Abstract
Arboretum virus (ABTV) and Puerto Almendras virus (PTAMV) are two mosquito-associated rhabdoviruses isolated from pools of Psorophora albigenu and Ochlerotattus fulvus mosquitoes, respectively, collected in the Department of Loreto, Peru, in 2009. Initial tests suggested that both viruses were novel rhabdoviruses and this was confirmed by complete genome sequencing. Analysis of their 11 482 nt (ABTV) and 11 876 (PTAMV) genomes indicates that they encode the five canonical rhabdovirus structural proteins (N, P, M, G and L) with an additional gene (U1) encoding a small hydrophobic protein. Evolutionary analysis of the L protein indicates that ABTV and PTAMV are novel and phylogenetically distinct rhabdoviruses that cannot be classified as members of any of the eight currently recognized genera within the family Rhabdoviridae, highlighting the vast diversity of this virus family.
Rhabdoviridae is a diverse family of non-segmented, negative-sense ssRNA viruses that infect a wide range of vertebrates, invertebrates and plants (Dietzgen et al., 2012). Here we report the characterization of two novel rhabdoviruses, collected in Puerto Almendras (3° 50′ S, 73° 22′ W) in the Loreto Department of north-eastern Peru. Arboretum virus (ABTV; strain LO-121) and Puerto Almendras virus (PTAMV; strain LO-39) were isolated from pools of female Ochlerotattus fulvus and Psorophora albigenu mosquitoes, collected by light trap on 22 February and 25 March 2009, respectively, in the grounds of the botanical garden located in the town. After collection, the mosquitoes were transported on dry ice to the US Naval Medical Research Unit no. 6 in Lima, where they were initially processed, and an aliquot was forwarded to the University of Texas Medical Branch for virus isolation. Here, we demonstrate that ABTV and PTAMV are novel rhabdoviruses with a distinctive morphology and genome organization, and are phylogenetically divergent from known rhabdoviruses.
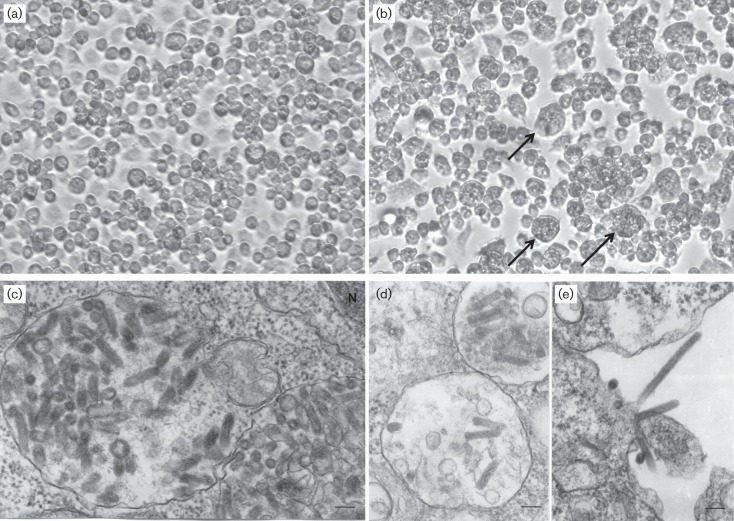
On initial inoculation of ABTV and PTAMV into flask cultures of C6/36 (Aedes albopictus) (Igarashi, 1978) cells maintained at 28 °C, viral cytopathic effect (CPE) was observed in 4–5 days. The infected C6/36 cells detached from the plastic surface and displayed clumping; some cells were enlarged or bloated in appearance (Fig. 1a, b). In contrast, neither virus produced detectable CPE in Vero E6 (African green monkey kidney) or BHK-21 (baby hamster kidney) cell cultures maintained at 37 °C. Likewise, they did not produce illness or death when inoculated intracranially in newborn (1–2 day old) mice. A reverse transcription PCR (RT-PCR) assay was used to assess further whether the mammalian cell lines could support replication of the viruses in the absence of CPE. MacVector version 12.7.3 was used to design specific primers Lo-121FW, 5′-AGGCGAACCAACTTACACCC-3′, and Lo-121REV, 5′-TCGTGCGGATACTGTAGTG-3′ (targeting nucleotides 644–663 and 941–923, respectively, in the ABTV genome), and Lo-39FW, 5′-CGGTGCTCATAAATCGGTTTC-3′, and Lo-39REV, 5′-TGTATTGTGGGTCTCCTGTC-3′ (targeting nucleotides 285–305 and 661–642, respectively, in the PTAMV genome) (Rastogi, 2000). Cell-free supernatants of the Vero and BHK-21 cells infected with ABTV or PTAMV were collected at 1, 2, 3, 4, 6 and 8 days post-infection (p.i.) and viral genetic load was evaluated by RT-PCR assay. Viral RNA was extracted using a QIAamp viral RNA mini kit (Qiagen), and amplicons were generated using the Titan one step RT-PCR kit (Roche) and visualized in 1.5 % TAE (Tris-acetate EDTA) agarose gels (Fig. S1, available in the online Supplementary Material). This assay demonstrated that ABTV and PTAMV were detected in the supernatants of each vertebrate cell line, although ABTV was only detected at 1 day p.i. in BHK-21 cells (Fig. S1). Each cell line is permissive for replication of a wide range of other arthropod-transmitted viruses.
Fig. 1.
Cytopathology and ultrastructure of the viruses ABTV and PTAMV. (a) Control (non-infected) C6/36 cells. Magnification, ×200. (b) CPE seen in C6/36 cells 4 days after ABTV infection; cells are clumping together and detaching; some cells are enlarged and bloated in appearance (arrows). Magnification, ×200. (c, d) PTAMV virions inside intracytoplasmic vacuoles. Bars, 100 nm. N, Nucleus. (e) ABTV virions budding from the cell surface. Bar, 100 nm.
In ultrathin sections of infected C6/36 cells, PTAMV virions were found predominantly within large intracytoplasmic vacuoles (Fig. 1c, d). They displayed typical bullet-shaped morphology with particles 40–55 nm in diameter and up to 190 nm long. ABTV virions formed at the cell surface (Fig. 1e). They displayed a thin rod-like shape and were 40–45 nm in diameter and up to 460 nm long. Sections were processed as described previously (Vasilakis et al., 2013), and were cut on a Leica EM UC7 ultramicrotome (Leica Microsystems), stained with lead citrate and examined in a Philips 201 transmission electron microscope at 60 kV.
To further characterize ABTV and PTAMV, the complete genome sequences were determined. Briefly, infected C6/36 cells were grown to 90 % confluence in T25 culture flasks, and virus harvest, isolation and processing of viral RNA (vRNA) for next generation sequencing (NGS) were as described previously (Vasilakis et al., 2013). The complete genome sequences were assigned GenBank accession numbers KC994644 (ABTV) and KF534749 (PTAMV); accession numbers for the genome sequences of rhabdoviruses used in the phylogenetic analyses are listed in Table S1.
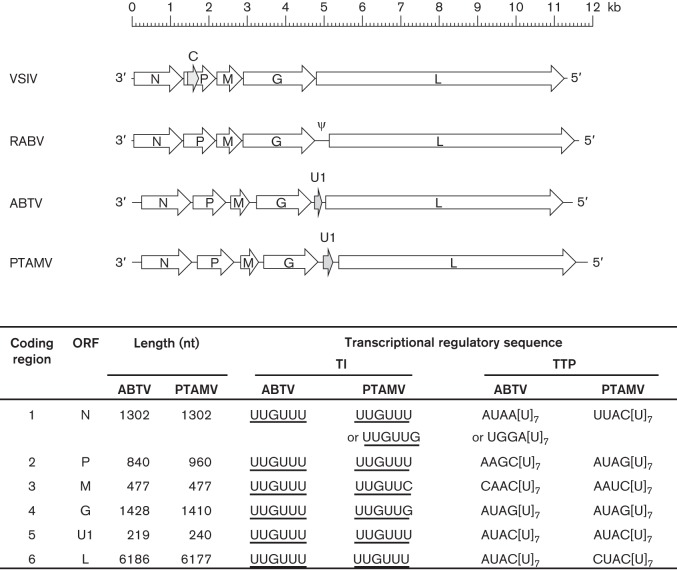
The (−) ssRNA ABTV and PTAMV genomes have a similar organization, each containing long leader (l) and trailer (t) regions and six long ORFs arranged in the order 3′-l-N-P-M-G-U1-L-t-5′ (Fig. 2). The PTAMV genome (11 876 nt) is larger than the ABTV genome (11 482 nt), primarily due to longer intergenic regions and a longer P ORF. Each ORF is bounded by sequences similar to the transcription initiation (TI) and transcription termination-polyadenylation (TTP) sequences of other animal rhabdoviruses. The putative TI sequence is conserved (UUGUU); the TTP sequences feature the consensus motif AC[U]7, which occurs in some genes as other variations of the dinucleotide preceding the septa-uridine run. Unlike some other rhabdoviruses, both contain no small alternative or overlapping ORFs; each of the six genes appears to be transcribed as a monocistronic mRNA.
Fig. 2.
Organization of ABTV and PTAMV genomes. Schematic representation of the ABTV and PTAMV genome organization, where block arrows indicate the location of long ORFs, including the U1 ORF (grey shading). Shown below are the ARBV and PTAMV coding regions, designated ORFs and putative transcription regulatory sequences. The genomes of Vesicular stomatitis Indiana virus (VSIV) and Rabies virus (RABV) are also shown.
The N ORFs each encode 434 aa mildly acidic polypeptides (ABTV, 49.7 kDa; PTAMV, 49.9 kDa) that share 38.6 % overall amino acid sequence identity. A clustal_x alignment of their N proteins with that of Vesicular stomatitis Indiana virus (VSIV) indicated preservation of several known conserved motifs, as well as each of the eight basic residues located in the RNA-binding cavity that are known to coordinate binding to viral genomic RNA in the ribonucleoprotein (RNP) (Green et al., 2006; Luo et al., 2007) (Fig. S2).
The P ORFs encode acidic 280 aa (32.0 kDa) and 320 aa (36.5 kDa) polypeptides, in ABTV and PTAMV, respectively, which, although displaying no significant sequence identity, are similar to other rhabdovirus P proteins in size and net charge. A pairwise alignment of the ABTV and PTAMV P proteins indicated a relatively high level of sequence identity in the N-terminal and C-terminal regions, and that three indels (9, 16 and 10 aa) in the N-terminal accounted for half the size difference between the proteins (data not shown). Rhabdovirus P proteins generally share low sequence identity and are intrinsically disordered with a modular organization comprising structured domains alternating with disordered regions (Karlin et al., 2003).
The M ORFs each encode 159 aa basic proteins in ABTV and PTAMV (19.3 kDa and 19.1 kDa, respectively), which share 49.1 % pairwise amino acid identity, but show no match to other known rhabdovirus M proteins in a pBLAST search. However, like other rhabdovirus M proteins, each contains a sequence in the N-terminal region that resembles a late budding domain motif that mediates the interaction of M proteins with cellular proteins during the budding process (Freed, 2002). This motif occurs as PPxS in ABTV and PSxS in PTAMV, rather than PPxY and/or P(S/T)AP, which are characteristic of lyssaviruses, vesiculoviruses and ephemeroviruses (Harty et al., 1999; Irie et al., 2004; Wirblich et al., 2008). The PPxS motif has also been identified recently in Kolente rhabdovirus (Ghedin et al., 2013).
The G ORFs encode 476 aa and 472 aa polypeptides, in ABTV and PTAMV, respectively. Each is predicted to contain six potential N-glycosylation sites in the ectodomain (NetNGlyc 1.0 server), four at similar positions. On this basis, the mature G proteins would each have a molecular mass of ~71 kDa following cleavage at the predicted signal peptidase sites (SignalP). A pairwise alignment with the VSIV G protein indicated preservation of each of the highly conserved cysteine residues (CI–CXII) that are characteristic of rhabdovirus G proteins, forming six disulphide bridges (Fig. S3a). As reported for some other rhabdovirus G proteins, two additional cysteine residues also occur in the downstream lateral domain of ABTV and PTAMV, and these are likely to form a seventh disulphide bridge (Walker & Kongsuwan, 1999).
The U1 ORFs encode, in ABTV and PTAMV, respectively, 73 aa (8.9 kDa) and 80 aa (9.5 kDa) small hydrophobic proteins, each with a predicted transmembrane domain, short N-terminal ectodomain and long C-terminal endodomain (49 aa and 44 aa) featuring a high proportion of basic residues (K, R, H) and large aromatic residues (F, Y, W) (Fig. S3b). This structural arrangement is typical of class IA viroporins such as the influenza A virus M2 protein and human immunodeficiency virus 1 Vpu, in which the basic and aromatic residues interact with the membrane and participate in ion channel formation (Nieva et al., 2012). Small hydrophobic proteins are encoded in the genomes of many other animal rhabdoviruses, either between the M and G genes (Tupaia rhabdovirus, Durham virus and Oak Vale rhabdovirus) or in the region downstream of the G gene (ephemeroviruses, tibroviruses and the Hart Park group) (Walker et al., 2011). Those located after the G gene generally have the structural features of class IA viroporins, and the viroporin-like properties have recently been confirmed experimentally for the bovine ephemeral fever virus α1 protein (Joubert et al., 2013).
The L ORFs encode 2062 aa (241 kDa) and 2060 aa (240 kDa) polypeptides, in ABTV and PTAMV, respectively, which share overall 36.1 % amino acid sequence identity and contain all the structural motifs of the rhabdovirus RNA-dependent RNA polymerase (L protein).
Lastly, we generated a phylogenetic tree to reveal the relationship of ABTV and PTAMV to other members of the diverse family Rhabdoviridae. The ABTV and PTAMV L protein sequences were compared with those of 46 other rhabdoviruses (members of the genera Cytorhabdovirus, Novirhabdovirus and Nucleorhabdovirus were excluded because their excessive divergence reduced phylogenetic resolution). The GenBank accession numbers for the genome sequences of select rhabdoviruses used in the phylogenetic analyses are listed in Table S1. Protein sequences were aligned using muscle (Edgar, 2004), and ambiguously aligned regions were removed using the Gblocks program (Talavera & Castresana, 2007), resulting in a sequence alignment of 1091 aa residues. The phylogenetic relationships were determined using the maximum-likelihood (ML) method available in PhyML 3.0 (Guindon et al., 2010), employing the WAG+Γ model of amino acid substitution, and subtree pruning and regrafting (SPR) branch-swapping. The phylogenetic robustness of each node was determined using 1000 bootstrap replicates.
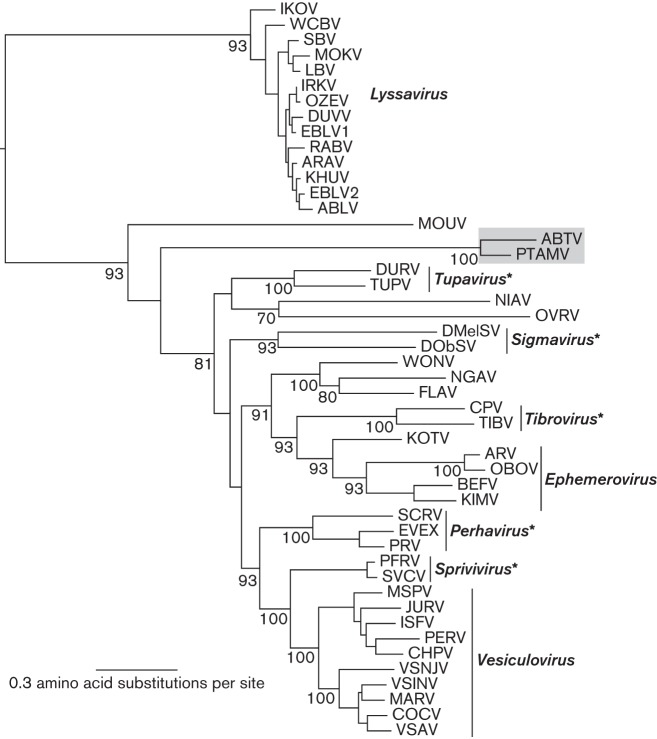
The ML tree of 48 rhabdovirus L protein sequences indicates that ABTV and PTAMV cluster together as a distinct lineage within the major groups of animal rhabdoviruses, which include members of the genera Vesiculovirus and Ephemerovirus (Fig. 3). While both cluster in this phylogeny between Moussa virus (MOUV) and the other animal rhabdoviruses, the long branch lengths and low bootstrap support indicate that these viruses are divergent, experiencing a long evolutionary separation from other rhabdoviruses described to date. Although fine-scale resolution of the evolutionary history of ABTV and PTAMV is not possible based on these data, their divergent nature strongly suggests that they represent novel virus species.
Fig. 3.
ML phylogenetic tree of 48 rhabdovirus L protein sequences. ABTV and PTAMV are shaded and bootstrap support values (>70 %) are shown for key nodes. All horizontal branch lengths are drawn to a scale of amino acid substitutions per site, and the tree is rooted in the position observed in a broader analysis of the Rhabdoviridae. *Genera not yet formally approved by the International Committee on Taxonomy of Viruses.
Collectively the data indicate that ABTV and PTAMV are two novel rhabdoviruses isolated from different mosquito species living in close proximity, but little is known about their arthropod and vertebrate host range and epidemiology. Both viruses failed to produce detectable CPE in vertebrate cell lines. Similarly, both viruses failed to produce illness and/or death in newborn mice, suggesting that they may infect vertebrate hosts in the absence of clinical signs of illness, as is also true of Tibrogargan and coastal plains rhabdoviruses (Cybinski et al., 1980; Gubala et al., 2011). However, we acknowledge that cultured cells, particularly Vero cells, which are deficient in the IFN response (Mosca & Pitha, 1986), are imperfect models to establish host susceptibility. Phylogenetically, ABTV and PTAMV sit between MOUV and a diverse array of rhabdoviruses isolated from mammals (Kurz et al., 1986), birds (Allison et al., 2011) and insects (Quan et al., 2011; Vasilakis et al., 2013), highlighting the wide diversity of rhabdovirus hosts and the potential for human exposure. Indeed, both P. albigenu (Watts et al., 1998) and O. fulvus (Turell et al., 2008) mosquitoes have been incriminated as competent vectors of several arboviruses of human importance (such as Venezuelan equine encephalitis virus, Eastern equine encephalitis virus and others) and both have been observed to feed on humans (Jones et al., 2004). However, since both ABTV and PTAMV have so far only been detected once in mosquito pools, a comprehensive and accurate assessment of their full geographical range, prevalence and host range remains to be determined through comprehensive surveillance studies.
The ABTV and PTAMV genomes have similar size and organization, encoding the five canonical structural proteins and a small hydrophobic protein (U1) in a single additional ORF located between the G and L genes. The structural characteristics of U1 suggest that it may function as a viroporin. ORFs encoding structurally similar proteins have been reported in the region between the G and L genes in several other rhabdoviruses, including ephemeroviruses, tibroviruses and members of the Hart Park serogroup, but these viruses have somewhat more complex genome organizations than ABTV and PTAMV, with multiple ORFs encoding additional accessory proteins (Walker et al., 2011). Furthermore, the L protein phylogeny indicates that ABTV and PTAMV are relatively distantly related to all other known rhabdoviruses. The lack of available data about the host range and prevalence of these viruses underscores the necessity of further studies to decipher this diverse and complex family of viruses.
Acknowledgements
We thank Anibal Huayanay for fieldwork assistance. This work was supported in part by a grant from the Institute for Human Infections and Immunity, University of Texas Medical Branch (N. V.), and NIH contract HHSN272201000040I/HHSN27200004/D04 (R. B. T., N. V.). E. C. H. is supported by an NHMRC Australia Fellowship. A. J. A. is supported by the J. W. McLaughlin Fellowship. This study was supported by the United States Department of Defense Global Emerging Infections Surveillance and Response System, a Division of the Armed Forces Health Surveillance Center, work unit no. 847705.82000.25GB.B0016. The sponsors had no role in this study other than providing funding. The opinions expressed in this work are those of the authors and do not reflect the official policy or position of the Department of the Navy, Department of Defense or US Government. None of the authors has a financial or personal conflict of interest related to this study. K. M. and T. J. K. are military service members, and F. C., C. G. and R. F. are employees of the US Government. This work was prepared as part of their official duties.
Footnotes
One supplementary table and three supplementary figures are available with the online version of this paper.
References
- Allison A. B., Palacios G., Travassos da Rosa A., Popov V. L., Lu L., Xiao S. Y., DeToy K., Briese T., Lipkin W. I. & other authors (2011). Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Res 155, 112–122 10.1016/j.virusres.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybinski D. H., St George T. D., Standfast H. A., McGregor A. (1980). Isolation of Tibrogargan, a new Australian rhabdovirus, from Culicoides brevitarsis. Vet Microbiol 5, 301–308 10.1016/0378-1135(80)90029-2 [DOI] [Google Scholar]
- Dietzgen R. G., Calisher C. H., Kurath G., Kuzmin I. V., Rodriguez L. L., Stone D. M., Tesh R. B., Tordo N., Walker P. J. & other authors (2012). Rhabdoviridae. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses, pp. 686–713 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. London: Elsevier/Academic Press [Google Scholar]
- Edgar R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E. O. (2002). Viral late domains. J Virol 76, 4679–4687 10.1128/JVI.76.10.4679-4687.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Rogers M. B., Widen S. G., Guzman H., Travassos da Rosa A. P., Wood T. G., Fitch A., Popov V., Holmes E. C. & other authors (2013). Kolente virus, a rhabdovirus species isolated from ticks and bats in the Republic of Guinea. J Gen Virol 94, 2609–2615 10.1099/vir.0.055939-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T. J., Zhang X., Wertz G. W., Luo M. (2006). Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313, 357–360 10.1126/science.1126953 [DOI] [PubMed] [Google Scholar]
- Gubala A., Davis S., Weir R., Melville L., Cowled C., Boyle D. (2011). Tibrogargan and Coastal Plains rhabdoviruses: genomic characterization, evolution of novel genes and seroprevalence in Australian livestock. J Gen Virol 92, 2160–2170 10.1099/vir.0.026120-0 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59, 307–321 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Harty R. N., Paragas J., Sudol M., Palese P. (1999). A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol 73, 2921–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi A. (1978). Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J Gen Virol 40, 531–544 10.1099/0022-1317-40-3-531 [DOI] [PubMed] [Google Scholar]
- Irie T., Licata J. M., McGettigan J. P., Schnell M. J., Harty R. N. (2004). Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J Virol 78, 2657–2665 10.1128/JVI.78.6.2657-2665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. W., Turell M. J., Sardelis M. R., Watts D. M., Coleman R. E., Fernandez R., Carbajal F., Pecor J. E., Calampa C., Klein T. A. (2004). Seasonal distribution, biology, and human attraction patterns of culicine mosquitoes (Diptera: Culicidae) in a forest near Puerto Almendras, Iquitos, Peru. J Med Entomol 41, 349–360 10.1603/0022-2585-41.3.349 [DOI] [PubMed] [Google Scholar]
- Joubert D. A., Blasdell K. R., Audsley M. D., Trinidad L., Monaghan P., Dave K. A., Lieu K., Amos-Ritchie R., Jans D. A. & other authors (2013). Bovine ephemeral fever rhabdovirus α1 protein has viroporin-like properties and binds importin β1 and importin 7. J Virol 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin D., Ferron F., Canard B., Longhi S. (2003). Structural disorder and modular organization in Paramyxovirinae N and P. J Gen Virol 84, 3239–3252 10.1099/vir.0.19451-0 [DOI] [PubMed] [Google Scholar]
- Kurz W., Gelderblom H., Flügel R. M., Darai G. (1986). Isolation and characterization of a tupaia rhabdovirus. Intervirology 25, 88–96 10.1159/000149661 [DOI] [PubMed] [Google Scholar]
- Luo M., Green T. J., Zhang X., Tsao J., Qiu S. (2007). Conserved characteristics of the rhabdovirus nucleoprotein. Virus Res 129, 246–251 10.1016/j.virusres.2007.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca J. D., Pitha P. M. (1986). Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol Cell Biol 6, 2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J. L., Madan V., Carrasco L. (2012). Viroporins: structure and biological functions. Nat Rev Microbiol 10, 563–574 10.1038/nrmicro2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan P. L., Williams D. T., Johansen C. A., Jain K., Petrosov A., Diviney S. M., Tashmukhamedova A., Hutchison S. K., Tesh R. B. & other authors (2011). Genetic characterization of K13965, a strain of Oak Vale virus from Western Australia. Virus Res 160, 206–213 10.1016/j.virusres.2011.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi P. A. (2000). MacVector: integrated sequence analysis for the Macintosh. Methods Mol Biol 132, 47–69 [DOI] [PubMed] [Google Scholar]
- Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56, 564–577 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Turell M. J., O’Guinn M. L., Dohm D., Zyzak M., Watts D., Fernandez R., Calampa C., Klein T. A., Jones J. W. (2008). Susceptibility of Peruvian mosquitoes to eastern equine encephalitis virus. J Med Entomol 45, 720–725 10.1603/0022-2585(2008)45[720:SOPMTE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Vasilakis N., Widen S., Mayer S. V., Seymour R., Wood T. G., Popov V., Guzman H., Travassos da Rosa A. P., Ghedin E. & other authors (2013). Niakha virus: a novel member of the family Rhabdoviridae isolated from phlebotomine sandflies in Senegal. Virology 444, 80–89 10.1016/j.virol.2013.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P. J., Kongsuwan K. (1999). Deduced structural model for animal rhabdovirus glycoproteins. J Gen Virol 80, 1211–1220 [DOI] [PubMed] [Google Scholar]
- Walker P. J., Dietzgen R. G., Joubert D. A., Blasdell K. R. (2011). Rhabdovirus accessory genes. Virus Res 162, 110–125 10.1016/j.virusres.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D. M., Ramirez G., Cabezas C., Wooster M. T., Carrillo C., Chuy M., Gentrau E. J., Hayes C. G. (1998). Arthropod-borne viral diseases in Peru. In An Overview of Arbovirology in Brazil and Neighbouring Countries, pp. 193–218 Edited by da Rosa A. P. A. T., Vasconcelos P. F. C., da Rosa J. F. S. T. Belem: Instituto Evandro Chagas [Google Scholar]
- Wirblich C., Tan G. S., Papaneri A., Godlewski P. J., Orenstein J. M., Harty R. N., Schnell M. J. (2008). PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J Virol 82, 9730–9738 10.1128/JVI.00889-08 [DOI] [PMC free article] [PubMed] [Google Scholar]