-W. is funded by a Biotechnology and Biological Sciences Research Council (BBSRC) grant (BB/I019464/1) and T. B. K. was a recipient of a BBSRC doctoral training grant (BB/D526161/1).
Abbreviations:
- B2H
bacterial two-hybrid
- EPS
exopolysaccharide
Footnotes
Two supplementary tables and four supplementary figures are available with the online version of this paper.
References
- Aguilar C., Vlamakis H., Guzman A., Losick R., Kolter R. (2010). KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. MBio 1, e00035–e10. 10.1128/mBio.00035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M., Chastanet A., Débarbouillé M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70, 6887–6891. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet E., Gruszczyk J., Terreux R., Gueguen-Chaignon V., Vigouroux A., Obadia B., Cozzone A. J., Nessler S., Grangeasse C. (2010). Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol Microbiol 77, 1315–1325. 10.1111/j.1365-2958.2010.07291.x [DOI] [PubMed] [Google Scholar]
- Blair K. M., Turner L., Winkelman J. T., Berg H. C., Kearns D. B. (2008). A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320, 1636–1638. 10.1126/science.1157877 [DOI] [PubMed] [Google Scholar]
- Branda S. S., González-Pastor J. E., Ben-Yehuda S., Losick R., Kolter R. (2001). Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98, 11621–11626. 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Kolter R., Losick R. (2009). Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis. Mol Microbiol 74, 876–887. 10.1111/j.1365-2958.2009.06900.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Norman T., Kolter R., Losick R. (2010). An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev 24, 754–765. 10.1101/gad.1915010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F., Kearns D. B., Branda S. S., Kolter R., Losick R. (2006). Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59, 1216–1228. 10.1111/j.1365-2958.2005.05019.x [DOI] [PubMed] [Google Scholar]
- Commichau F. M., Herzberg C., Tripal P., Valerius O., Stülke J. (2007). A regulatory protein–protein interaction governs glutamate biosynthesis in Bacillus subtilis: the glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65, 642–654. 10.1111/j.1365-2958.2007.05816.x [DOI] [PubMed] [Google Scholar]
- Derouiche A., Bidnenko V., Grenha R., Pigonneau N., Ventroux M., Franz-Wachtel M., Nessler S., Noirot-Gros M.-F., Mijakovic I. (2013). Interaction of bacterial fatty-acid-displaced regulators with DNA is interrupted by tyrosine phosphorylation in the helix-turn-helix domain. Nucleic Acids Res 41, 9371–9381. 10.1093/nar/gkt709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier C., Pietack N., Gunka K., Wrede C., Lehnik-Habrink M., Herzberg C., Hübner S., Stülke J. (2011). A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J Bacteriol 193, 5997–6007. 10.1128/JB.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier C., Newman J. A., Kovács A. T., Kaever V., Herzberg C., Rodrigues C., Boonstra M., Kuipers O. P., Lewis R. J., Stülke J. (2014). The YmdB phosphodiesterase is a global regulator of late adaptive responses in Bacillus subtilis. J Bacteriol 196, 265–275. 10.1128/JB.00826-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeasse C., Nessler S., Mijakovic I. (2012). Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos Trans R Soc Lond B Biol Sci 367, 2640–2655. 10.1098/rstb.2011.0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A. M., Shazand K., Frandsen N., Stragier P. (1995). Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167, 335–336. 10.1016/0378-1119(95)00652-4 [DOI] [PubMed] [Google Scholar]
- Guttenplan S. B., Blair K. M., Kearns D. B. (2010). The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet 6, e1001243. 10.1371/journal.pgen.1001243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg C., Weidinger L. A., Dörrbecker B., Hübner S., Stülke J., Commichau F. M. (2007). SPINE: a method for the rapid detection and analysis of protein–protein interactions in vivo. Proteomics 7, 4032–4035. 10.1002/pmic.200700491 [DOI] [PubMed] [Google Scholar]
- Jers C., Pedersen M. M., Paspaliari D. K., Schütz W., Johnsson C., Soufi B., Macek B., Jensen P. R., Mijakovic I. (2010). Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol Microbiol 77, 287–299. 10.1111/j.1365-2958.2010.07227.x [DOI] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95, 5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D. B., Losick R. (2003). Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49, 581–590. 10.1046/j.1365-2958.2003.03584.x [DOI] [PubMed] [Google Scholar]
- Kiley T. B., Stanley-Wall N. R. (2010). Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation. Mol Microbiol 78, 947–963. 10.1111/j.1365-2958.2010.07382.x [DOI] [PubMed] [Google Scholar]
- Kunst F., Rapoport G. (1995). Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol 177, 2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M., Pförtner H., Rempeters L., Pietack N., Herzberg C., Stülke J. (2010). The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol Microbiol 77, 958–971. [DOI] [PubMed] [Google Scholar]
- Lewis R. J., Brannigan J. A., Offen W. A., Smith I., Wilkinson A. J. (1998). An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J Mol Biol 283, 907–912. 10.1006/jmbi.1998.2163 [DOI] [PubMed] [Google Scholar]
- Lopez D., Vlamakis H., Kolter R. (2009). Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33, 152–163. 10.1111/j.1574-6976.2008.00148.x [DOI] [PubMed] [Google Scholar]
- Ludwig H., Meinken C., Matin A., Stülke J. (2002). Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J Bacteriol 184, 5174–5178. 10.1128/JB.184.18.5174-5178.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoon A. L., Guttenplan S. B., Kearns D. B., Kolter R., Losick R. (2011). Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193, 2027–2034. 10.1128/JB.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michna R. H., Commichau F. M., Tödter D., Zschiedrich C. P., Stülke J. (2014). SubtiWiki–a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res 42 (D1), D692–D698. 10.1093/nar/gkt1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I., Poncet S., Boël G., Mazé A., Gillet S., Jamet E., Decottignies P., Grangeasse C., Doublet P. & other authors (2003). Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J 22, 4709–4718. 10.1093/emboj/cdg458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona J. K., Morona R., Miller D. C., Paton J. C. (2003). Mutational analysis of the carboxy-terminal (YGX)4 repeat domain of CpsD, an autophosphorylating tyrosine kinase required for capsule biosynthesis in Streptococcus pneumoniae. J Bacteriol 185, 3009–3019. 10.1128/JB.185.10.3009-3019.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. A., Rodrigues C., Lewis R. J. (2013). Molecular basis of the activity of SinR protein, the master regulator of biofilm formation in Bacillus subtilis. J Biol Chem 288, 10766–10778. 10.1074/jbc.M113.455592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski A., Mehert A., Prescott A., Kiley T. B., Stanley-Wall N. R. (2011). YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol 193, 4821–4831. 10.1128/JB.00223-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic D., Michelsen O., Zahradka K., Silva C., Petranovic M., Jensen P. R., Mijakovic I. (2007). Bacillus subtilis strain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol Microbiol 63, 1797–1805. 10.1111/j.1365-2958.2007.05625.x [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Soulat D., Grangeasse C., Vaganay E., Cozzone A. J., Duclos B. (2007). UDP-acetyl-mannosamine dehydrogenase is an endogenous protein substrate of Staphylococcus aureus protein-tyrosine kinase activity. J Mol Microbiol Biotechnol 13, 45–54. 10.1159/000103596 [DOI] [PubMed] [Google Scholar]
- Veening J. W., Smits W. K., Hamoen L. W., Jongbloed J. D., Kuipers O. P. (2004). Visualization of differential gene expression by improved cyan fluorescent protein and yellow fluorescent protein production in Bacillus subtilis. Appl Environ Microbiol 70, 6809–6815. 10.1128/AEM.70.11.6809-6815.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H., Aguilar C., Losick R., Kolter R. (2008). Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22, 945–953. 10.1101/gad.1645008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11, 157–168. 10.1038/nrmicro2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. (1996). PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265. [DOI] [PubMed] [Google Scholar]
- Whitfield C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75, 39–68. 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- Winkelman J. T., Bree A. C., Bate A. R., Eichenberger P., Gourse R. L., Kearns D. B. (2013). RemA is a DNA-binding protein that activates biofilm matrix gene expression in Bacillus subtilis. Mol Microbiol 88, 984–997. 10.1111/mmi.12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wugeditsch T., Paiment A., Hocking J., Drummelsmith J., Forrester C., Whitfield C. (2001). Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem 276, 2361–2371. 10.1074/jbc.M009092200 [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Young F. E. (1974). Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]