Abstract
Internal Ribosome Entry sites (IRESs) are RNA sequences that can recruit the translation machinery independently of the 5′ end of the messenger RNA. IRESs are found in both viral and cellular RNAs and are important for regulating gene expression. There is great diversity in the mechanisms used by IRESs to recruit the ribosome and this is reflected in a variety of RNA sequences that function as IRESs. The ability of an RNA sequence to function as an IRES is conferred by structures operating at multiple levels from primary sequence through higher-order three-dimensional structures within dynamic RNPs. When these diverse structures are compared, some trends are apparent, but overall it is not possible to find universal rules to describe IRES structure and mechanism. Clearly, many different sequences and structures have evolved to perform the function of recruiting, positioning, and activating a ribosome without using the canonical cap-dependent mechanism. However, as our understanding of the specific sequences, structures, and mechanisms behind IRES function improves, more common features may emerge to link these diverse RNAs.
Keywords: internal ribosome entry sites, translation initiation, RNA structure, viral RNAs, translation regulation
Introduction
The process of translating an mRNA molecule to produce the encoded protein is divided into four phases, the first being translation initiation. During initiation, the ribosome is recruited to the mRNA and placed at the correct start codon, establishing the reading frame. Initiation must be accurate to avoid starting protein synthesis at the incorrect codon and must also be rapid to allow timely production of proteins when needed. Indeed, initiation is considered the rate-limiting step in translation and is highly regulated in eukaryotes.1 Understanding the regulated mechanisms by which ribosomes are recruited, positioned, and activated on eukaryotic cellular mRNAs is an area of active study, with implications for human disease. Currently, ribosome recruitment to the message and subsequent translation initiation in eukaryotic cells is described by two broad mechanisms: the canonical cap- and scanning-dependent process and internal initiation of translation.
Comparing cap-dependent and internal translation initiation
In the canonical cap-dependent mechanism, a modified nucleotide cap on the 5′ end of the mRNA sequence (usually a 7-methyl guanosine; m7G) binds the eukaryotic initiation factor (eIF) 4F complex. The eIF4F complex is composed of eIF4E (the cap binding protein), eIF4A (an RNA helicase) and eIF4G (a scaffolding protein). Binding of the eIF4F complex leads to the recruitment of the 43S complex (comprised of the 40S ribosomal subunit, eIF2, eIF3 and initiator tRNA) (Fig. 1A). The 43S complex scans the message in a 5′ to 3′ direction until the start codon in good “context” is located (assisted by eIF1 and eIF1A).2, 3 Start codon recognition halts scanning, and GTP hydrolysis results in dissociation of a subset of eIF proteins and joining of the 60S subunit to the 40S to result in a translationally competent 80S ribosome at the start codon. This cap-dependent process is the predominant mechanism used by cellular mRNAs to recruit and position the ribosome and it is regulated in a number of ways; for a more detailed description of canonical initiation and its regulation the reader is directed to two excellent reviews.4, 5
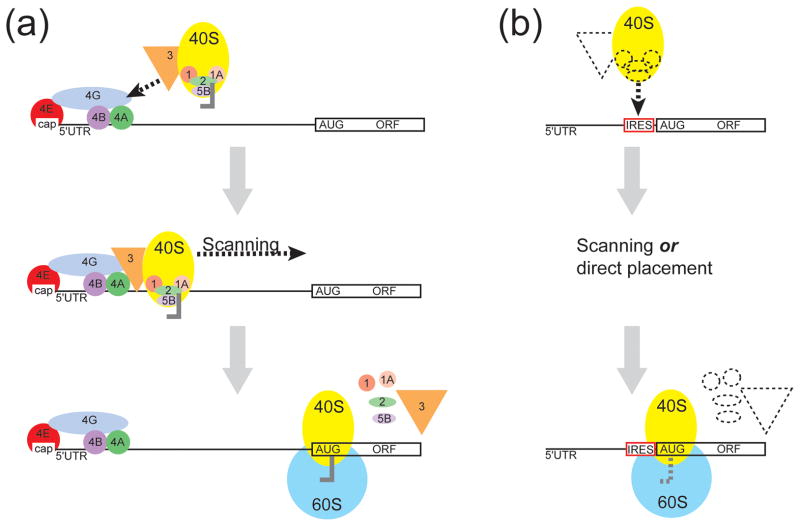
Figure 1.
Comparison of the mechanism of cap-dependent and cap-independent translation initiation in eukaryotes. (a) In cap-dependent translation initiation, the m7G cap (cap) is bound by eIF4E (red) and this leads to the binding of additional initiation factors. Recruitment of the 40S ribosomal subunit (yellow) and associated factors occurs through the interaction of eIF3 (orange). The 40S subunit-containing preinitiation complex then scans to the start codon. Start codon recognition and GTP hydrolysis allows the initiation factors to dissociate and the 60S ribosomal subunit joins to create a translationally competent 80S ribosome. (b) In cap-independent internal translation initiation, RNA sequences called internal ribosome entry sites (IRESs) recruit the 40S subunit independently of the cap. The mechanism behind 40S recruitment varies between different IRES RNAs and may or may not require the use of additional initiation factors (dashed shapes). Depending on the IRES RNA, the 40S is either recruited directly or scans to the start codon. Once the start codon is recognized, the initiation factors dissociate (if used) and the 60S ribosomal subunit joins.
Alternatively, mRNAs may recruit ribosomes for translation initiation in a cap-independent, internal process.6 As the name implies, cap-independent initiation does not require the 5′ cap structure to recruit the ribosome to the message (Fig. 1B). Rather, in this mechanism it is the mRNA sequence itself that confers the ability to bind the translational machinery. This RNA-dependent mechanism was first discovered in the 5′ untranslated region (5′UTR) of two picornaviruses,7, 8 and the fact that these RNA sequences could initiate translation independently of the 5′ end of the RNA led to the term “internal ribosome entry sites” (IRESs). For the purposes of this review, IRESs are defined as RNAs which initiate translation through a cap-and end-independent process. Although not considered IRESs, there are examples of RNAs which can initiate translation through a cap-independent but not end-independent process. For more information on this concept, the reader is directed to another review.9
Since their discovery in two picornaviruses, IRESs have been found in a number of other viral RNAs and also in cellular mRNAs.10, 11 IRESs are mostly found in the 5′UTRs upstream of the open reading frame they control, but there are exceptions. Some are found between open reading frames12, 13 and some are found within an open reading frame.14 Not surprisingly, the purpose for IRES activity in different mRNAs varies. For example, in some viruses, the IRES drives translation of the viral genomic RNA to produce viral proteins under conditions when host cell cap-dependent initiation is repressed15 while in some cellular mRNAs the IRES drives translation of mRNAs involved in the cellular response to stress. The presence of IRESs in cellular mRNAs has been proposed as a mechanism for precise translational regulation of specific messages, allowing the cell to adjust its proteome in response to certain conditions such as stress, stimuli or the cell cycle.11, 16 Taken together, IRES-driven translation initiation is an important strategy used by cells to regulate gene expression and is critical for the life cycle of many viruses.
The continued discovery of RNAs containing IRESs has revealed remarkable diversity in terms of the apparent mechanisms used (Fig. 2).17 Some IRESs can recruit both ribosomal subunits without using any eIFs, whereas others use a subset of the eIFs to initiate translation. Still others not only use a subset of the eIFs, but also co-opt proteins not normally associated with translation as part of their mechanism (called IRES trans-acting factors; ITAFs).16, 18, 19 For most IRESs, the complement of proteins that are associated with and used by the RNA remains unknown. Identifying these factors and how they affect translation from specific IRES RNAs remains an area of active research.
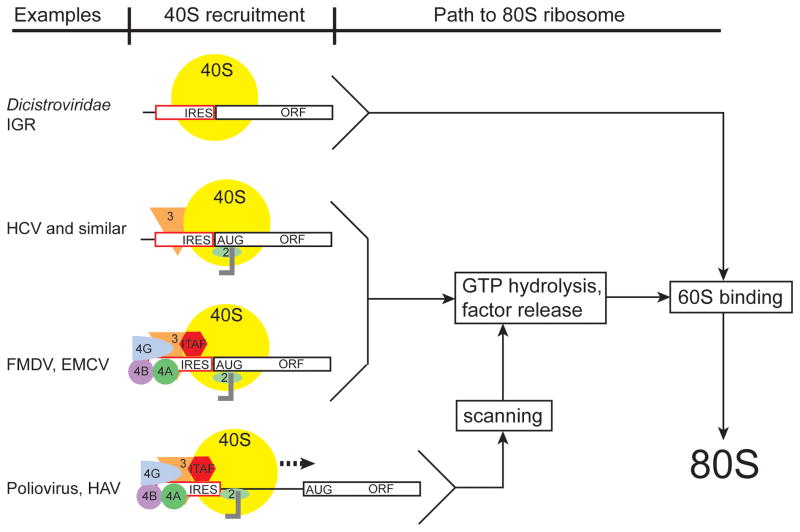
Figure 2.
Examples of the diversity of mechanisms for ribosome recruitment by viral IRES RNAs. Viral IRESs for which mechanistic information is available has allowed them to be placed in several classes. In the Dicistroviridae IGR IRESs, the 40S subunit is recruited directly to the initiator codon, without the need for additional initiation factors or tRNA. Although not depicted in this figure, one report has shown that the IGR IRESs can bind directly to pre-formed 80S ribosomes119. In other IRESs (HCV and similar) the 40S subunit directly binds the IRES but requires additional factors (eIF3, 2) for function. In still other IRESs (FMDV, EMCV), additional initiation factors are required for recruitment of the 40S subunit directly to the initiator codon. Finally, another set of IRESs (Poliovirus, HAV), not only require additional initiation factors to recruit the 40S subunit to the RNA but utilize a scanning mechanism for identification of the AUG start codon. A simplified pathway is shown, depicting how these different pathways compare. A possible relationship between the need for factors and the ability of an IRES RNA to form a stable 3-D fold is discussed elsewhere.79 Note that for many other IRESs, their mechanisms are unknown and thus they cannot yet be placed in a class. It also remains possible that some will have mechanisms different from anyxia of these.
The ability of an RNA to function as an IRES is encoded entirely in its sequence and it is this sequence that determines the structure of the IRES RNA and the binding sites for interacting proteins. Thus it is not surprising that the various contexts in which IRESs are found, the differences in their purposes, and the variety of the proteins they bind is reflected in a diversity of sequences. There is no single RNA sequence that defines an IRES; rather, many different sequences and structures have evolved to perform this function.17 This fact presents questions, including: What are the primary, secondary, and tertiary structural determinants of IRES RNA function? How are these different levels of RNA structure used by IRES RNAs? Can we learn anything about universal modes of IRES function by studying the structures of the RNAs that drive this process? In this review, we present an overview of what is known of how RNA structure drives IRES function, considering RNA structure at several levels and how those structural features may drive function. We do not attempt to discuss all known IRESs, but rather use a set of examples to illustrate key points of our understanding, with particular emphasis on current research.
How IRESs are studied functionally
Before discussing the relationship between different levels of RNA structure and IRES function, it is worthwhile to note some of the ways in which IRES RNAs are identified, characterized and studied. Often, an RNA sequence is suspected to contain an IRES if the mRNA it is within is translated under conditions where cap-dependent translation is inhibited, such as during mitosis,20 poliovirus infection,21 or hypoxia22 or if it is contained within a RNA virus that does not contain a canonical m7G cap. There is great diversity in RNA sequences and structures used by IRES RNAs, which has made identification of an IRES by bioinformatic approaches such as homology searches and secondary structure predictions technically challenging.23–25 Therefore identification of an IRES must be demonstrated experimentally. There are many assays available to identify potential IRESs and each assay has associated caveats and alternative explanations. Here, we do not attempt to fully review all of these assays; this would be a review in itself. Instead, we will present the reader with a few important examples to assist in evaluation of the IRES literature. For a more comprehensive analysis of these assays and the caveats associated with them, the reader is directed to several excellent resources.9, 26, 27
A widely-used method for identifying an IRES involves placing the putative IRES RNA sequence upstream of a reporter gene and then measuring the levels of protein expressed from the IRES RNA relative to a non-IRES control (the 5′ leader from β-globin is widely used)(Fig. 3A). In addition to a negative control, a known IRES is often used as a positive control (commonly the Hepatitis C virus (HCV) IRES, or the Encephalomyocarditis virus (EMCV) IRES). Different versions of the reporter construct are used. Specifically, the putative IRES can be positioned between two different reporters (dual- or bi-cistronic), or placed upstream of a single reporter (monocistronic). In the bi-cistronic test, the sequence of interest is inserted between two reporter ORFs on a single mRNA. Expression of the upstream reporter occurs through a cap- and end-dependent mechanism, while expression of the downstream reporter only occurs if ribosomes are recruited internally to the message. If expression of the downstream reporter is greater from the putative IRES element than the non-IRES control, the candidate RNA is considered a potential IRES. In the monocistronic test, the candidate sequence is placed upstream of the ORF for a single reporter protein and expression of the protein is compared to a negative control. Unlike the case of the bi-cistronic reporter, expression of the monocistronic reporter is not exclusively cap-independent and thus must be studied under conditions where cap-dependent translation is inhibited, or studied using an in-vitro transcribed RNA construct containing a modified cap structure that is not capable of recruiting the translation machinery. If expression of the reporter construct is maintained under these conditions relative to the non-IRES control, the sequence of interest is tentatively considered to be able to initiate translation in a cap-independent process. It should be noted that the monocistronic test does not per se demonstrate end- independent (or internal) initiation and thus must be complemented with other approaches to demonstrate the presence of an IRES. The tests described above can be performed in either cell-free extracts or in cell culture. Advantages of cell-free extracts are that the conditions of the lysate can be varied readily. In cell culture, the RNA can be in vitro transcribed and directly transfected into the cells, or produced in the cell’s nucleus by transfection of the cells with a DNA vector containing the IRES-reporter sequence. Finally, it is worth noting that a circularized RNA can also be used to identify an IRES.28, 30 This type of construct prevents any end-dependent ribosome recruitment 28 and challenges previous views that eukaryotic ribosomes cannot bind circular RNAs.29 However, the circularization approach has only been used a few times,30 likely due to the technical challenge of efficiently circularizing large RNAs.
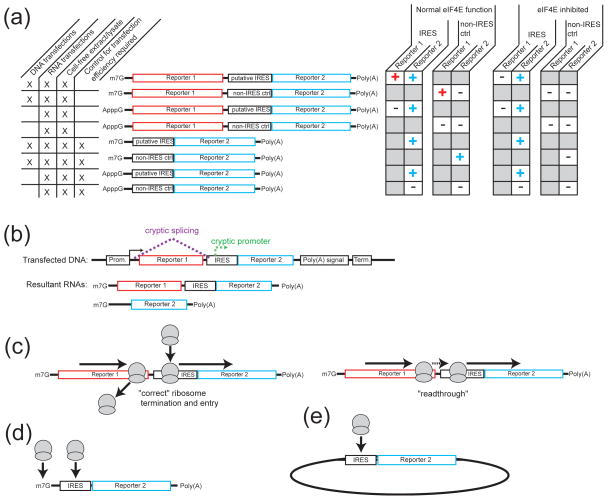
Figure 3.
How IRESs are identified and functionally studied and the caveats and considerations in these experiments. (a) IRESs typically are identified by placing the sequence of interest in front of a reporter construct and expression of the reporter is compared to a non-IRES control construct. Examples of the types of constructs used in these studies, the conditions under which they can be used, and the expected results of expression studies are depicted. For example, dual-cistronic reporter constructs can be used in both RNA and DNA transfections of cells, as well as in-vitro transcribed and programmed into lysate. The upstream reporter serves as an internal control for transfection efficiency and unlike their monocistronic counterparts, these constructs do not require a transfection control. Under normal eIF4E function, both reporters should be expressed if the insert contains an IRES. However, if the insert does not contain an IRES (such as in the non-IRES control construct), the downstream reporter should not be expressed. In contrast, under conditions where eIF4E is inhibited, the upstream reporter should not be expressed in either construct, while the downstream reporter will be expressed if the insert contains an IRES. (b) Transfection of dual-cistronic DNA constructs into cells can be subjected to nuclear processing events (dashed lines). These events include cryptic splicing (magenta) and cryptic promoter activity (green). If present, these result in the production of capped mono-cistronic RNA species (shown as the resultant RNAs) and can lead to false positive IRES reporter levels. (c) The use of dual-cistronic DNA constructs also can lead to ribosomal readthrough on the message (right) rather than correct termination and re-entry on the downstream reporter (left). This would yield false positive signal of IRES activity. (d) Capped, mono-cistronic constructs may recruit ribosomes through both a cap-dependent and cap-independent process, confounding clear delineation of the level of IRES activity. In this case, IRES identification must occur under conditions where cap-dependent translation is inhibited. (e) Diagram of a circular RNA containing an IRES upstream of a reporter sequence. Assuming the RNA remains intact during the experiment (e.g. – is not nicked), ribosomes can only enter in a cap- and end-independent mechanism; hence, expression of the reporter indicates an IRES.
Robust studies of a putative IRES generally couple the translation assays mentioned above with control experiments designed to detect alternate explanations for apparent IRES activity.26 Given some debate surrounding the concept of IRESs in cellular mRNAs,9, 31, 32 these experiments have become especially important. For example, if a putative IRES sequence contains a cryptic promoter, then transfection of cells with a plasmid containing a dual luciferase reporter might give rise to capped monocistronic RNAs containing only the second reporter and a portion of the IRES. As a result, subsequent measurements of IRES activity based on this reporter would be artificially high and would not reflect true IRES-driven initiation (Fig. 3B). Cryptic splicing can also occur, giving rise to undesired monocistronic constructs. Control experiments to detect these events include RT-PCR-based methods, Northern blot analysis, promoter-less assays and shRNA-based approaches.26 A way of avoiding cryptic events during reporter transcription and processing is to directly transfect the cells with reporter mRNAs, although this method does not allow the reporter to interact with nuclear ITAFs which may be important for the activity of some IRESs (especially cellular).33, 34 RNA transfection techniques also necessitate measurement of the chemical stability of the RNAs within the cells to control for different degradation rates. Other concerns include read-through of ribosomes from the upstream reporter (rather than authentic internal entry) giving rise to production of downstream reporters (in dual-cistronic constructs) (Fig 3C), or a combination of cap-dependent and independent entry of ribosomes on monocistronic RNAs (Fig. 3D). Collectively, it is clear that establishing the presence of an IRES in a specific RNA sequence requires careful consideration of the natural context of the RNA, careful choice of the reporter systems (often more than one), thorough control experiments, and stepwise elimination of alternate explanations.
Taken together, there is no single “gold-standard” experiment that can be used to establish the presence of an IRES within an RNA sequence. Rather, robust identification of an authentic IRES requires a set of carefully controlled experiments that are designed to eliminate alternate explanations. However, once a set of suitable assays and controls are established to measure protein production from a putative IRES sequence, the IRES sequence can be mutated, portions deleted, truncations made, etc. to establish the sequences that comprise the IRES activity and the conditions under which it is active. It should be noted that some RNAs may control translation initiation by a combination of mechanisms and thus possess both IRES and non-IRES characteristics. These possibilities and some pitfalls of some IRES assays are discussed in recent reviews.27, 35
IRES activity based on primary structure (sequence)
As of 2010, there were at least 68 viral and 115 cellular IRESs reported.36 What, if any, commonalities can be drawn between these IRES RNAs in terms of primary structure? Traditionally, long, GC-rich 5′UTRs were thought to be good candidate IRESs, as in vitro experiments suggested that such qualities made a 5′UTR an inefficient substrate for ribosome scanning in cap-dependent translation.37 However, a comparison of the GC content of all published 5′UTRs containing IRESs (as of 2006) to all 5′UTRs in the genome indicate that there is no difference in their GC content.24 This observation is corroborated by a study comparing the translational efficiencies of the GC-rich 5′UTRs from Apaf-1 and c-myc mRNAs which indicated that these UTRs were translated efficiently by a cap-dependent mechanism and the translational efficiency was comparable to that produced by the shorter, less GC-rich, 5′UTR from β-globin.38 Interestingly, these 5′UTRs are also reported to initiate translation through an IRES,39–41 suggesting that these mRNAs may use a combination of initiation strategies. In addition, the intergenic region (IGR) IRESs from the Dicistroviridae are known to fold into complex three-dimensional (3-D) RNA structures (see below) and yet are AU-rich. For example, the cricket paralysis virus (CrPV) IGR IRES is 64% AU. Together, these data indicate that in general, the base composition of a particular RNA is not directly correlated to, or predictive of, IRES function.
On the other hand, it does appear that UTRs containing IRES elements tend to be longer than the average 5′UTR length of ~150 nts.24 However, the actual RNA sequences which contribute to IRES function can be quite diverse in length and generally tend to be smaller than the UTRs which harbor them. For example, IRES-containing 5′UTRs from picornaviruses (including viruses such as poliovirus and EMCV) range in size from 610 to 1450 nts, however in almost all cases of this type of IRES activity is conferred by an ~450 nt segment within the 5′UTR. HCV contains a 5′UTR of 342 nts and an IRES of ~300 nt, while the aforementioned Dicistroviridae IGR IRESs are around 190 nt. Smaller IRES sequences also have been reported. For example, a 90 nt segment of the 1,040 nt 5′UTR from the cellular type 1 insulin-like growth factor (IGF1R) mRNA42 and a 9 nt element from the 196 nt 5′UTR of the Gtx mRNA43 both function as IRESs. The diversity in IRES sequence lengths correlates with a diversity in mechanism. For example, in the case of the picornavirus IRESs, maintenance of the global architecture of the RNA is critical for recruiting a subset of eIFs, ITAFs and the 40S ribosomal subunit. In comparison, in the IGF1R and Gtx IRESs accessibility of a small RNA element appears to be critical for IRES function presumably through direct base-pairing to the 40S subunit, although this hypothesis has not been directly tested with binding assays. Thus, in these cases, the primary structure of the RNA appears to underlie IRES activity.
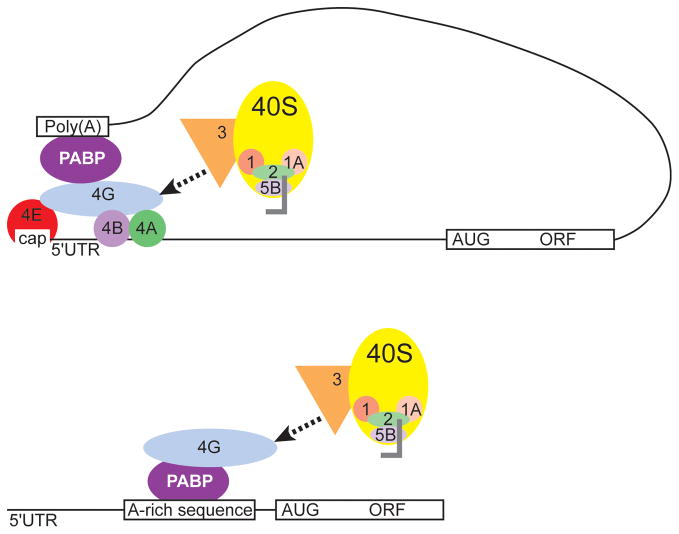
The IGF1R IRES is not the only IRES where small segments of RNA are important to IRES function. Another example comes from the 298 nt 5′UTR of the yeast invasive growth gene, YMR181c. Based on a series of well-controlled experiments, this 5′UTR was identified as containing an IRES whose activity was dependent on a poly-A tract within a 60 nt unstructured region of the RNA (Fig. 4). The poly-A tract serves as a binding site for the yeast poly-A binding protein 1 (pab1), suggesting a mechanism for IRES function where pab1 binding to the 5′UTR could lead to internal initiation from the IRES by functionally substituting for the cap and eIF4E in recruiting eIF4G.44 The identification of short RNA elements contributing to cap-independent translation in these and other 5′UTRs presents a possible hypothesis for some IRES function where RNA primary structure (sequence) provides non-canonical binding sites for components of the translation machinery to initiate translation in a cap-independent fashion.
Figure 4.
Model for the mechanism of internal ribosome recruitment by the YMR181c IRES. In cap-dependent translation initiation, PABP binds to the poly-A tail at the 3′ end of the mRNA as well as to eIF4G at the 5′ end of the message (top). This 5′-3′ crosstalk leads to circularization of the message and efficient recycling of ribosomes. In contrast, PABP binds an A-rich sequence the 5′UTR of YMR181c, leading to recruitment of eIF4G and the downstream translation machinery (bottom). Thus, in YMR181c, binding of the PABP to the A-rich primary structure upstream of the AUG leads to recruitment of eIF4G independently of the cap and 4E interaction.
IRES activity based on secondary structure
The fact that there is no overall sequence similarity between identified IRES RNAs implies that the ability of many IRESs to form specific secondary structures is critical to IRES function. This hypothesis is highlighted by phylogenetic comparisons of picornavirus IRESs, where overall secondary structure, rather than primary sequence, is well conserved.45 At first glance, it is apparent that IRES RNA secondary structures are quite diverse (Fig. 5A). For example, the Plautia stali intestine virus (PSIV) contains two IRESs, one at the 5′UTR of the ORF1 gene,46 and a second, intergenic (IGR) IRES between ORF1 and ORF2 (Fig. 6A).47 The 5′UTR IRES structure is extended and composed of several RNA hairpins and a putative pseudoknot, while in contrast the IGR IRES is more compact, and comprised of several pseudoknot structures48 (for the purposes of this review, we are including discussions of pseudoknots with secondary structure as their presence can be shown within a two-dimensional diagram of base-pairing; review of pseudoknot folding: 49). While less is known about the 5′UTR IRES than the IGR IRES, it is clear that the two function differently in in vitro translation extracts, suggesting a very different mechanisms between these two IRESs.46 This mechanistic difference may be related to differences in their secondary structures and how these structures drive initiation.
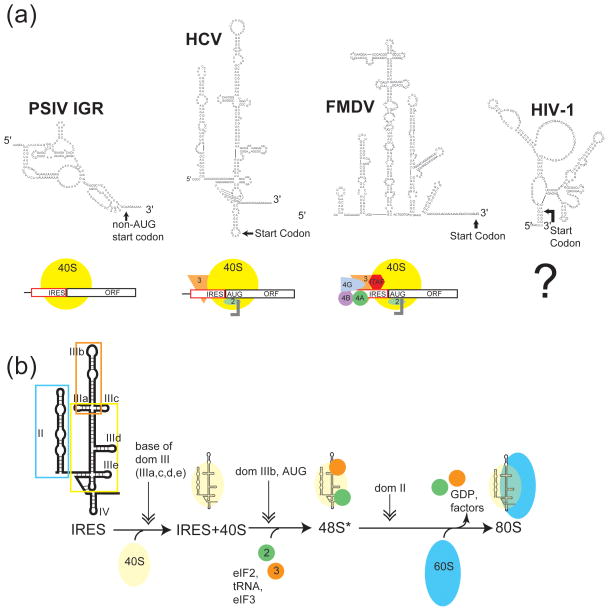
Figure 5.
Examples in the diversity of IRES RNA secondary structures and the role of secondary structure in mechanism. (a) Secondary structure models of several viral IRESs and diagrams of their factor requirements for 40S recruitment. In the case of HIV-1 5′UTR IRES, the secondary structure is known,118, 120 while the mechanism behind internal 40S recruitment is still unknown. (b) Detailed diagram of the mechanism for RNA structure-based ribosome recruitment in the HCV-like IRESs. HCV-like IRESs bind the 40S subunit (yellow circle) directly using the base of domain III (boxed in yellow on the secondary structure diagram). Domain IIIb (orange box) recruits eIF2, leading to tRNA and eIF3 (green and orange circles) binding. The resulting complex is called the 48S*, asterisk denoting the deffierence between it and canonically assembled 48S.57 Domain II (blue box) triggers GTP hydrolysis and 60S recruitment (blue oval).
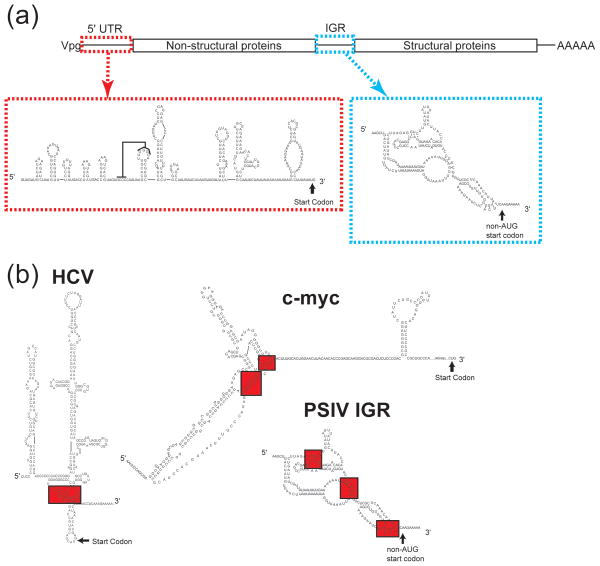
Figure 6.
Further examples in the diversity of IRES RNA secondary structures. (a) Diagram of the Dicistroviridae genome, with the 5′UTR and intergenic region (IGR) IRESs highlighted (red and blue dashed boxes, respectively). Secondary structure diagrams of PSIV IRESs are shown. A proposed pseudoknot interaction is indicated with a line on the 5′ IRES diagram. (b) Secondary structure diagrams of the HCV, c-myc and PSIV IGR IRESs with pseudoknot structures depicted (shaded in red).
Despite the overall diversity in IRES secondary structure, some IRESs do have similar secondary structures, which in turn correlate with a common mechanism (review:17). For example, IRESs from members of the Flaviviridae family of single stranded positive sense RNA viruses (including viruses such as HCV, GB virus B, Bovine Viral Diarrhea Virus (BVDV) and classical swine fever virus (CSFV)) and many picornaviruses50 can form similar extended RNA secondary structures with three major structural domains numbered II to IV (Fig. 5B). The HCV IRES domains are comprised of several hairpin structures with loops, bulges, and helical junctions as well as a pseudoknot structure. Extensive mutational analysis of the HCV IRES indicates that secondary structure is critical to IRES function, and through biochemical and functional assays, a structure-based mechanism for ribosome recruitment has been deduced (reviews:51, 52) (Fig. 5B). In summary, domains IV, IIb, IIIa, IIIc-f and the pseudoknot bind the 40S directly, while domains IIIa-b are important for eIF3 binding and recruitment of the ternary complex.53–57 Domain II and the apical parts of domain III are important for 80S ribosome formation.52, 56–58 Recent mutational and biochemical studies have elucidated additional roles for the pseudoknot structure and domain IIb.59–61 Taken together, it is clear that the HCV-like IRESs use a specific secondary structure to recruit and properly position components of the translation machinery to initiate translation non-canonically. Intriguingly, a recent study of the cellular IRES c-Src determined that this IRES also could bind directly to the 40S subunit in an initiation factor-independent fashion, features which are similar to the HCV-like IRESs.62 To our knowledge, this study is the first example of a cellular IRES binding directly to the 40S subunit. Given the commonalities between the factor requirements of the HCV-like IRESs and the c-Src IRES, it is interesting to speculate that the c-Src IRES may adopt a secondary structure similar to the HCV-like IRESs, however this remains to be determined.
While the overall secondary structure of HCV-like IRESs is conserved and this structure relates to a common mechanism, certain structural elements also are found in other IRESs, although their functional roles may differ. For example, pseudoknots are found in the IGR IRESs of the Dicistroviridae family of viruses as well as in the cellular IRESs c-myc and l-myc (Fig 6B). The IGR IRESs contain three pseudoknot structures (PK1–3) which all have critical roles in binding the ribosome and initiating cap-independent translation.12, 63–67 In contrast, the pseudoknot of the c-myc IRES is inhibitory to IRES function, although how it does so has not been explored.68 In the case of the l-myc IRES, the role of the pseudoknot to IRES function has not been investigated.69 The identification of common RNA structures with very different roles in IRES function highlights the diversity of structure-based mechanisms.
Currently, much more is known about the contribution of RNA secondary structure to the function of viral IRESs than to cellular IRESs, although this is starting to change. The growing body of evidence suggests that in the case of many cellular IRESs, the requirement for specific RNA secondary structures is not as critical for function. This hypothesis is based on the following experimental observations. First, mutations in cellular IRESs predicted to disrupt secondary structure often have no effect or may even increase IRES activity.68 Second, deletion analysis of cellular IRESs indicates that small fragments of RNA are capable of initiating internal translation initiation as well, or sometimes better, than the full length 5′UTR.42–44 Third, a comparison of the known secondary structures of cellular IRESs indicates very little similarity, even between IRESs of closely related genes.19, 25, 70 Fourth, many cellular IRESs require ITAFs for function, potentially to remodel the RNA.71 While these observations explain an overall trend for cellular IRESs, there are certainly exceptions. For example, studies on the VEGF cellular IRES have identified an RNA G-quadruplex structure, that when modified to disrupt the structure completely abolished IRES activity.72 This study provides one of the rare examples where local mutations to a cellular IRES structure completely disrupt function. Taken together, it is clear that the contribution of specific RNA secondary structures to IRES function is quite diverse amongst IRES RNAs and no global generalizations can be made.
IRES activity based on tertiary and three-dimensional folded structures
RNA, like proteins, can fold into defined 3-D structures that are critical for function, and this suggests that the function of some IRESs may be conferred by higher-order RNA structures. Evidence for this is found in the fact that many IRESs not only have extensive, conserved, and functionally important secondary structures but also have conserved sequences in parts of the RNA predicted to be single-stranded. These regions may be involved in long-range RNA-RNA interactions that stabilize the 3-D conformation. In addition, the aforementioned presence of pseudoknots in many IRESs suggests that folded higher-order structures may be present, as pseudoknots can often fold into very compact and stable structures with important biological functions.73
Despite evidence that folded 3-D RNA structures may be important determinants of function for many IRESs, direct observation and characterization of these structures has been accomplished for only two IRESs: the HCV IRES and members of the IGR IRESs from the Dicistroviridae. Both are known to bind directly to the 40S subunit without requiring any other factors,67, 74 and for both disruption of the 3-D folded structure leads to loss of ribosome affinity and of function.53, 66 In both, there are regions of the RNA backbone that are packed together tightly enough to exclude solvent,66, 75 a characteristic observed in other RNAs including tRNA and catalytic RNAs such as the group I intron.76, 77 The results are folded structures in which the parts of the IRES RNA that interact directly with the ribosome are positioned correctly in 3-D space to make their specific contacts.
Although they both adopt higher-order folds that interact directly with the ribosome, there are major differences in the global conformations of the HCV IRES and the IGR IRESs.78, 79 The IGR IRESs assume an overall compact structure with two independently folded domains, and cryo-EM reconstructions show that this compact structure docks into the space between the large and small subunits of the ribosome where it is sufficient to induce joining of the two subunits without the aid of any protein factors (Fig 7).80, 81 In the case of the HCV IRES, the RNA forms an overall extended structure (consistent with its extended secondary structure) organized, in part, around two compactly folded junction regions (Fig. 7). This extended structure makes contacts to ribosomal proteins on several widely separated parts of the small subunit, including both the space between the subunits and the solvent-exposed part.82, 83 This architecture likely reflects the mechanistic requirement of the HCV IRES to interact with eIF3 in addition to the 40S subunit,84 an idea borne out in cryo-EM based models of this ternary complex.85 The fact that the IRES from HIV-286 and the aforementioned c-Src62 have been shown to bind directly to the ribosome without the aid of other factors raises the question of whether these RNAs might have regions of compactly folded RNAs, pre-folded conformations, and a higher order fold. To date, studies exploring this question have not been reported.
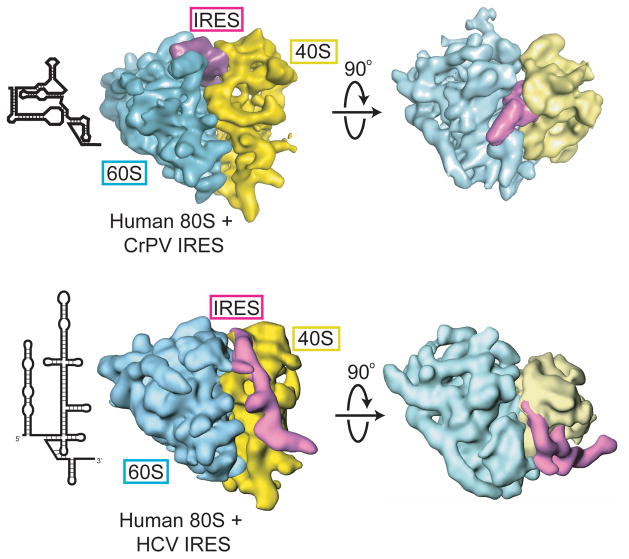
Figure 7.
Cryo-EM reconstructions of the CrPV IGR IRES (top) and HCV IRES (bottom) bound to human 80S ribosomes. The 60S subunit is in cyan, the IRES RNA in purple and 40S subunit in yellow. Two views are shown for each, with the secondary structure cartoon of each IRES to the left. The overall differences in the global architectures of these two IRESs is obvious, as is the difference in their locations on the ribosome.
The structures of the IGR IRESs and the HCV IRESs have been studied using both nuclear magnetic resonance (NMR) and x-ray crystallography. For the IGR IRESs crystal structures of both independently folded domains and the structure of the smaller domain bound to a 70S ribosome has been solved using x-ray crystallography.65, 87, 88 These structures show the detailed intramolecular contacts that stabilize the compact structure, and also show that these IRESs mimic the codon-anticodon interaction that normally forms between an mRNA and a tRNA. In addition, these high-resolution structures can now be combined with cryo-EM reconstructions and biochemical and functional studies to develop the first complete models for the interaction of the IGR IRES with the ribosome (Fig. 8). These structures also allow the development of new mechanistic models which include manipulation of the ribosome and its conformational states. In the case of the HCV IRES, NMR and x-ray crystallography have been used to solve the structures of isolated secondary structure domains,89–96 as the extended (and likely somewhat dynamic) conformation of the IRES RNA overall has precluded solving the structure of the IRES as a whole. Nonetheless, this “divide and conquer” approach yields structures that can be fit into cryo-EM reconstructions.97 Although the crystal structures of an 80S ribosome and 40S subunit were recently reported,98, 99 high-resolution structures of IRES RNAs bound to eukaryotic ribosomes have not yet been solved.
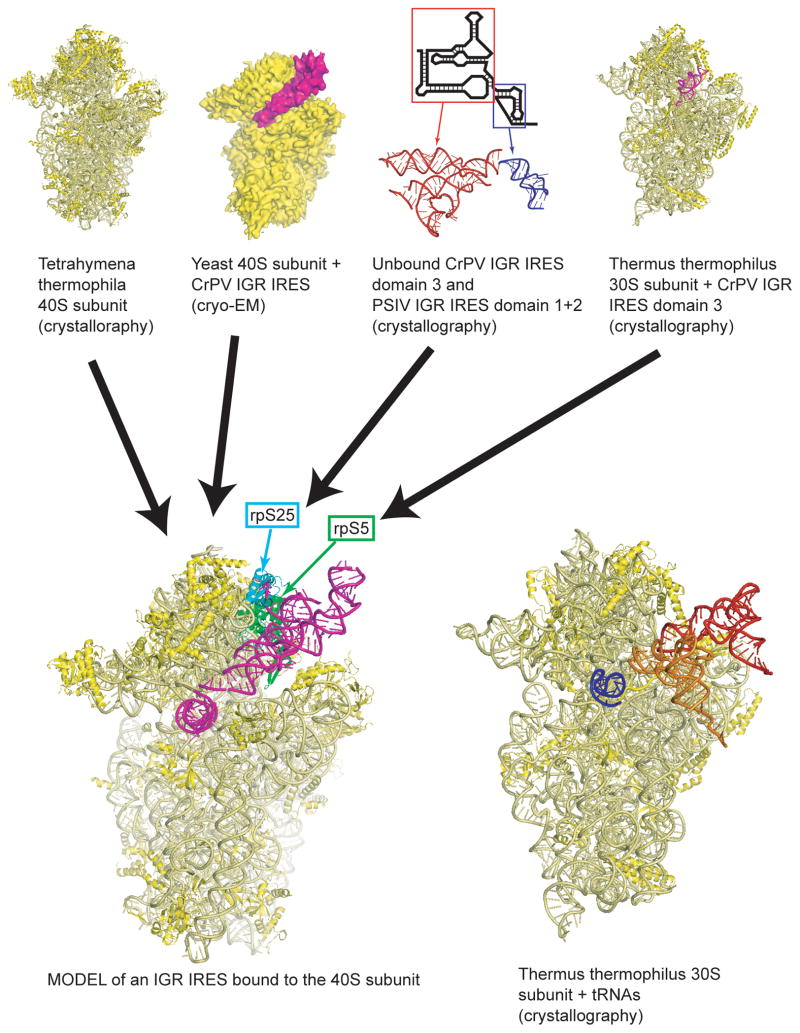
Figure 8.
Example of how multiple sets of structural data can lead to a model. In this example, an x-ray crystal structure of the 40S subunit from Tetrahymena thermophila (top left, eIF1A is not shown),98 a cryo-EM structure of the CrPV IGR IRES bound to the ribosome (second from left, the 60S subunit is not shown),81 x-ray crystal structures of unbound IGR IRES domains (second from right, with secondary structure cartoon,65, 88 and the x-ray crystal structure of an IGR IRES domain bound to a 70S ribosome (top right, 50S subunit not shown)87 were combined to yield the model at lower left. In this model, the IGR IRES is shown in magenta, and two proteins known to interact with the IGR IRES are shown in cyan (rpS25) and green (rpS5). To the right of the IRES structure is the crystal structure of a 70S ribosome with bound tRNAs (50S subunit not shown).121 Note how the IGR IRES spans the binding sites of all three tRNAs.
The IGR IRESs and HCV IRES are the best studied IRESs in terms of their 3-D folded conformation, but evidence suggests that other IRESs form 3-D structures that are important for function. One example comes from the foot-and-mouth disease virus (FMDV), a member of the picornavirus family and an IRES with extensive secondary structure. Evidence suggests that distal regions of RNA within the IRES interact directly, indicative of a higher-order fold that is important for IRES function.100–102 However, although the FMDV IRES RNA (and other IRES RNAs) may have a higher-order fold and perhaps regions of compact RNA packing, a picture of the structure and the overall conformation at the detail of the IGR and HCV IRES has not yet been reported. In many cases this may be due, in part, to a requirement for ITAF proteins that stabilize the active 3-D conformation of the RNA and that have been shown to be required for full activity of many IRESs, both cellular and viral. Indeed, it has been shown that binding of both ITAF proteins as well as canonical initiation factors to the FMDV IRES RNA induces structural changes in the IRES that might reflect formation of the active conformation.103–105 Thus, for many IRESs, formation of a specific 3-D structure may be important for function, but the RNA alone may not have the ability to form that structure without bound ITAFs.
IRES activity based on interactions with IRES trans acting factors (ITAFs)
Despite the lack of direct structural information regarding IRES-ITAF interactions, biochemical and functional assays indicate that ITAFs play an important role in the function of many IRES RNAs (review:106). IRES RNAs are very diverse in their ITAF requirements and this correlates with functional diversity (review:107). For example, the Apaf-1 IRES requires PTB and unr to function108 and is active during apoptosis,109 presumably to maintain Apaf-1 protein levels for its role in apoptotic signalling. In contrast, the Bag-1 IRES requires PTB and poly(rC) binding protein 1110 and is active during chemotoxic stress rather than apoptosis.111 In the case of the Bag-1 IRES, evidence suggests that the relocalization of its cognate ITAFs allows for activation under stress,111 indicating an intricate mechanism for the regulation of protein synthesis. Studies such as these indicate that the regulation of IRES RNAs by ITAFs may present the cell with a complex mechanism to regulate protein synthesis in response to external stimuli (review:16). It is believed that the ITAF proteins contribute to IRES function by remodeling the RNA into the proper conformation for internal initiation. Evidence for this comes from chemical probing data of the Apaf-1 and Bag-1 cellular IRESs demonstrating conformational changes in the ribosomal entry window of the IRES RNA in the presence of its cognate ITAFs.71, 112
Despite their diversity in ITAF requirements, in some cases very different ITAFs can cause similar conformational changes in their cognate IRES RNAs. Evidence for this comes from studies of the type II picornavirus IRESs EMCV and FMDV. As described earlier, these IRESs are structurally similar, and this correlates with a common overall mechanism to IRES function. These IRESs require the same components of the canonical translation machinery, but differ in their ITAF requirements. The ITAF for EMCV is the pyrimidine tract binding protein (PTB), while the FMDV IRES requires ITAF45 in addition to PTB. Despite their differences in ITAF requirements, studies have demonstrated that these proteins induce similar conformational changes in the IRES in which two domains are brought closer together in three-dimensional space.113 This conformational change potentially stabilizes the overall three-dimensional structure of the IRES,114 and may place the IRES in a biologically active conformation. In contrast, PTB binding to the poliovirus IRES induces a much more localized change in IRES RNA conformation, suggesting that in the case of this IRES, PTB may serve a completely different role in IRES function.115 Taken together, it is clear that IRES RNAs are quite diverse in their ITAF requirements. In some cases a single ITAF may be required (as in the case of EMCV), while in others a set of ITAFs may cooperatively be required (as in FMDV, Bag-1 and Apaf-1). This observation presents a view of some IRESs as intricate ribonucleoprotein (RNP) complexes, allowing for dynamic regulation and modulation of IRES function.
The structural basis of IRES function – future directions
We have presented examples of how the RNA structures at all levels: primary, secondary, tertiary, and within RNPs, are determinants of IRES activity. Below, we suggest several areas of research that we believe are among the most important for understanding how IRES structure drives function.
First, there is a need to determine the secondary and tertiary structures of many more IRESs (particularly of cellular origin) using not just thermodynamic base-pairing predictions, but detailed structural probing coupled with mutagenesis. Knowledge of the secondary structures of more IRESs will directly address outstanding questions in the field, to include: Do these RNAs adopt a single secondary structure, or an ensemble of structures? When diverse IRESs are examined, how important is secondary structure to the function of each IRES? Do cellular IRESs possess any similarities to their viral counterparts and are there common IRES secondary structures yet to be discovered? A larger database of the secondary structures of IRESs might lead to the identification of conserved IRES-specific secondary structural motifs, and this in turn could enable successful bioinformatics-based searches for novel IRES RNAs. In addition, once the secondary structure of a given IRES is known, mutants can be generated to target parts of the secondary structure, and these can be tested in functional, structural, and biochemical assays. Such structure-based mutation strategies can confirm the secondary structures, demonstrate if a given secondary structure element is functionally important, and ultimately determine the specific steps in the initiation process the secondary structure contributes to (i.e. initiation factor recruitment, 40S binding, 80S formation, etc.). The tools to undertake these studies continue to improve, most notably in the area of high-throughput probing methods that have been used to determine the secondary structures of RNAs as large as the HIV-1 genome.116–118 Application of these and other methods should lead to a larger dataset of IRES secondary and tertiary structural information to guide the design of new experiments, to include high-resolution structural studies.
Another area ripe for more input is structural studies of ITAFs and initiation factors bound to IRES RNAs. ITAFs appear to be ubiquitous, and we have some insight into the structural changes that can occur in an IRES when factors bind in specific locations. However, a complete structural picture of an IRES RNA decorated with its necessary factors has yet to be reported. In other words, we have yet to see the structure of a complete IRES-containing RNP. This type of structure would allow us to observe the specific contacts that are made between the IRES RNA and ITAFs, and when coupled with mutational and functional assays, would reveal how ITAFs contribute to the function of IRES RNAs. For example, once a contact between a given IRES and ITAF is identified, this region of the IRES could be mutated, and its effect on ITAF binding and IRES function tested and correlated. It is likely that the contributions of ITAFs to IRES structure and function will vary between IRESs, but some themes might emerge when these IRES RNP structures are seen in high-resolution.
Ultimately, high-resolution crystal structures and cryo-EM reconstructions of IRES RNPs bound to eukaryotic ribosomes will reveal important intermolecular contacts between IRES RNAs, ITAFs, initiation factors, and the ribosome. This will give direct insight into the role of each macromolecular component in the initiation process. For example, it is unclear if all IRESs make direct contact to the ribosome or if the ribosome is contacted by IRES-bound factors, or a combination of both; high resolution structures would address this. The absence of current structural data regarding IRES-ITAF complexes and larger ribosome-containing complexes is likely due to the challenge of isolating or reconstituting these complexes and applying structural methods to potentially dynamic assemblies. These are challenging goals, but as methods improve such studies become more feasible.
Finally, although we know translation initiation is a process involving structural transitions and conformationally dynamic states, the pictures that we have so far of IRES function are largely static. Through cryo-EM studies, we know that some IRESs can induce structural changes in the ribosome, however, these data provide only a snapshot of an IRES-ribosome complex. IRES RNAs and RNPs may be inducing conformational changes in the ribosome (and potentially other components of the translation machinery) throughout the initiation process, from preinitiation complex formation to elongation. These movements and conformational changes may vary between IRESs, but by having a complete picture of IRES and preinitiation complexes from initiation to elongation we can relate IRES mechanism to the more general mechanisms common to all translation. Such studies are likely to require a combination of cutting-edge biochemical and biophysical studies in which the translation initiation process can be carefully controlled, monitored, and structurally interrogated at each step.
Conclusion
In conclusion, it is clear that the ability of IRES RNAs to bypass the need for a cap and drive an alternate mechanism of translation initiation is conferred by RNA structures operating at multiple levels from primary (sequence) through higher-order three-dimensional structures within dynamic RNPs. Hence, overall generalizations about the structures of IRES RNAs and their modes of action cannot be made. Although some IRESs are clearly similar to one another there is no established set of IRES consensus motifs that applies to all IRESs and defines them as a whole. Although some viral IRESs have been placed in mechanistic or structural classes, when viewed overall the diversity of IRES structure and mechanism in most cases precludes easy classification, at least based on the information currently available.
Acknowledgments
The authors wish to thank Jennifer Doudna, Sandy Martin, and David Costantino for critical reading of this manuscript. IRES research in the Kieft Lab is currently funded by grant GM081346 from the National Institute of Health. Terra-Dawn Plank is a Predoctoral Fellow of the American Heart Association. Jeffrey Kieft is an Early Career Scientist of the Howard Hughes Medical Institute.
Contributor Information
Terra-Dawn M. Plank, Department of Biochemistry and Molecular Genetics, University of Colorado Denver School of Medicine, Aurora, Colorado, 80045, USA
Jeffrey S. Kieft, Email: Jeffrey.kieft@ucdenver.edu, Howard Hughes Medical Institute and Department of Biochemistry and Molecular Genetics, University of Colorado Denver School of Medicine, Aurora, Colorado, 80045, USA
References
- 1.Mathews MB, Sonenberg N, Hershey BJW. Origins and Principles of Translational Control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 1–40. [Google Scholar]
- 2.Kozak M. An analysis of vertebrate mRNA sequences: Intimations of translational control. Journal of Cell Biology. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 4.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestova T, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 6.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 7.Jang SK, et al. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virology. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 9.Shatsky IN, et al. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol Cells. 2010;30:285–293. doi: 10.1007/s10059-010-0149-1. [DOI] [PubMed] [Google Scholar]
- 10.Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 129–153. [Google Scholar]
- 11.Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 155–172. [Google Scholar]
- 12.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Hertz MI, Thompson SR. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology. 2011 doi: 10.1016/j.virol.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieleski L, Talbot SJ. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol. 2001;75:1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarnow P. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J Virol. 2003;77:2801–2806. doi: 10.1128/JVI.77.5.2801-2806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balvay L, et al. Structural and functional diversity of viral IRESes. Biochim Biophys Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 19.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, transacting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 20.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 21.Chen CY, Sarnow P. Internal ribosome entry sites tests with circular mRNAs. Methods Mol Biol. 1998;77:355–363. doi: 10.1385/0-89603-397-X:355. [DOI] [PubMed] [Google Scholar]
- 22.Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116–1131. doi: 10.1261/rna.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatakeyama Y, et al. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baird SD, et al. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird SD, et al. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664–4677. doi: 10.1093/nar/gkm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Eden ME, et al. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 29.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 30.Carter PS, Jarquin-Pardo M, De Benedetti A. Differential expression of Myc1 and Myc2 isoforms in cells transformed by eIF4E: evidence for internal ribosome repositioning in the human c-myc 5′UTR. Oncogene. 1999;18:4326–4335. doi: 10.1038/sj.onc.1202890. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider R, et al. New ways of initiating translation in eukaryotes. Mol Cell Biol. 2001;21:8238–8246. doi: 10.1128/MCB.21.23.8238-8246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh DC, V, Mauro P. Reconciling contradictory reports regarding translation of BACE1 mRNA: initiation mechanism is altered by different expression systems. RNA Biol. 2009;6:54–58. doi: 10.4161/rna.6.1.7567. [DOI] [PubMed] [Google Scholar]
- 34.Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Malys N, McCarthy JE. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci. 2011;68:991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mokrejs M, et al. IRESite--a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010;38:D131–136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagliocco FA, et al. The influence of 5′-secondary structures upon ribosome binding to mRNA during translation in yeast. J Biol Chem. 1993;268:26522–26530. [PubMed] [Google Scholar]
- 38.Andreev DE, et al. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanbru C, et al. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 40.Stoneley M, et al. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 41.Coldwell MJ, et al. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 42.Meng Z, et al. The human IGF1R IRES likely operates through a Shine-Dalgarno-like interaction with the G961 loop (E-site) of the 18S rRNA and is kinetically modulated by a naturally polymorphic polyU loop. J Cell Biochem. 2010;110:531–544. doi: 10.1002/jcb.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci U S A. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilbert WV, et al. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 45.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 46.Shibuya N, Nakashima N. Characterization of the 5′ internal ribosome entry site of Plautia stali intestine virus. J Gen Virol. 2006;87:3679–3686. doi: 10.1099/vir.0.82193-0. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pleij CW, Rietveld K, Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kieft JS, et al. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kieft JS, et al. Mechanisms of internal ribosome entry in translation initiation. Symposia on Qualitative Biology. 2001;66:277–283. doi: 10.1101/sqb.2001.66.277. [DOI] [PubMed] [Google Scholar]
- 55.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji H, et al. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci U S A. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 58.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA. 2011 doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allam H, Ali N. Initiation factor eIF2-independent mode of c-Src mRNA translation occurs via an internal ribosome entry site. J Biol Chem. 2010;285:5713–5725. doi: 10.1074/jbc.M109.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfingsten JS, Castile AE, Kieft JS. Mechanistic role of structurally dynamic regions in Dicistroviridae IGR IRESs. J Mol Biol. 2010;395:205–217. doi: 10.1016/j.jmb.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang CJ, Lo MC, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol. 2009;387:42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 65.Costantino DA, et al. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishiyama T, et al. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Quesne JP, et al. Derivation of a structural model for the c-myc IRES. J Mol Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 69.Jopling CL, et al. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10:287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li PW, et al. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition. Nucleic Acids Res. 2006;34:853–864. doi: 10.1093/nar/gkj490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mitchell SA, et al. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 72.Morris MJ, et al. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 73.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 75.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 76.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 77.Celander DW, Cech TR. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991;251:401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- 78.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spahn CM, et al. Cryo-EM Visualization of a Viral Internal Ribosome Entry Site Bound to Human Ribosomes; The IRES Functions as an RNA-Based Translation Factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 81.Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 82.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 83.Boehringer D, et al. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure (Camb) 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 84.Pestova TV, et al. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Siridechadilok B, et al. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 86.Locker N, Chamond N, Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu J, et al. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc Natl Acad Sci U S A. 2011;108:1839–1844. doi: 10.1073/pnas.1018582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lukavsky PJ, et al. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 90.Collier AJ, et al. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat Struct Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 91.Kim I, Lukavsky PJ, Puglisi JD. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J Am Chem Soc. 2002;124:9338–9339. doi: 10.1021/ja026647w. [DOI] [PubMed] [Google Scholar]
- 92.Lukavsky PJ, et al. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 93.Klinck R, et al. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kieft JS, et al. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 95.Zhao Q, et al. Structure of hepatitis C virus IRES subdomain IIa. Acta Crystallogr D Biol Crystallogr. 2008;64:436–443. doi: 10.1107/S0907444908002011. [DOI] [PubMed] [Google Scholar]
- 96.Dibrov SM, et al. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew Chem Int Ed Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 97.Stark MR, et al. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA. 2006;12:2014–2019. doi: 10.1261/rna.93506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rabl J, et al. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 99.Ben-Shem A, et al. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 100.Serrano P, Ramajo J, Martinez-Salas E. Rescue of internal initiation of translation by RNA complementation provides evidence for a distribution of functions between individual IRES domains. Virology. 2009;388:221–229. doi: 10.1016/j.virol.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 101.Fernandez-Miragall O, et al. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ramos R, Martinez-Salas E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5:1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pilipenko EV, et al. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 104.Yu Y, et al. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song Y, et al. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.King HA, Cobbold LC, Willis AE. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 107.Spriggs KA, et al. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 108.Mitchell SA, et al. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol Cell Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ungureanu NH, et al. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. J Biol Chem. 2006;281:15155–15163. doi: 10.1074/jbc.M511319200. [DOI] [PubMed] [Google Scholar]
- 110.Pickering BM, et al. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dobbyn HC, et al. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008;27:1167–1174. doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pickering BM, et al. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol Cell Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu Y, et al. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kafasla P, et al. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 115.Kafasla P, et al. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mitra S, et al. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res. 2008;36:e63. doi: 10.1093/nar/gkn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pestova TV, I, Lomakin B, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]