Abstract
PURPOSE
A recent genome-wide association study reported the novel finding that variants in diacylglycerol kinase kappa (DGKK) were associated with hypospadias. Our objectives were to determine whether this finding could be replicated in a more racially-ethnically diverse study population of California births and to provide a more comprehensive investigation of variants.
METHODS
We examined the association of 27 DGKK SNPs with hypospadias, relative to population-based non-malformed controls born in selected California counties from 1990-2003. Analyses included a maximum of 928 controls and 665 cases (91 mild, 336 moderate, 221 severe, 17 undetermined). Results for mild and moderate cases were similar so they were grouped together.
RESULTS
For mild and moderate cases, odds ratios (OR) for 15 of the 27 SNPs had p-values <0.05; two were <1, and the others ranged from 1.3 to 1.8. Among severe cases, ORs tended to be closer to one and none of the p-values were <0.05. Due to high LD across the SNPs, haplotype analyses were conducted, and two blocks were generated. These analyses identified a set of eight variants that was associated with a three- to four- fold increased risk, relative to the most common haplotype, regardless of severity of the phenotype (the OR was 4.1, p<10-4 for mild to moderate cases and 3.3, p=0.001 for severe cases).
CONCLUSIONS
This study confirms that DGKK variants are associated with hypospadias. Further studies are needed to enable a more thorough investigation of DGKK variability and to delineate the mechanism by which DGKK contributes to urethral development.
Keywords: HYPOSPADIAS, GENES
INTRODUCTION
Hypospadias occurs when the urethral opening is shifted toward the ventral side of the penis. Recently, the first genome-wide association study (GWAS) of hypospadias was published5. The study involved a GWAS of eight pooled DNA samples from 436 Dutch cases, which was followed by individual-level genotyping of 11 SNPs in the discovery and replication samples. All subjects were of European descent. The strongest finding was for the two SNPs that were individually genotyped for DGKK, which encodes diacylglycerol kinase kappa on the X chromosome. The minor alleles (i.e., less frequent) were associated with more than a two-fold increased risk of hypospadias and had a population attributable risk just over 30% in both the discovery and replication samples.
Little is known about DGKK6. Like related diacylglycerol kinases, DGKK phosphorylates diacylglycerol to generate phosphatidic acid within the plasma membrane, thus down-regulating diacylglycerol signaling. Evidence suggests it is expressed in the genitourinary system5-7, but embryonic expression in humans has not to our knowledge been examined.
Our objective was to determine whether the novel association between DGKK variants and hypospadias could be replicated in a racially-ethnically diverse study population of California births. We also aimed to provide a more comprehensive investigation of DGKK variation by including 27 SNPs, versus the two SNPs replicated in the original GWAS. Our hypothesis was that variant DGKK genotypes would be associated with increased hypospadias risk. In addition, we examined whether variants within CCNB3, which is just downstream of DGKK, were associated with hypospadias.
METHODS
The study population included all male infants born from 1990-2003 to mothers who were residents of eight California Central Valley counties and from 1990-1997 to residents of Los Angeles, San Francisco, and Santa Clara counties, reflecting counties where case ascertainment was actively conducted by the California Birth Defects Monitoring Program (CBDMP). CBDMP staff ascertained cases by reviewing medical records at hospitals and genetic centers in relevant counties8.
Cases were classified by severity, based on the reported anatomical position of the urethral opening. Mild cases had a meatus limited to the coronal or glanular penis (British Pediatric Association [BPA] codes 752.605, 752.625), moderate cases had a meatus on the penile shaft, and severe cases had a meatus at the peno-scrotal junction or perineal area (BPA codes 752.606, 752.607, 752.626, 752.627). Assignment of severity was finalized based on review by a medical geneticist (EJL or Dr. Cynthia Curry)9. Cases for which the anatomical position was not sufficiently described (codes 752.600, 752.620) were excluded. Cases with no other anomalies or only minor anomalies (e.g., sacral/pilonidal dimple) were considered “isolated”. Cases with major accompanying anomalies were considered “non-isolated”. Cases classified as having a known single gene disorder or chromosomal abnormality were excluded. Cases with accompanying genital anomalies (e.g., cryptorchidism, bifid scrotum, small penis, ambiguous genitalia, and “other” genital abnormalities, BPA codes 752) but no other major anomalies were classified as isolated.
Cases were linked with birth certificates using identifiers such as name, date of birth and delivery hospital from medical records. They were also linked with archived newborn bloodspots, which served as the source of DNA and available for 667 cases.
The underlying study population included 1,246,172 non-malformed live born male infants eligible for control selection. We randomly selected 931 controls with available bloodspots, in proportion to the underlying birth population for that year, to give an approximate 2:1 ratio of controls to cases from Central Valley counties and a 1:1 ratio from non-Central Valley counties. The ratio differed due to the presence of a secondary on-going study in the Central Valley that allowed for a larger control group.
For all subjects, information on the following covariates was derived from birth certificates: maternal race-ethnicity, education, age, and parity; plurality; and infant birthweight and gestational age at delivery.
Genomic DNA was extracted from dried bloodspots using MasterPure™ Complete DNA and RNA Purification Kit (Epicentre Biotechnologies Madison, WI) and 10 ng genomic DNA was then used for whole genome amplification (Qiagen Repli-g® kit). DGKK tagSNPs that assay the known common SNPs either directly or indirectly via linkage disequilibrium (i.e., via correlation of the occurrence of the SNPs) among measured and unmeasured SNPs were generated using the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). The program provided tagSNPs that cover common variation at r2>0.80 across the DGKK gene for a “cosmopolitan” population, including Hispanics. TagSNPs with minor allele frequencies (i.e., frequency of the less common allele in the study population) greater than 5% were selected. Initially, 37 tagSNPs were identified. To design PCR primers, DGKK sequence was obtained from GeneBank (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene), accession number AL357894.6. This sequence was imported to Sequenom Typer software to design primers to capture and amplify regions within DGKK. Two multiplex genotyping assays were designed that included 25 DGKK SNPs and nine CCNB3 SNPs for the Sequenom Mass ARRAY MALDI-TOF system (primer sequences and reaction conditions are available upon request). For DGKK, the most 5’ SNP (rs4074319) lies within the fifth intron, while 22 other SNPs are within other introns, together with two SNPs from DGKK exons (rs5961179 and rs4143304). Two additional SNPs (rs7063116 and rs1934179) were genotyped using the Illumina GoldenGate platform; they were included so that we could replicate the specific findings reported by the previous hypospadias GWAS5. rs7063116 is outside of the DGKK coding sequence, lying upstream of the coding region, and rs1934179 is intronic. A similar protocol was used to select nine tagSNPs for the MALDI-TOF assay from CCNB3, which is the next gene downstream from DGKK.
We first examined the association of each SNP with risk of hypospadias (analyzed as yes/no for each allele, since DGKK is on the X chromosome). In the interest of retaining as many subjects and SNPs as possible in analyses, we did not exclude SNPs or subjects based on call rates (SNP-specific call rates ranged from 88-98% for cases and 90-96% for controls; average subject-specific call rate was 90% for cases and 92% for controls), with the exception of one control with failed genotyping. We conducted analyses of all cases grouped together as well as separate analyses by severity of phenotype. Second, we examined haplotypes. We used Haploview to define haplotype blocks and their frequencies based on control genotypes 10. To assess which SNPs were driving haplotype associations, we undertook step-wise regression of the corresponding SNPs, using backward elimination and retaining SNPs with p-values <0.20. Maximum likelihood estimates of odds ratios (OR) and corresponding 95% confidence intervals (CI) were calculated from logistic regression models to estimate relative risks. ORs were adjusted for maternal race-ethnicity (Hispanic, non-Hispanic white, other) and for residence in the Central Valley (yes/no), the latter due to the differing case-control ratio based on this variable. We also examined results separately for Hispanics and non-Hispanic whites.
The study was approved by IRBs for the state of California, at Stanford University and Children’s Hospital Oakland.
RESULTS
Mothers of cases were more likely than controls to be non-Hispanic white, have higher education, and be older and nulliparous, and cases were more likely to be low birthweight and delivered early (Table 1). Two cases and two controls with missing race-ethnicity were excluded from SNP-based analyses, such that analyses included 928 controls and a maximum of 665 cases (91 mild, 336 moderate, 221 severe, 17 undetermined). Although we chose tagSNPs based on those with MAF>5% in the Genome Variation Server, 12/25 DGKK tagSNPs had MAF<5% among our California controls (Table 2).
Table 1.
Descriptive characteristics among cases and controls, overall and by phenotype severity.
| Percent (n) | |||||
|---|---|---|---|---|---|
| Controls (n=930) | All Cases (n=667) | Mild Cases (n=91) | Moderate Cases (n=337) | Severe Cases (n=222) | |
| Maternal race-ethnicity | |||||
| White | 30 (281) | 43 (289) | 56 (51) | 52 (174) | 25 (56) |
| Hispanic | 52 (484) | 35 (231) | 31 (28) | 30 (101) | 43 (96) |
| Others | 18 (163) | 22 (145) | 13 (12) | 18 (61) | 31 (69) |
| Unknown | 0 (2) | 0 (2) | 0 (0) | 0 (1) | 1 (1) |
| Maternal education | |||||
| < High school | 39 (363) | 25 (167) | 31 (28) | 23 (76) | 27 (59) |
| High school | 31 (288) | 27 (180) | 32 (29) | 27 (90) | 24 (54) |
| > High school | 29 (272) | 48 (317) | 36 (33) | 51 (170) | 49 (108) |
| Unknown | 1 (7) | 1 (3) | 1 (1) | 0 (1) | 1 (1) |
| Maternal age | |||||
| < 25 years | 46 (423) | 30 (197) | 37 (34) | 28 (93) | 29 (64) |
| 25-34 years | 43 (404) | 52 (349) | 52 (47) | 53 (178) | 52 (115) |
| 35 or more years | 11 (103) | 18 (121) | 11 (10) | 20 (66) | 19 (43) |
| Number of previous live births | |||||
| 0 | 36 (332) | 52 (350) | 43 (39) | 48 (163) | 62 (138) |
| 1 | 32 (299) | 26 (173) | 31 (28) | 28 (95) | 21 (47) |
| ≥ 2 | 32 (299) | 21 (142) | 26 (24) | 23 (78) | 16 (36) |
| Unknown | 0 (0) | 0 (2) | 0 (0) | 0 (1) | 1 (1) |
| Infant birthweight | |||||
| ≤ 2500 g | 5 (48) | 31 (205) | 21 (19) | 21 (69) | 49 (109) |
| > 2500 g | 95 (882) | 69 (462) | 79 (72) | 80 (268) | 51 (113) |
| Gestational age at delivery | |||||
| < 37 weeks | 8 (73) | 23 (152) | 22 (20) | 18 (60) | 31 (69) |
| ≥ 37 weeks | 88 (817) | 74 (492) | 75 (68) | 80 (268) | 64 (143) |
| Unknown | 4 (40) | 4 (23) | 3 (3) | 3 (9) | 5 (10) |
| Maternal residence in Central Valley | |||||
| No | 45 (415) | 62 (415) | 0 (0) | 73 (246) | 73 (163) |
| Yes | 55 (515) | 38 (252) | 100 (91) | 27 (91) | 27 (59) |
Table 2.
Association of DGKK SNPs with hypospadias, among all cases and by phenotype severity.
| All cases | Mild & moderate cases | Severe cases | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rs number a | Minor allele | MAF among controls | Percent of cases / controls with genotype (call rates) | No. of cases with minor / major allele | No. of controls with minor / major allele | OR (95% CI) a | P value | OR (95% CI) b | P value | OR (95% CI) b | P value |
| 1 | rs4074319 | T | 2.5 | 89/92 | 8 / 583 | 21 / 834 | 0.4 (0.2 -1.0) | 0.04 | 0.6 (0.2 -1.5) | 0.28 | 0.2 (0.1 -1.0) | 0.05 |
| 2 | rs7879090 | G | 0.5 | 89/92 | 2/589 | 4/849 | 0.5 (0.1 -2.9) | 0.45 | 1.1 (0.2 -6.0) | 0.95 | N.C | N.C |
| 3 | rs5961179 | G | 36.7 | 88/92 | 240/348 | 312/538 | 1.3 (1.1 -1.7) | 0.01 | 1.5 (1.1 -1.9) | 0.003 | 1.0 (0.7 -1.4) | 0.87 |
| 4 | rs7882950 | T | 35.3 | 90/93 | 229/367 | 305/559 | 1.3 (1.1 -1.7) | 0.02 | 1.4 (1.1 -1.9) | 0.01 | 1.0 (0.7 -1.5) | 0.87 |
| 5 | rs12556919 | T | 3.9 | 94/93 | 40/587 | 34/830 | 1.4 (0.9 -2.3) | 0.16 | 1.8 (1.1 -3.0) | 0.03 | 0.8 (0.3 -2.0) | 0.57 |
| 6 | rs12012084 | G | 1.6 | 90/92 | 6/590 | 14/844 | 0.5 (0.2 -1.3) | 0.14 | 0.6 (0.2 -1.9) | 0.40 | 0.3 (0.1 -1.5) | 0.15 |
| 7 | rs17003341 | T | 34.7 | 90/92 | 236/360 | 296/556 | 1.4 (1.1 -1.8) | 0.003 | 1.5 (1.2 -2.0) | 0.001 | 1.1 (0.8 -1.6) | 0.58 |
| 8 | rs1320573 | A | 1.3 | 89/93 | 8/585 | 11/848 | 0.8 (0.3 -2.2) | 0.73 | 0.8 (0.2 -2.6) | 0.73 | 0.9 (0.3 -3.0) | 0.87 |
| 9 | rs17003346 | G | 0.1 | 90/93 | 0/596 | 1/861 | N.C | N.C | N.C | N.C | N.C | N.C |
| 10 | rs1934190 | G | 46.7 | 89/92 | 302/288 | 400/456 | 1.4 (1.1 -1.8) | 0.003 | 1.6 (1.2 -2.1) | 0.0003 | 1.0 (0.7 -1.5) | 0.80 |
| 11 | rs4143304 | T | 47.4 | 89/92 | 322/273 | 406/450 | 1.5 (1.2 -1.8) | 0.001 | 1.8 (1.4 -2.3) | <10-4 | 1.0 (0.7 -1.4) | 0.83 |
| 12 | rs1934188 | T | 43.4 | 89/92 | 284/310 | 370/482 | 1.4 (1.1 -1.8) | 0.003 | 1.6 (1.2 -2.1) | 0.0004 | 1.1 (0.8 -1.6) | 0.54 |
| 13 | rs17003348 | C | 2.9 | 90/93 | 23/575 | 25/841 | 1.1 (0.6 -2.0) | 0.72 | 1.3 (0.7 -2.5) | 0.43 | 0.9 (0.4 -2.1) | 0.76 |
| 14 | rs7888440 | C | 0.7 | 92/91 | 2/609 | 6/834 | 0.4 (0.1 -2.0) | 0.25 | 0.3 (0.0 -2.9) | 0.32 | N.C | N.C |
| 15 | rs7877459 | G | 0.8 | 89/92 | 5/584 | 7/846 | 0.9 (0.3 -2.9) | 0.87 | 0.3 (0.0 -2.8) | 0.32 | 1.6 (0.4 -5.9) | 0.46 |
| 16 | rs17328236 | G | 30.9 | 89/92 | 211/381 | 263/587 | 1.5 (1.2 -1.9) | 0.001 | 1.7 (1.3 -2.2) | 0.0002 | 1.1 (0.8 -1.6) | 0.58 |
| 17 | rs9969978 | C | 43.8 | 88/91 | 286/296 | 370/474 | 1.4 (1.1 -1.8) | 0.002 | 1.7 (1.3 -2.2) | <10-4 | 1.1 (0.8 -1.5) | 0.69 |
| 18 | rs5961182 | G | 0.7 | 88/92 | 3/585 | 6/846 | 0.9 (0.2 -3.8) | 0.88 | 1.0 (0.2 -5.2) | 0.98 | 1.0 (0.1 -8.4) | 0.97 |
| 19 | rs12171755 | T | 36.7 | 89/93 | 201/390 | 316/545 | 1.0 (0.8 -1.3) | 0.93 | 1.1 (0.9 -1.4) | 0.43 | 0.8 (0.5 -1.1) | 0.18 |
| 20 | rs1934183 | T | 49.8 | 88/91 | 254/334 | 422/425 | 0.7 (0.6 -0.9) | 0.001 | 0.6 (0.4 -0.8) | <10-4 | 1.0 (0.7 -1.4) | 0.91 |
| 21 | rs1934170 | T | 0.9 | 94/93 | 6/618 | 8/852 | 0.9 (0.3 -2.6) | 0.81 | 0.9 (0.2 -3.3) | 0.82 | 0.9 (0.2 -3.6) | 0.88 |
| 22 | rs1934179 c | T | 48.3 | 97/93 | 353/289 | 418/447 | 1.4 (1.2 -1.8) | 0.001 | 1.8 (1.4 -2.3) | <10-4 | 0.9 (0.7 -1.3) | 0.74 |
| 23 | rs6614511 | T | 6.5 | 93/92 | 20/600 | 56/798 | 0.5 (0.3 -0.9) | 0.01 | 0.3 (0.1 -0.7) | 0.004 | 0.8 (0.4 -1.6) | 0.60 |
| 24 | rs1934184 | G | 0.6 | 89/93 | 1/594 | 5/855 | 0.2 (0.0 -2.1) | 0.20 | 0.4 (0.0 -3.8) | 0.44 | N.C | N.C |
| 25 | rs5961183 | C | 44.3 | 92/92 | 308/307 | 378/476 | 1.4 (1.1 -1.7) | 0.005 | 1.6 (1.2 -2.0) | 0.0004 | 1.0 (0.7 -1.4) | 0.99 |
| 26 | rs7876567 | T | 43.0 | 89/91 | 283/309 | 363/481 | 1.3 (1.1 -1.7) | 0.01 | 1.5 (1.2 -1.9) | 0.002 | 1.0 (0.7 -1.4) | 0.90 |
| 27 | rs7063116 c | A | 42.5 | 97/96 | 290/358 | 379/512 | 1.2 (1.0 -1.5) | 0.07 | 1.3 (1.0 -1.7) | 0.02 | 0.9 (0.7 -1.3) | 0.73 |
SNPs are ordered from 5’ to 3’ across the gene.
ORs compare the minor allele with the major allele as reference and were adjusted for maternal race-ethnicity and residence in the Central Valley.
Two SNPs selected from van der Zanden et al. 5.
MAF = minor allele frequency (i.e., frequency of the allele that is less common in the study population), NC = not calculated
With all cases grouped together, ORs for 14/27 DGKK SNPs had p-values <0.05; three of these ORs were less than one, and the other 11 ranged from 1.3-1.5 (Table 2). Among these 14 SNPs with p-values <0.05, all but one had MAF>5%. Results for mild and moderate cases were similar, so they were analyzed together (data not shown). Among mild/moderate cases, the pattern was similar to that overall, but with ORs tending to be slightly farther from 1.0; i.e., of 15 SNPs with p-values <0.05, two were <1, and the others ranged from 1.3-1.8. For severe cases, ORs tended to be closer to one, and no p-values were <0.05 (see Table 2).
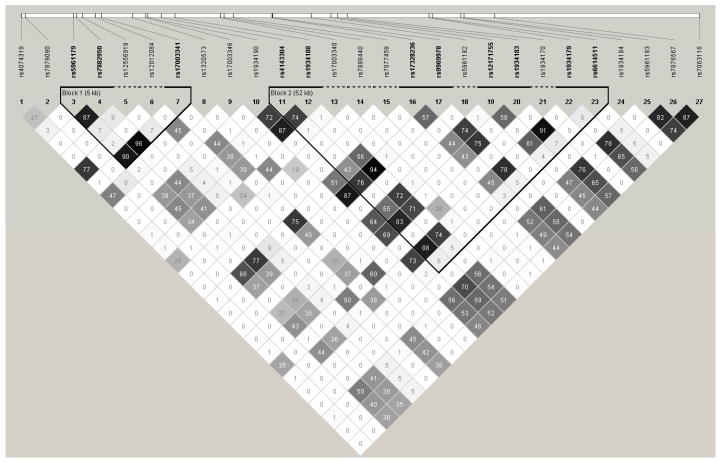
The similarity in results across SNPs reflected their being in high linkage disequilibrium: 21 of the R-squared values were >0.8 (Figure 1). Based on this, two haplotype blocks were generated. Block 1 contained three SNPs (Table 3, Figure 1). Relative to the ACC haplotype (i.e., the most common haplotype, which reflects having the major allele for all three SNPs in the block), the GTT haplotype was associated with modestly increased risk of mild and moderate hypospadias (OR 1.5, p=0.004); the OR for severe cases was 1.0. Results suggested an increased risk for the GCT haplotype among mild to moderate cases (OR 2.9, p=0.07).
Figure 1.
Pair-wise R-squared values for DGKK SNPs and indication of SNPs in haplotype blocks. a
a The squares indicate the R2 values between pairs of SNPs (×100), among control subjects only; the rs numbers in bold and above the solid lines were part of the noted haplotype blocks.
Table 3.
Association of DGKK haplotypes with hypospadias, among all cases and by phenotype severity. a
| Controls (n=841) | All cases (n=587) | Mild and moderate cases (n=382) | Severe cases (n=190) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Block 1 Haplotypes b | Percent | Percent | OR (95% CI) | P value | Percent | OR (95% CI) | P value | Percent | OR (95% CI) | P value |
| ACC | 62.9 | 58.8 | Reference | – | 56.0 | Reference | – | 64.2 | Reference | |
| GTT | 34.0 | 37.7 | 1.4 (1.1-1.7) | 0.01 | 40.1 | 1.5 (1.1-1.9) | 0.004 | 32.6 | 1.0 (0.7-1.5) | 0.81 |
| GCC | 2.3 | 1.5 | 0.6 (0.2-1.3) | 0.17 | 1.8 | 0.9 (0.3-2.2) | 0.76 | 1.1 | 0.2 (0.1-1.1) | 0.06 |
| GCT | 0.7 | 1.7 | 2.1 (0.7-6.2) | 0.17 | 1.8 | 2.9 (0.9-9.1) | 0.07 | 1.6 | 1.5 (0.3-6.7) | 0.61 |
| Others c | 0.1 | 0.3 | N.C. | N.C. | 0.3 | N.C. | N.C. | 0.5 | N.C. | N.C. |
| Controls (n=734) | All cases (n=528) | Mild and moderate cases (n=342) | Severe cases (n=174) | |||||||
| Block 2 Haplotypes d | Percent | Percent | OR (95% CI) | P value | Percent | OR (95% CI) | P value | Percent | OR (95% CI) | P value |
| 01-CCAGCTCA | 42.1 | 39.4 | Reference | – | 36.6 | Reference | – | 44.3 | Reference | |
| 02-TTGCTGTA | 25.3 | 24.4 | 1.2 (0.9-1.7) | 0.17 | 26.9 | 1.5 (1.0-2.1) | 0.03 | 19.5 | 0.8 (0.5-1.3) | 0.41 |
| 03-TTACTGTA | 9.8 | 8.9 | 1.1 (0.7-1.6) | 0.73 | 9.1 | 1.2 (0.8-2.0) | 0.40 | 8.6 | 0.9 (0.5-1.7) | 0.78 |
| 04-CCAGCTCT | 7.1 | 3.2 | 0.5 (0.3-1.0) | 0.05 | 1.2 | 0.3 (0.1-0.8) | 0.01 | 7.5 | 0.8 (0.4-1.6) | 0.53 |
| 05-TCAGCGTA | 4.5 | 5.1 | 1.0 (0.6-1.8) | 0.90 | 5.3 | 1.4 (0.7-2.6) | 0.35 | 4.6 | 0.5 (0.2-1.3) | 0.17 |
| 06-TTGCCGTA | 4.1 | 11.4 | 3.7 (2.2-6.1) | <10-4 | 12.6 | 4.1 (2.4-7.1) | <10-4 | 9.8 | 3.3 (1.6-6.6) | 0.001 |
| 07-TTACCGTA | 1.9 | 2.8 | 1.8 (0.8-3.9) | 0.15 | 3.2 | 2.4 (1.0-5.8) | 0.04 | 2.3 | 1.2 (0.4-4.1) | 0.77 |
| 08-CCAGCGCA | 1.5 | 1.3 | 0.9 (0.3-2.5) | 0.86 | 1.8 | 1.4 (0.5-3.9) | 0.57 | 0.6 | 0.4 (0.1-3.2) | 0.37 |
| Others c | 3.7 | 3.4 | N.C. | N.C. | 3.5 | N.C. | N.C. | 2.9 | N.C. | N.C. |
ORs were adjusted for maternal race-ethnicity and residence in the Central Valley.
Block 1 SNPs: 3, 4, 7 (see Table 2).
“Others” includes any haplotype for which the frequency was less than 1% among cases and controls.
Block 2 SNPs: 11, 12, 16, 17, 19, 20, 22, 23 (see Table 2).
N.C. = not calculated
Block 2 contained eight SNPs (its haplotypes are referred to as HT01-08 for simplicity) (Table 3, Figure 1). HT01 was the most common and reflected the major allele for each SNP except SNP20 (rs1934183). HT02 was the next most common and reflected the minor allele for each SNP except SNP20 and SNP23 (rs6614511); in SNP-specific analyses, the minor alleles for these two SNPs were associated with ORs <1.0 (Table 2). HT02 was associated with modestly increased risk of mild to moderate hypospadias (OR 1.5, p=0.03). HT04 was associated with a decreased risk for mild to moderate hypospadias, (OR 0.3, p=0.01). It differed from HT01 on only one SNP, SNP23. HT06 was associated with increased risk (OR 4.1, p<10-4 for mild to moderate cases and 3.3, p=0.001 for severe cases).
We conducted a step-wise regression analysis of the SNPs in each haplotype block. After backward elimination, the minor allele of SNP7 from Block 1 (rs17003341) was associated with increased risk of hypospadias, and SNP19 from Block 2 (rs12171755) was associated with reduced risk, regardless of phenotype severity (Table 4). In addition, from Block 2, SNP12 was associated with increased risk of severe hypospadias, and SNP23 was associated with reduced risk of mild to moderate hypospadias.
Table 4.
Association of DGKK haplotype block SNPs with hypospadias, among all cases and by phenotype severity, based on results from backward step-wise regression models. a
| SNP | rs number | All cases | Moderate & mild cases | Severe cases | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-value | OR (95% CI) | p-value | OR (95% CI) | p-value | |||
| Block 1c | 3 | rs5961179 | 0.5 (0.2-1.2) | 0.13 | removed | 0.3 (0.1-1.0) | 0.05 | |
| 4 | rs7882950 | removed | 0.4 (0.1-1.4) | 0.16 | removed | |||
| 7 | rs17003341 | 2.5 (1.1-5.7) | 0.02 | 3.3 (1.1-10.0) | 0.03 | 3.9 (1.1-14.0) | 0.04 | |
| Block 2c | 11 | rs4143304 | removed | removed | removed | |||
| 12 | rs1934188 | 1.6 (0.9-2.8) | 0.14 | removed | 2.3 (1.3-4.0) | 0.004 | ||
| 16 | rs17328236 | 1.3 (0.9-2.0) | 0.17 | removed | removed | |||
| 17 | rs9969978 | removed | 1.9 (1.0-3.5) | 0.05 | removed | |||
| 19 | rs12171755 | 0.5 (0.3-0.7) | 0.0001 | 0.4 (0.3-0.7) | 0.0003 | 0.4 (0.2-0.7) | 0.001 | |
| 20 | rs1934183 | removed | removed | removed | ||||
| 22 | rs1934179 | 1.4 (0.9-2.2) | 0.19 | 1.7 (1.0-3.0) | 0.07 | removed | ||
| 23 | rs6614511 | 0.6 (0.3-1.0) | 0.06 | 0.3 (0.1-0.8) | 0.02 | removed | ||
SNPs were selected from backward step-wise logistic regression models conducted separately for Block 1 and Block 2 SNPs; SNPs were removed if their p-value was >0.20; models included maternal race-ethnicity and residence in the Central Valley; ORs represent risks associated with the minor allele versus the major (most common) allele (see Table 2).
See Table 2 for explanation of blocks.
We also examined results separately for Hispanics and non-Hispanic whites (Supplemental Tables 1-3). SNP-specific results tended to be somewhat stronger for whites than Hispanics when examining mild and moderate cases, and haplotype results were generally similar.
Analyses of nine variants in CCNB3, a gene just downstream of DGKK, did not show association with hypospadias (Supplemental Table 4).
DISCUSSION
This study confirmed the previous novel finding of an association of DGKK with hypospadias, in a more racially/ethnically diverse study population, and with a more in-depth assessment of variation across the gene. Our study suggested that several DGKK SNPs were associated with modestly increased risk of hypospadias (<2-fold), especially mild to moderate phenotypes. Haplotype analyses identified eight variants associated with closer to a three- to four- fold increased risk, relative to the most common haplotype, regardless of severity of the phenotype. Thus, our study confirms that DGKK variants are associated with hypospadias.
The GWAS that first discovered an association of hypospadias with DGKK investigated only two SNPs in its replication phase, rs1934179 (SNP22, intronic) and rs7063116 (SNP27, in the 5’ upstream region) 5. The association of these two SNPs was more modest for mild to moderate cases in our study population (odds ratios <1.5) than in the previous study (odds ratios >2). These two SNPs were associated with severe hypospadias in only one of their two replication samples, and they were not associated with severe hypospadias in our study population (ORs were 0.9). Our study did find that other SNPs had somewhat stronger associations with hypospadias (some >1.5), especially after adjustment for selected other SNPs (as part of the step-wise regression).
In light of the high linkage disequilibrium across the gene, we conducted haplotype analyses and then a step-wise regression analysis of SNPs from the haplotype blocks in an attempt to identify smaller sets of SNPs predictive of risk. As noted, associations with certain haplotypes were stronger than associations with individual SNPs. Final models from the step-wise regression, which included multiple SNPs adjusted for each other, also yielded associations with certain SNPs that were stronger than the single SNP analysis. For example, SNP7 was modestly associated with mild to moderate but not severe hypospadias in the single SNP analysis (ORs were 1.5 and 1.1, respectively), but it was associated with 2- to 3-fold increased risks in the adjusted analysis, regardless of phenotype severity. Thus, it appears that going beyond a single-SNP analysis was an important part of understanding associations of the selected tagSNPs with hypospadias.
Our study relied on measurement of relatively common tagSNPs. None of the SNPs were predicted to have functional consequences. Thus, it is likely that the observed associations are driven by linkage disequilibrium with other, less common unmeasured variants.
Biologic explanation for the observed associations is uncertain, in part because very little is known about the functions of DGKK during embryogenesis. In adult tissues, DGKK mRNA expression was limited to testis and placenta6. There is also evidence that DGKK is expressed in murine male reproductive tract tissues during embryonic development relevant to the occurrence of hypospadias7. DGKK expression data are not available from human embryos, but using real time PCR, van der Zanden and colleagues showed that DGKK was expressed in preputial skin of 24 newborn healthy boys5. Little is known about the functional roles of DGKK. Imai et al. showed that tyrosine phosphorylation of DGKK was induced by H2O2-treated cells in a dose-dependent manner, and that DGKK was unique among DGK family members in this response to oxidative stress6. Until more is known about DGKK, further speculation about its involvement in urethral development seems premature.
Our study represents a robust population-based replication of a previous novel finding from a modest-sized GWAS. Our study is advantaged by its more comprehensive investigation of SNPs, its large sample size, racial-ethnic diversity, and use of population-based controls. Although our sample size was relatively large, several SNPs had low minor allele frequencies (7 were <1%), and power was limited for their analyses. Sample size was also limited for mild cases, but results suggested a similar pattern of findings for mild and moderate cases. Although SNP-based call rates were good in general, variable call rates across SNPs resulted in more limited sample size, and thus reduced power, for haplotype analyses. Another limitation is that we were unable to identify with certainty whether cases had cryptorchidism,and thus could not analyze those cases separately.
In general, evidence for a genetic contribution to hypospadias is strong, for example based on heritability studies 3, but the specific involvement of particular genes or pathways has not been well delineated. Most of our current knowledge stems from relatively small studies of specific candidate genes, with attempts at replication being the exception rather than the norm. Until the recent GWAS, discovery approaches consisted mainly of expression studies11, 12 and one small array comparative genomic hybridization study13. Our findings support further investigation of DGKK. Further evaluation of genetic variation in and around DGKK via sequencing or imputation to 1000 genomes data will are needed to enable a more thorough investigation of this gene, variability, and functional and mechanistic studies are needed to discover how DGKK contributes to urethral development.
Acknowledgments
We thank the California Department of Public Health Maternal Child and Adolescent Health Division for providing data. We also thank Dr. Cynthia Curry for her contributions to case review for some of the study subjects. This work was supported by the National Institute of Health [R01ES017060] and the Centers for Disease Control and Prevention [6U01DD000489]. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the California Department of Public Health.
Footnotes
SUPPLEMENTARY DATA DESCRIPTION
Four supplementary tables are included.
All authors approve the manuscript and report no conflict of interest. There are no previous publications of this manuscript.
References
- 1.Dolk H, Vrijheid M, Scott JE, et al. Toward the effective surveillance of hypospadias. Environ Health Perspect. 2004;112:398. doi: 10.1289/ehp.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulozzi LJ. International trends in rates of hypospadias and cryptorchidism. Environ Health Perspect. 1999;107:297. doi: 10.1289/ehp.99107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnack TH, Zdravkovic S, Myrup C, et al. Familial aggregation of hypospadias: a cohort study. Am J Epidemiol. 2008;167:251. doi: 10.1093/aje/kwm317. [DOI] [PubMed] [Google Scholar]
- 4.Kalfa N, Sultan C, Baskin LS. Hypospadias: etiology and current research. Urol Clin North Am. 2010;37:159. doi: 10.1016/j.ucl.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 5.van der Zanden LF, van Rooij IA, Feitz WF, et al. Common variants in DGKK are strongly associated with risk of hypospadias. Nat Genet. 2010;43:48. doi: 10.1038/ng.721. [DOI] [PubMed] [Google Scholar]
- 6.Imai S, Kai M, Yasuda S, et al. Identification and characterization of a novel human type II diacylglycerol kinase, DGK kappa. J Biol Chem. 2005;280:39870. doi: 10.1074/jbc.M500669200. [DOI] [PubMed] [Google Scholar]
- 7.McMahon AP, Aronow BJ, Davidson DR, et al. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 8.Croen LA, Shaw GM, Jensvold NJ, et al. Birth defects monitoring in California: a resource for epidemiologic research. Paediatr Perinat Epidemiol. 1991;5:423. doi: 10.1111/j.1365-3016.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 9.Carmichael SL, Shaw GM, Nelson V, et al. Hypospadias: trends and descriptive epidemiology in a California population. Epidemiol. 2003;14:701. doi: 10.1097/01.ede.0000091603.43531.d0. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Liu BC, Lin GT, et al. Up-regulation of estrogen responsive genes in hypospadias: microarray analysis. J Urol. 2007;177:1939. doi: 10.1016/j.juro.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Willingham E, Baskin LS. Gene expression profiles in mouse urethral development. BJU Int. 2006;98:880. doi: 10.1111/j.1464-410X.2006.06435.x. [DOI] [PubMed] [Google Scholar]
- 13.Tannour-Louet M, Han S, Corbett ST, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]