Abstract
Some respiratory tract infections remain unexplained despite extensive testing for common pathogens. Nasopharyngeal aspirates (NPAs) from 120 Chilean infants from Santiago with acute lower respiratory tract infections were analysed by viral metagenomics, revealing the presence of nucleic acids from anelloviruses, adenovirus-associated virus and 12 known respiratory viral pathogens. A single sequence read showed translated protein similarity to cycloviruses. We used inverse PCR to amplify the complete circular ssDNA genome of a novel cyclovirus we named CyCV-ChileNPA1. Closely related variants were detected using PCR in the NPAs of three other affected children that also contained anelloviruses. This report increases the current knowledge of the genetic diversity of cycloviruses whose detection in multiple NPAs may reflect a tropism for human respiratory tissues.
Cycloviruses, members of a proposed genus within the family Circoviridae, have a circular ssDNA genome of approximately 2 kb (Li et al., 2010). Genetically highly diverse cycloviruses were initially found in the faeces of Pakistani children with and without acute flaccid paralysis (Victoria et al., 2009), in wild chimpanzees (Li et al., 2010) and in tissues of farm animals including cows, goats, bats and chickens (Ge et al., 2011; Li et al., 2010, 2011). Unexpectedly, other cyclovirus species have also been detected in insects, namely dragonflies and cockroaches (Dayaram et al., 2013; Padilla-Rodriguez et al., 2013; Rosario et al., 2011). In 2013, a cyclovirus species (CyCV-CN) was found initially using viral metagenomics and then by PCR in 4 % of cerebrospinal fluid (CSF) specimens from Vietnamese children with unexplained central nervous system disorder, but not in CSF from patients with non-neurological problems, as well as in 4.2 % of faeces from healthy Vietnamese children (Tan et al., 2013). CyCV-CN DNA was also detected in a throat swab (Tan et al., 2013). In this study, 58 % of faecal specimens from pigs and poultry in Vietnam were also positive for the same cyclovirus, suggesting possible sources of human infection (Tan et al., 2013). A related cyclovirus was also detected in 10 % of CSF samples and 15 % of serum samples from adult patients with paraplegia (leg paralysis) from Malawi (Smits et al., 2013).
Nasopharyngeal aspirates (NPAs) from Chilean children less than 2 years old with acute lower respiratory infections were tested for respiratory syncytial virus (RSV), adenovirus, parainfluenza virus 1–3 and influenza A and B viruses by indirect immunofluorescence assays and virus isolation (Avendaño et al., 2003). From 1998 to 2000, a mean of 29 % of acute lower respiratory infections samples were positive for RSV (Avendaño et al., 2003). To initiate the characterization of the viruses in non-reactive NPA samples, viral particles were enriched by filtration, and unprotected DNA and RNA were digested using a combination of nuclease enzymes (Victoria et al., 2009). The remaining nucleic acids were then extracted using a MagMAX Viral RNA Isolation kit (Life Technologies), which recovers both RNA and DNA. A DNA library was constructed using a ScriptSeq v2 RNA-Seq Library Preparation kit (Epicentre), which amplifies both RNA and DNA, and sequenced using the Illumina MiSeq platform. Viral sequences were identified using translated protein sequence similarity searches to annotated viral proteins available in GenBank (using blastx) and results were mapped using the NCBI Virus Taxonomy browser (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?name=Viruses). The study was approved by the University of California at San Francisco committee on human research.
A total of 120 respiratory specimens from Chilean infants were analysed in 12 pools of 10 specimens using one Illumina MiSeq run of 250 base paired-ends. Viral sequence reads were identified with amino acid similarity >95 % to known viruses. Most numerous sequences were from anelloviruses, followed by enterovirus C, betacoronavirus, bocavirus 1, RSV, human adenovirus 3, enterovirus B, human rhinoviruses A and C, human parainfluenza 3, adeno-associated virus, human pneumovirus, human rhinovirus B and human parechovirus sequences (Table 1). Anelloviruses were identified in all but one pool (Table 1). Anelloviruses have been reported previously in human respiratory secretions (Burián et al., 2011; Jartti et al., 2012). Anelloviruses are usually considered commensal viruses (Okamoto, 2009a), although increased prevalence was found in bronchoalveolar lavage of children with acute exacerbation idiopathic pulmonary fibrosis (Wootton et al., 2011) and acute respiratory diseases (Maggi et al., 2003), and in lung tissues of pigs infected with known respiratory pathogens (Rammohan et al., 2012). Anellovirus plasma load is also ncreased in advanced AIDS (Li et al., 2013) and in immunosuppressed patients following organ transplantation (De Vlaminck et al., 2013). Increased anellovirus loads may reflect increased replication in immune cells stimulated by chronic inflammation, rather than indicating a direct pathogenic role (De Vlaminck et al., 2013). Detection of RSV, parainfluenza 3 and adenovirus in four, two and one pools, respectively, was probably the result of viral loads being too low for detection by immunofluorescence assays and cell culture (Avendaño et al., 2003). Except for anelloviruses and adeno-associated virus, all other viruses found have been associated with respiratory symptoms.
Table 1. Distribution of sequence reads to different viral types/species in 12 NPA pools from Chile.
| Virus | NPA pool no. and no. of reads | Total reads | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| 1 246 305 | 1 381 713 | 1 561 246 | 1 775 509 | 1 136 293 | 1 383 673 | 1 327 383 | 1 139 854 | 664 165 | 621 987 | 1 033 709 | 1 462 032 | ||
| Anelloviruses | 247 | 1115 | 605 | 43 | 81 | 907 | 950 | 575 | 58 | 123 | 645 | 5347 | |
| Enterovirus C | 1759 | 772 | 2531 | ||||||||||
| Beta coronavirus | 1198 | 1198 | |||||||||||
| Bocavirus 1 | 326 | 12 | 116 | 454 | |||||||||
| Respiratory syncytial virus | 10 | 13 | 13 | 119 | 4 | 159 | |||||||
| Human adenovirus 3 | 93 | 93 | |||||||||||
| Enterovirus B | 85 | 85 | |||||||||||
| Human rhinovirus A | 5 | 10 | 11 | 11 | 37 | ||||||||
| Human rhinovirus C | 8 | 2 | 2 | 12 | |||||||||
| Human parainfluenza 3 | 2 | 8 | 10 | ||||||||||
| Adeno-associated virus | 10 | 10 | |||||||||||
| Human metapneumovirus | 6 | 2 | 8 | ||||||||||
| Human rhinovirus B | 4 | 4 | |||||||||||
| Human parechovirus | 2 | 2 | 4 | ||||||||||
| Cyclovirus | 1 | 1 | |||||||||||
One sequence from one sample pool showed significant similarity to cyclovirus proteins (blastx E-score of 2×10−7 to dragonfly cyclovirus, GenBank accession no. KC512919). The full circular cyclovirus genome, referred to as CyCV-ChileNPA1, was then amplified using inverse PCR with specific primers designed from the Illumina-derived short sequence and directly Sanger sequenced by primer walking. Putative ORFs in the cyclovirus genome were predicted using the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/) requiring an initiation Met codon.
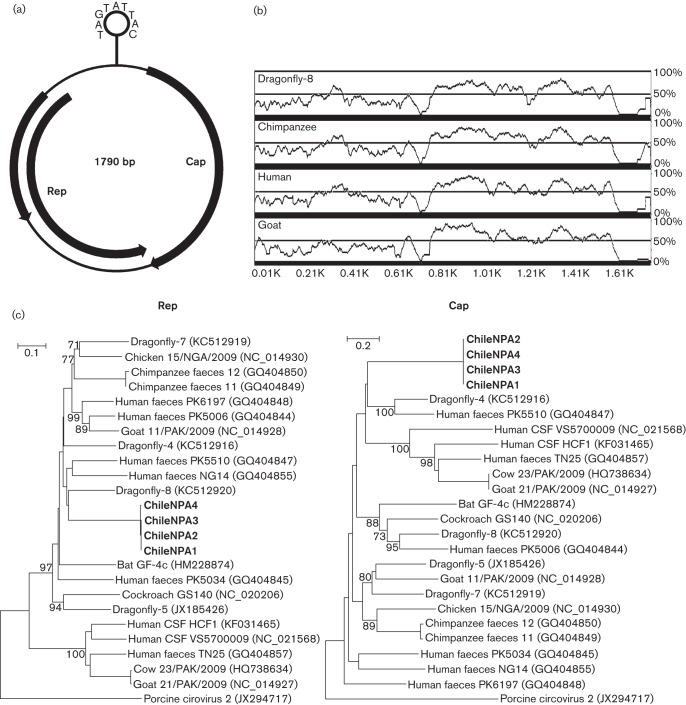
The complete circular genome of CyCV-ChileNPA1 was 1790 bases, with a G+C content of 43 mol%. This genome consisted of two major ORFs encoding replication-associated protein (Rep) and capsid protein (Cap). The intergenic region was 247 bases and encoded a putative stem–loop structure with a stem length of 13 bases, predicted by the Mfold program (Zuker, 2003). Similarly to other cycloviruses, the highly conserved nonamer (TAGTATTAC) was found in the loop (Fig. 1a).
Fig. 1.
Details of the novel cyclovirus CyCV-ChileNPA1. (a) Genome organization and its stem–loop structure. The locations of the putative rep and cap genes are indicated by arrows. (b) Pairwise sequence alignments of CyCV-ChileNPA1 with its closest relatives. The sequence nucleotide similarity (%) is indicated by the height of each point along the y-axis. The x-axis shows the nucleotide positions in the complete genome. (c) Phylogenetic trees generated with Rep and Cap proteins (concatenated) of cycloviruses. Bars, amino acid substitutions per position.
The International Committee on the Taxonomy of Viruses has also proposed a threshold of 75 % nucleotide identity over the entire genome and 70 % amino acid identity for the capsid protein. Rep showed the closest match (65 %) to that of dragonfly CyV-8, whilst Cap shared a lower identity of 30 % with that of the same virus. The higher level of nucleotide divergence of cap relative to rep was also observed with sequence alignments of CyCV-ChileNPA1 with its closest relatives (Fig. 1b). Such a high level of sequence divergence indicated that CyCV-ChileNPA1 may be considered a new species within the genus Cyclovirus.
CyCV-ChileNPA1 shared conserved Rep motifs (Dayaram et al., 2013; Rosario et al., 2012). Analysis of the deduced amino acid Rep sequences of CyCV-ChileNPA1 revealed three rolling-circle replication motifs I–III: FTxNN (FTWHD), YCSKxGX (YCSKSGE) and HLQGxxNL (HLQGFCSL), respectively (Fig. 2). In the N terminus of cyclovirus Rep, two consensus high-affinity DNA-binding specificity determinants (SPDs), TxR for SPD-region 1 and PxR for SPD-region 2, were present (Dayaram et al., 2013; Londoño et al., 2010). CyCV-ChileNPA1 showed a mutated VxR for SPD-region 1 of unknown functional consequence (Fig. 2). The C-terminal region of the CyCV-ChileNPA1 Rep protein possessed ATP-dependent helicase motifs Walker A, B and C, or GxxGTGKS (GPPGTGKS), VIIDDFYGW and ITSN, respectively (Fig. 2).
Fig. 2.

Alignment of Rep proteins of the newly identified CyCV-ChileNPA1 and dragonfly cyclovirus-8. Conserved motifs are shown within boxes. SPD-r, specificity determinant region. White boxes show RCR and shaded boxes show helicase motifs.
Sequence alignment was performed using clustal_x (Saitou & Nei, 1987). A phylogenetic tree with 100 bootstrap resamples of the alignment datasets was generated using mega5 and the neighbour-joining method (Tamura et al., 2011). Bootstrap values (based on 100 replicates) for each node are given for values >70 % (Fig. 1c). Phylogenetic analysis confirmed the presence of a highly diverse cyclovirus species.
To determine the prevalence of this virus, a nested PCR assay was designed and used to test all 120 NPA samples. Primers ChileNPA-F1 (5′-TGGGTCAGGCTATTACTGGGAG-3′) and ChileNPA-R1 (5′-ACTGAATGTCCGTCCGTTGTCC-3′) were used for the first round of PCR, and primers ChileNPA-F2 (5′-CAGTGCCATAGTACAGAGTGCCCA-3′) and ChileNPA-R2 (5′-CTCCCCTACTCAAAGAACTCGCCT-3′) for the second round of PCR, resulting in an expected amplicon of ~310 bp. The PCR conditions were as follows: denaturation at 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 53 or 55 °C (for the first or second round, respectively) for 30 s and 72 °C for 1 min, a final extension at 72 °C for 10 min, and then held at 4 °C. Amplicons were then sequenced directly for identification. Three additional cases (CyCV-ChileNPA2–4) were positive for the new cyclovirus, yielding a prevalence of 3.3 % in the studied population (4/120). The full genomes of these three viruses were then acquired by overlapping PCR. The genomic sequences of CyCV-ChileNPA2–4 shared a high nucleotide identity of >99 %, showing two, five and six nucleotide mutations compared with the CyCV-ChileNPA1 genome, respectively. All of these point mutations were synonymous except R25S and K93E in the ORF of CyCV-ChileNPA4 Rep.
The four CyCV-ChileNPA PCR-positive samples were then reanalysed using the same metagenomics approach but individually tagged to identify other viruses in these four samples. A total of 2697 unique sequence reads were generated. Three unique CyCV-ChileNPA1 reads were generated from the NPA sample in which it was originally detected. No other viral sequences were detected. A total of 6362 unique reads were also generated from the other three samples positive only by PCR for the cyclovirus. All three samples contained anellovirus sequences (a total of 456 reads) and no other close matches to mammalian viruses. Anelloviruses are highly prevalent viruses present in many anatomical sites of different mammals (Okamoto, 2009b) and are generally considered commensal infections. Anelloviruses have also been found in a significant minority of cases of idiopathic pulmonary fibrosis and in cases of acute lung injury (Wootton et al., 2011), are at higher prevalence in plasma and nasopharyngeal samples of febrile versus non-febrile cases (McElvania TeKippe et al., 2012) and are generally increased in the plasma of immunosuppressed individuals such as advanced AIDS patients (Li et al., 2013) or transplant recipients (De Vlaminck et al., 2013). A porcine anellovirus (torque teno sus virus species 1) has also been associated with porcine respiratory disease complex where it might exacerbate infections caused by porcine circovirus 2 and the arterivirus porcine reproductive and respiratory disease symptom virus (Rammohan et al., 2012).
The detection of cyclovirus DNA in different human samples, including faeces, blood and CSF, and in the muscle tissues of farm animals suggests that cycloviruses may cause systemic infections in mammals (Li et al., 2010; Smits et al., 2013; Tan et al., 2013). The detection of cyclovirus DNA in NPAs (upper respiratory tract) of children with lower tract respiratory problems raises the possibility of a role for these viruses in respiratory illnesses. Further investigations of the host and tissue tropism, the transmission route(s) and any physiological consequences of human cyclovirus infections and possible interactions with anelloviruses are required.
Acknowledgements
We acknowledge NHLBI grant R01 HL105770 to E. L. D. and the Blood Systems Research Institute for sustained support.
References
- Avendaño L. F., Palomino M. A., Larrañaga C. (2003). Surveillance for respiratory syncytial virus in infants hospitalized for acute lower respiratory infection in Chile (1989 to 2000). J Clin Microbiol 41, 4879–4882 10.1128/JCM.41.10.4879-4882.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burián Z., Szabó H., Székely G., Gyurkovits K., Pankovics P., Farkas T., Reuter G. (2011). Detection and follow-up of torque teno midi virus (‘small anelloviruses’) in nasopharyngeal aspirates and three other human body fluids in children. Arch Virol 156, 1537–1541 10.1007/s00705-011-1021-0 [DOI] [PubMed] [Google Scholar]
- Dayaram A., Potter K. A., Moline A. B., Rosenstein D. D., Marinov M., Thomas J. E., Breitbart M., Rosario K., Argüello-Astorga G. R., Varsani A. (2013). High global diversity of cycloviruses amongst dragonflies. J Gen Virol 94, 1827–1840 10.1099/vir.0.052654-0 [DOI] [PubMed] [Google Scholar]
- De Vlaminck I., Khush K. K., Strehl C., Kohli B., Luikart H., Neff N. F., Okamoto J., Snyder T. M., Cornfield D. N. & other authors (2013). Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155, 1178–1187 10.1016/j.cell.2013.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li J., Peng C., Wu L., Yang X., Wu Y., Zhang Y., Shi Z. (2011). Genetic diversity of novel circular ssDNA viruses in bats in China. J Gen Virol 92, 2646–2653 10.1099/vir.0.034108-0 [DOI] [PubMed] [Google Scholar]
- Jartti T., Jartti L., Ruuskanen O., Söderlund-Venermo M. (2012). New respiratory viral infections. Curr Opin Pulm Med 18, 271–278 10.1097/MCP.0b013e328351f8d4 [DOI] [PubMed] [Google Scholar]
- Li L., Kapoor A., Slikas B., Bamidele O. S., Wang C., Shaukat S., Masroor M. A., Wilson M. L., Ndjango J. B. & other authors (2010). Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84, 1674–1682 10.1128/JVI.02109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shan T., Soji O. B., Alam M. M., Kunz T. H., Zaidi S. Z., Delwart E. (2011). Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol 92, 768–772 10.1099/vir.0.028704-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Deng X., Linsuwanon P., Bangsberg D., Bwana M. B., Hunt P., Martin J. N., Deeks S. G., Delwart E. (2013). AIDS alters the commensal plasma virome. J Virol 87, 10912–10915 10.1128/JVI.01839-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño A., Riego-Ruiz L., Argüello-Astorga G. R. (2010). DNA-binding specificity determinants of replication proteins encoded by eukaryotic ssDNA viruses are adjacent to widely separated RCR conserved motifs. Arch Virol 155, 1033–1046 10.1007/s00705-010-0674-4 [DOI] [PubMed] [Google Scholar]
- Maggi F., Pifferi M., Tempestini E., Fornai C., Lanini L., Andreoli E., Vatteroni M., Presciuttini S., Pietrobelli A. & other authors (2003). TT virus loads and lymphocyte subpopulations in children with acute respiratory diseases. J Virol 77, 9081–9083 10.1128/JVI.77.16.9081-9083.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElvania TeKippe E., Wylie K. M., Deych E., Sodergren E., Weinstock G., Storch G. A. (2012). Increased prevalence of anellovirus in pediatric patients with fever. PLoS ONE 7, e50937 10.1371/journal.pone.0050937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. (2009a). History of discoveries and pathogenicity of TT viruses. Curr Top Microbiol Immunol 331, 1–20 [DOI] [PubMed] [Google Scholar]
- Okamoto H. (2009b). TT viruses in animals. Curr Top Microbiol Immunol 331, 35–52 [DOI] [PubMed] [Google Scholar]
- Padilla-Rodriguez M., Rosario K., Breitbart M. (2013). Novel cyclovirus discovered in the Florida woods cockroach Eurycotis floridana (Walker). Arch Virol 158, 1389–1392 10.1007/s00705-013-1606-x [DOI] [PubMed] [Google Scholar]
- Rammohan L., Xue L., Wang C., Chittick W., Ganesan S., Ramamoorthy S. (2012). Increased prevalence of torque teno viruses in porcine respiratory disease complex affected pigs. Vet Microbiol 157, 61–68 10.1016/j.vetmic.2011.12.013 [DOI] [PubMed] [Google Scholar]
- Rosario K., Marinov M., Stainton D., Kraberger S., Wiltshire E. J., Collings D. A., Walters M., Martin D. P., Breitbart M., Varsani A. (2011). Dragonfly cyclovirus, a novel single-stranded DNA virus discovered in dragonflies (Odonata: Anisoptera). J Gen Virol 92, 1302–1308 10.1099/vir.0.030338-0 [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. (2012). A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol 157, 1851–1871 10.1007/s00705-012-1391-y [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Smits S. L., Zijlstra E. E., van Hellemond J. J., Schapendonk C. M., Bodewes R., Schürch A. C., Haagmans B. L., Osterhaus A. D. (2013). Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010-2011. Emerg Infect Dis [Internet] 19 10.3201/eid1909.130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. V., van Doorn H. R., Nghia H. D., Chau T. T., Tu L. T., de Vries M., Canuti M., Deijs M., Jebbink M. F. & other authors (2013). Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. mBio 4, e00231-13 10.1128/mBio.00231-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651 10.1128/JVI.02301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton S. C., Kim D. S., Kondoh Y., Chen E., Lee J. S., Song J. W., Huh J. W., Taniguchi H., Chiu C. & other authors (2011). Viral infection in acute exacerbation of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 183, 1698–1702 10.1164/rccm.201010-1752OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31, 3406–3415 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]