Abstract
Posttranslational modification by ubiquitination determines intracellular location and fate of numerous proteins thus impacting a diverse array of physiologic functions. Past dogma has been that ubiquitin was only coupled to substrates by isopeptide bonds to internal lysine residues or less frequently peptide bonds to the N-terminus. Enigmatically, however, several proteins lacking lysines had been reported to retain ubiquitin dependent fates. Resolution of this paradox was afforded by recent observations that ubiquitination of substrates can also occur on cysteine or serine and threonine residues by thio- or oxy-ester bond formation, respectively (collectively called esterification). Although chemically possible, these bonds were considered too labile to be of physiological relevance. In this review we discuss recent evidence for the ubiquitination of protein substrates by esterification and speculate on its mechanism and its physiological importance.
Keywords: ubiquitin, ubiquitination, esterification, thioester bond, oxyester bond, ubiquitin-dependent degradation, ER-associated degradation, ubiquitin conjugating enzyme and ubiquitin protein ligase
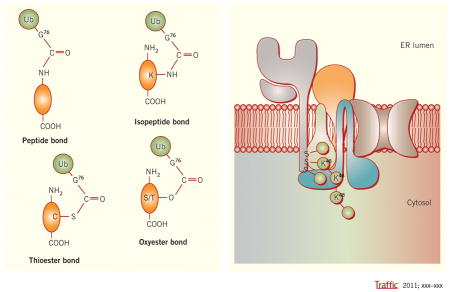
Ubiquitination is a process by which proteins are covalently modified by a 76aa ubiquitin moiety (Ub). This process is carried out by the orchestrated action of three types of enzymes—Ub activating enzyme (E1), Ub conjugating enzyme (E2) and Ub protein ligase (E3). Initially, the C-terminal glycine (Gly76) of Ub is activated by E1 in an ATP dependent manner. The activated Ub is then transferred to the active site cysteine (Cys) of an E2. Finally, the E3 interacts with the E2 to facilitate the transfer of the Ub from the E2 to the substrate. The attachment of Ub typically is achieved by formation of an amide bond between the G76 of Ub and the ε-NH2 of an internal lysine (Lys) residue, or less frequently, the free α-NH2 group of the N terminus of substrate (1) (Fig 1A, upper panel). Unexpectedly, recent studies have shown that Ub can be linked to substrates via Cys residues or serine (Ser)/threonine (Thr) residues by formation of thio- or oxy-ester bonds, respectively (Fig 1A, lower panel). In addition, substrates can be modified by one Ub moiety (mono-ubiquitination) or an Ub chain (poly-ubiquitination). Poly-Ub chains are typically linked by amide bonds to one of the 7 Lys residues or the N-termini of Ub moieties. Importantly the extent and linkage of the ubiquitination has a profound impact on the fate of the modified proteins. Thus when substrates are ubiquitinated there are two decisions an E3/E2 pair makes: which substrate residue to modify and which Ub linkage to use to build the chain (if substrates are polyubiquitinated). The precise mechanism by which E3/E2 makes these two decisions is incompletely understood (2–4). We speculate in this review article that future studies of substrate ubiquitination by esterification will provide key insights into the precise molecular and chemical mechanisms by which E3 and E2 enzymes combine to ubiquitinate proteins with appropriate modifications to control their function.
Figure 1.
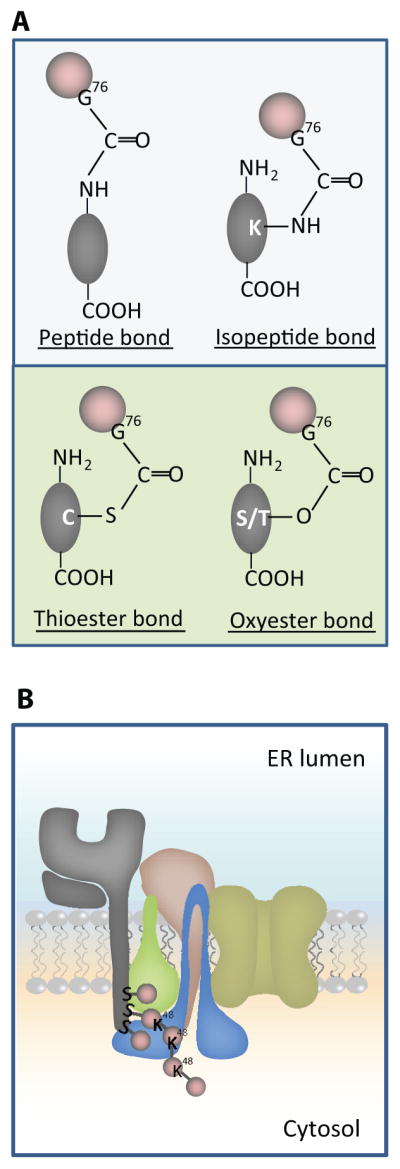
A. Schema c showing different chemical bonds between Ub and substrates. The upper panel shows pep de and isopep de linkages between Ub (Pink balls) and protein substrates (Gray ovals). Ub coupling on the N-terminus is shown on the le# and Ub coupling to an internal K residue is shown on the right. The lower panel of A shows thio- and oxy-ester linkages of Ub to C or S/T residues of substrates, respec vely. B. Schema c model showing how viral ligase mK3 (blue) interacts with Ube2j2 (green) to ubiqui nate S residues on the cytosolic tail of MHCI heavy chain (HC) (gray), then build a Lys48 (K48) linked chain. The adaptor proteins TAP/tapasin that confer substrate specificity by orien ng mK3/Ube2j2 with the HC tail are shown in yellow/tan.
The initial evidence for ubiquitination of internal non-Lys residues of proteins came from studies of viral MARCH (membrane-associated RING-CH) E3 ligases. Viral MARCH proteins are type III integral membrane proteins in which their N- and C-termini are cytosolic. The RING-CH motif in their N-terminal domains confers E3 activity (5). Based on the detection of highly conserved cellular homologs in man and mouse, viral MARCH ligases are presumed to have been stolen from the host and evolved in the virus to function in immune evasion (6–10). Since their discovery, viral MARCH ligases have turned out to be potent probes of Ub-dependent degradation pathways due to their potency and their limited number of substrates. For example, the viral ligase kK3 encoded by KSHV specifically targets MHC class I molecules for endocytosis and lysosome degradation by ubiquitinating the cytoplasmic tail of MHCI heavy chain (HC) (11,12). Surprisingly, an engineered mutant of HC with a Lys-less tail was still ubiquitinated and downregulated by kK3 (13). The ubiquitination of this Lys-less HC was dependent upon a Cys residue on the tail and was sensitive to reducing reagent β-mercaptoethanol indicating a thioester linkage. Although the active sites of E2s bind Ub via thioester bonds, these findings with kK3 provided the first evidence that substrates could also be coupled to Ub by thioester bonds. Interestingly, mK3, a homolog of kK3 that is expressed by murine γherpesvirus 68 was found to ubiquitinate Ser or Thr and not Cys residues on the tails of HC substrates (14). Furthermore, it was demonstrated that an HC tail with only Ser residues as potential Ub sites was resistant to denaturing with SDS but sensitive to mild alkaline treatment, thus chemically confirming an ester bond linkage. What is perhaps more surprising is that mK3 preferentially promoted ubiquitination of Ser residues on wildtype HCs even when Lys residues were in close proximity. More recently, using a semipermeabilized cell system, Ube2j2 was identified as the cognate E2 interacting with mK3 to preferentially catalyze conjugation of Ser residues (15). Thus viral ligase mK3 provided the first evidence that Ser/Thr on substrates could be coupled with Ub by ester bonds.
Several recent papers have also implicated cellular ligases in ubiquitination by esterification using common methods (Box1). Observations from representative papers will be briefly summarized here because their similarities and differences with each other and the above findings with viral ligases have intriguing mechanistic implications. These combined reports also accentuate the diversity of fundamental cellular functions where thioester- and ester-linked substrate ubiquitination has been implicated.
Box1. Current methods to distinguish different type of ubiquitination.
Genetic mutagenesis. Mutation of all Lys residues to Arg has been used to implicate the ubiquitination of non-Lys residues on substrates (as used in most studies reviewed here). Caveat: this approach provides only indirect evident for non-Lys conjugation and substitutions of Lys to Arg can cause secondary structural alterations potentially affecting required molecular interactions.
Chemical treatment. Unlike amide bonds, Ub moieties attached to the Cys residues of substrates via a thioester bond are susceptible to reductant, whereas Ub moieties attached by oxyester bonds to Ser or Thr residues are sensitive to mild alkaline hydrolysis (60). Based on these properties, the linkage of Ub conjugates can be distinguished by treating them with a reductant (dithiothreitol or β-mercaptoethanol), or mild alkaline conditions (such as 1M hydrozylamine, pH9.0 or 50–100mM NaOH). Typically, upon such treatment, the Ub attachment on a substrate via Cys or Ser/Thr residues visualized by western blot will disappear or be largely reduced. Caveat: resistance vs. sensitivity to the chemical treatment is relative (e.g. if treated too long in alkaline conditions even an amide bond can be affected). Thus a conclusion of non-Lys ubiquitination drawn based on such treatment should be cautious and a known Lys ubiquitination sample should be included as a control.
Mass spectrometry. This is the obvious gold standard to directly determine which residues are Ub conjugated. This method has been used to identify specific sites of ubiquitination via Lys residues based on the fact that trypsin proteolysis of an ubiquitinated protein produces a signature peptide. This signature peptide is characterized by a Gly-Gly remnant from the C-terminus of Ub that attached to a Lys residue of the substrate, which can be identified by a mass shift in the modified residue (61,62). Caveat: due to the labile nature of thio-/oxy-ester bonds Mass spectrometry approaches to demonstrate non-Lys ubiquitination have been a major challenge for us and others (33).
Apoptosis by Bid
In mammalian cells, induction of apoptosis by Bid requires cleavage in its unstructured loop, and subsequent release of its C-terminal proapoptotic Bcl-2 homology 3 (BH3) domain that in the absence of apoptotic stimuli is sequestered by the N-terminal region (tBid-N). It was found that upon apoptotic stimulus-induced Bid cleavage, the tBid-N is directly ubiquitinated and degraded, thus freeing the BH3-containing C-terminal fragment. tBid-N has no Lys residues and N-terminal ubiquitination was ruled out. However, mutation of multiple Cys, Ser and Thr residues within the N-terminal fragment in tBid-N abrogated ubiquitination, suggesting linkage of Ub by both thio-and oxy-ester bonds was critical (16). In support of this conclusion Ub conjugates on tBid-N were found to be susceptible to alkaline hydrolysis and reducing conditions.
Endocytic trafficking by HIV protein Vpu
Surface expression of innate immune restriction factor BST-2/tetherin on the surface is down-regulated by HIV-1 protein Vpu that presumably blocks recycling of endocytic BST-2 back to the surface. This blockage is partially mediated by interaction of Vpu with β-TrCP, the substrate adaptor of an SCF (Skp-Cullin1-F box) E3 ligase complex which in turn induces ubiquitination of BST-2. It was recently found that mutation of all potential ubiquitination sites in the cytoplasmic domain of BST-2, including Lys, Cys, Ser and Thr residues, was necessary to abrogate Vpu-mediated ubiquitination, suggesting conventional and non-conventional modes of ubiquitination may be carried out by the SCF complex (17).
Import of cargo proteins into peroxisomes by Pex5p
In yeast and mammals mono-ubiquitination of the peroxisome import receptor Pex5p on a conserved Cys is required for its release from the peroxisome membrane to recycle for another round of import (18–20). This conjugation is carried out by Pex4p as the E2 and Pex10 and/or Pex12p as the E3 (21–25). Two ATPases, Pex1p and Pex6p associated with the peroxisome membrane, function as dislocases for removal of mono-ubiquitinated Pex5p from the membrane (26). When the release process is blocked, Pex5p is polyubiquitinated on two conserved Lys residues by RING finger E3 ligase Pex2p paired with the E2 Ubc4, which leads to ERAD of Pex5p (25). Thus, the fate of whether Pex5p undergoes recycling vs. degradation is determined by distinct ubiquitination machineries using non-conventional or conventional Ub conjugation, respectively.
ERAD of orphan TCRα
The T-cell antigen receptor α-chain requires assembly as a heterodimeric complex to be expressed on the T cell surface for specific antigen recognition. Unassembled TCRα is subjected to ERAD (27,28). However, its short tail lacks Lys residues and mutation of all Lys residues in its extracellular domain to arginine (Arg) did not affect the degradation of the protein, raising the question of the nature of the ubiquitination (29). Recently, the Bonifacino laboratory reported that substitution of two evolutionarily conserved Ser residues on the tail of TCRα to Ala residues remarkably reduced the extent of ubiquitination as well as the rate of degradation of TCRα (30). Ubiquitination of TCRα, however, was not impaired by replacement of 2 Ser residues with Cys, Thr or Lys residues. These findings suggest that the two Ser residues on the tail of TCRα very likely function as the Ub acceptor sites and Cys, Lys and Thr can serve as alternatives (30). In a similar story the Bonifacino laboratory used a mutagenesis approach to show Ser/Thr as well as Lys residues on the tail of CD4 contribute to its ERAD induced by the HIV-1 protein Vpu (31).
ERAD of non-secreted NS-1 Ig light chain
Partially oxidized NS-1 immunoglobulin expressed in mammalian cells is ubiquitinated on its VL domain and rapidly degraded by ERAD (32). Using mutagenesis and biochemical approaches, Shimizu et al. showed that ubiquitination on NS-1 was sensitive to high pH but not affected by mutation of all Lys residues of the VL domain. These findings suggested that ubiquitination of NS-1 preferentially occurs on Ser or Thr residues (33). However, when all Ser and Thr residues were mutated, Ub was conjugated to Lys residues. Both types of ubiquitination (Ser/Thr or Lys) resulted in ERAD (33). Significantly, HRD1 (a major ERAD-associated E3 ligase in yeast and mammals (34–37)) was implicated in the ubiquitination of NS-1 via either Ser/Thr or Lys residues. Shimizu et al. also found that Ub conjugates of two other known HRD1 substrates, the mini-immunoglobulin heavy chain and the MHK α1-anti-trypsin mutant, were partially sensitive to alkaline treatment, underscoring the pervasiveness of Ser/Thr conjugation of ERAD substrates by HRD1. Of note, these authors also reported their failure to detect ester bonds on substrates by mass spectrometry, consistent with the labile nature of this linkage.
Based on the above reported examples of substrate ubiquitination by esterification, four general mechanistic conclusions can be drawn. First, there is flexibility in whether Lys vs. non-Lys residues on substrates are ubiquitinated implying common Ub components are used. Non-Lys ubiquitination has been observed on both experimentally-derived Lys-less mutant substrates as well as native substrates lacking Lys residues. Interestingly, non-conventional residues can be primary Ub acceptor sites even when Lys residues are available. For example, Ser/Thr ubiquitination can be the preferred mechanism of modification like ubiquitination of HC by mK3 or NS-1 by HRD1 (14,33). In both cases, non-primary conjugation residues can serve as alternatives, suggesting that ubiquitination of Lys vs. non-Lys residues may be carried out by the same Ub machinery. Indeed several lines of evidence clearly support this notion. In all of the above examples where the E3 was identified, it is the same E3 that facilitates both Lys and non-Lys conjugations. In addition, in a semipermeabilized cell system, mK3 interacted with solubilized Ube2j2 to support Lys-ubiquitination when Ser residues were not present. However, in live cells, Ube2j2 and mK3 preferentially ubiquitinated Ser/Thr and not Lys residues (15). Mechanistically the implication of this finding is that Ube2j2 is not specialized for Ser/Thr conjugation. In other words, the E2 is not the only determinant of non-Lys ubiquitination. Of note, studies of Pex4p-dependent ubiquitination of a conserved Cys residue on Pex5p support the same conclusion. In mammals, the functional homolog of Pex4p is Ube2d1/2/3 (38). Ube2d members consist exclusively of the core catalytic domain (39). They have been shown to have broad E3 interacting activity ranging from HECT type, RING type to U box type E3s to promote Ub conjugation on Lys residues of substrates (40). Thus, Ube2d members are also not specialized for non-Lys ubiquitination.
Second, the ability to conjugate Ub on non-Lys residues requires efficacious E2 and appropriate E3/E2 pairs
Studies of viral ligases have demonstrated that specific sequences surrounding the Ub acceptor residues (Lys or non-Lys) are not required for Ub conjugation of HC. For example, viral ligase mK3 ubiquitinated engineered HC substrates with a poly-Gly tail containing either two Lys or Ser/Thr residues (41). Similarly, viral ligase kK3 ubiquitinated engineered HC tails with a single Cys residue and randomly arranged Gly and Ala residues (42). Although surrounding sequence was not important, these viral ligases were found to be highly selective in tail location of the Ub acceptor site; and this optimal location was the same whether Lys or non-Lys residues were Ub conjugated. Thus selection of Ub sites in these examples is likely based on their proximity to the active site of E2 positioned by E3, as previously proposed in certain conventional ubiquitination models (43–45). In the specific case of viral ligase mK3, binding to adaptor proteins TAP/tapasin confers mK3 specific detection of HC (Fig. 1B). In this model, the binding to TAP/tapasin positions the mK3/Ube2j2 complex in proximity to the C-terminus of the HC tail (Fig 1B). If similar proximity models are generalizable to other situations where non-Lys ubiquitination has been implicated, then juxtaposition of the substrate and catalytic site of the E2 by E3 is likely a key factor in determining which substrate residue, Lys or non-Lys is ubiquitinated. However, proximity is not the only determining factor since not all E3/E2 pairs can support non-Lys ubiquitination. For instance, mK3/Ube2j2 facilitates both Ser/Thr- and Lys-ubiquitination, whereas mK3/Ube2d1 only facilitates Lys ubuiquitination (15). Interestingly, however, Ube2d1 also mediates Cys-ubiquitination on PEX5 (38). Thus non-Lys ubiquitination requires a selective E2 and a specific E3/E2 interaction. In strong support of this notion several recent studies have demonstrated that E3s can induce a conformational change that activates E2s (46–48). To define the functional importance of E3/E2 interaction, a recent in vitro study by Wenzel et al. used free Lys to discharge Ub from different human E2s tested alone or with various E3s. This assay was designed to mimic the nucleophilic attack of substrate Lys residues on the charged E2. As a positive control, free Cys was used to disrupt the thioester-linked Ub from the E2. They found that UBE2K, UBC13 and UBCH5C, E2s known to function with RING-type E3s, could react with both Cys and Lys, whereas UBCH7 that has specificity for HECT-type E3s could only react with Cys, but not Lys to release the charged Ub in the absence of the E3 (49). Their findings suggest that E2s have both intrinsic and E3-dependent properties that determine their capacity to Ub conjugate different types of substrate residues. The success of the Wenzel et al. study highlights the future possibility of using similar approaches to predict or define functional E2/E3 pairs which facilitate non-Lys ubiquitination.
Third, ubiquitination of multiple closely located acceptor sites (Lys and/or Ser/Thr) may be important for poly-Ub chain assembly
For most substrates listed above more than one residue, and sometimes more than one type of residue may be initially conjugated. This is evidenced by the findings that when multiple sites are available only mutation of all the potential sites can abrogate ubiquitination of the substrate, e.g. as seen above in studies of tBid-N, NS-1 and Vpu. In mK3-induced HC ubiquitination, more Ub moieties were found to be associated with the HCs when multiple Ser residues were appropriately clustered on the tail (15). Furthermore, wild type HC tails with multiple Ser residues were degraded more rapidly than engineered single Ser containing tails, suggesting multi-mono Ub on substrate may assist in the building a poly-Ub chain long enough for proteasome recognition (14). Multi-mono Ub could promote poly-Ub chain assembly by providing multiple choices for where to build a chain or by increasing affinity between the E3 ligase and substrate to improve processivity of chain assembly. It should be noted that this multi-initial Ub conjugation phenomenon has also been observed in conventional ubiquitination such as substrate ubiquitination mediated by multi-subunit E3 ligases, APC and SCF (48,50). This similarity implies that the mechanism involved in Ub chain elongation may be the same no matter which residue is used for initial Ub conjugation.
Fourth, non-Lys ubiquitination occurs only on substrate and not on a poly-Ub chain
To address how initial conjugation of Ub on non-Lys residues may influence chain assembly, a semipermeabilized cell assay was used to probe HC ubiquitination by mK3-Ube2j2 (i.e. an ERAD model system in which a specific E2/E3 pair was known to couple Ub to Ser or Lys residues on HC substrates) (15). In these studies, mutants of each of the 7 Lys residues in Ub were tested for their ability to assemble a poly-Ub chain on the HCs with either one Ser or one Lys acceptor site. These comparisons indicated that only a Lys48 linkage on the chain was utilized. Furthermore, to determine whether a non-Lys residue at position 48 could function as an alternative acceptor site, a Lys48Ser Ub mutant was also tested and found to lack the ability to form a poly-Ub chain (X.W. unpublished). These findings demonstrate that an E3/E2 complex that has the potential to ubiquitinate either a Ser or Lys residue on the substrate, then builds a poly-Ub chain with exclusively Lys specific linkage. Furthermore, this conclusion is likely true for the other models where non-Lys Ub conjugation has been implicated. For example, it is probably the case for HRD1-induced non-conventional ubiquitinations because the two well-characterized cognate E2s of HRD1, Ube2k (Ubc1) and Ube2g2 (Ubc7), identified in yeast and mammals are both Lys48 specific (51,52). The physiological significance of this disparity in ubiquitination of substrate and formation of an Ub chain remains enigmatic. Recent findings show that orientation of the acceptor Ub and the interaction between the residues surrounding the acceptor Lys and the active site of E2 have a great impact on Lys selection of chain assembly (53–55). Thus, it is tempting to speculate that the first Ub conjugated on the substrate may create a new interaction face between Ub and Ube2j2 that specifically restricts a nucleophilic attack from K48. It should also be noted that in other model systems, different E2s may be sequentially recruited by E3 ligases to respectively conjugate the substrate and build a poly-Ub chain (51,56–58).
In summary, ubiquitination of substrates by esterification has clearly been conserved in evolution and implicated in diverse cell biological pathways. Although, the physiological significance has yet to be defined, evidence thus far suggests several possibilities. For example, the ability to conjugate Ser, Thr and Cys residues in addition to Lys residues might provide quality control and other Ub-dependent functions to proteins lacking Lys residues. Alternatively, additional conjugation sites may promote poly-Ub chain assembly. The chemically labile nature of non-Lys ubiquitination may also be required by certain cellular process, as evidenced by the Pex5p model where the lability of thioester bonds presumably leads to a more rapid recycling of peroxisomal import machinery. In addition, thio- and/or oxy-ester bonds may have differential sensitivity to DUB mediated deubiquitination facilitating its unique utility in biological processes. It is also possible that non-Lys Ub conjugation may provide another layer of regulation between different post-posttranslational pathways. For instance, Ser or Thr residues are also sites of phosphorylation and Cys residues can be modified in various ways including oxidation, glutathionylation, nitrosylation and acylation, all of which would alter the protein activity and/or localization. And finally, ubiquitination by esterification may provide a means for the host to counteract pathogen escape from degradation. For instance, several AB-toxins of bacteria subvert the ERAD pathway to enter target cells and lack internal Lys residues for Lys-linked ubiquitination (59). Regardless of its physiologic function, future studies of ubiquitination of substrates by esterification will likely provide unique molecular and structural insights into how E2s are selected by E3s, and how E3/E2 pairs combine to ubiquitinate specific sites on substrates and form Ub poly-chains.
Acknowledgments
We thank Dr. Janet Connolly for critical reading of this manuscript and Ms. Lusi Wang for art work in Fig 1. Funding for our research was provided by NIH grant AI019687.
Reference List
- 1.Ciechanover A, Ben Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem–J. 2010;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coscoy L, Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003;13:7–12. doi: 10.1016/s0962-8924(02)00005-3. [DOI] [PubMed] [Google Scholar]
- 6.Holzerlandt R, Orengo C, Kellam P, Alba MM. Identification of new herpesvirus gene homologs in the human genome. Genome Res. 2002;12:1739–1748. doi: 10.1101/gr.334302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto E, Ishido S, Sato Y, Ohgimoto S, Ohgimoto K, Nagano-Fujii M, Hotta H. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J Biol Chem. 2003;278:14657–14668. doi: 10.1074/jbc.M211285200. [DOI] [PubMed] [Google Scholar]
- 8.Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. 2004;78:1109–1120. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehner PJ, Hoer S, Dodd R, Duncan LM. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev. 2005;207:112–125. doi: 10.1111/j.0105-2896.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- 10.Ohmura-Hoshino M, Goto E, Matsuki Y, Aoki M, Mito M, Uematsu M, Hotta H, Ishido S. A novel family of membrane-bound E3 ubiquitin ligases. J Biochem. 2006;140:147–154. doi: 10.1093/jb/mvj160. [DOI] [PubMed] [Google Scholar]
- 11.Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155:1265–1273. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJ, Hansen TH. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol. 2009;187:655–668. doi: 10.1083/jcb.200908036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tait SW, de Vries E, Maas C, Keller AM, D’Santos CS, Borst J. Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J Cell Biol. 2007;179:1453–1466. doi: 10.1083/jcb.200707063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokarev AA, Munguia J, Guatelli JC. Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J Virol. 2011;85:51–63. doi: 10.1128/JVI.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho AF, Pinto MP, Grou CP, Alencastre IS, Fransen M, Sa-Miranda C, Azevedo JE. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J Biol Chem. 2007;282:31267–31272. doi: 10.1074/jbc.M706325200. [DOI] [PubMed] [Google Scholar]
- 19.Williams C, van den BM, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem. 2007;282:22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 20.Okumoto K, Misono S, Miyata N, Matsumoto Y, Mukai S, Fujiki Y. Cysteine Ubiquitination of PTS1 Receptor Pex5p Regulates Pex5p Recycling. Traffic. 2011;12:1067–1083. doi: 10.1111/j.1600-0854.2011.01217.x. [DOI] [PubMed] [Google Scholar]
- 21.Platta HW, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiel JA, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 23.Kragt A, Voorn-Brouwer T, van den BM, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 24.Williams C, van den BM, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 25.Platta HW, El Magraoui F, Baumer BE, Schlee D, Girzalsky W, Erdmann R. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platta HW, Debelyy MO, El Magraoui F, Erdmann R. The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans. 2008;36:99–104. doi: 10.1042/BST0360099. [DOI] [PubMed] [Google Scholar]
- 27.Lippincott-Schwartz J, Bonifacino JS, Yuan LC, Klausner RD. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- 28.Bonifacino JS, Suzuki CK, Lippincott-Schwartz J, Weissman AM, Klausner RD. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J Cell Biol. 1989;109:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 30.Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem. 2010;285:23916–23924. doi: 10.1074/jbc.M110.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell. 2010;40:917–926. doi: 10.1016/j.molcel.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 35.Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko M, Ishiguro M, Niinuma Y, Uesugi M, Nomura Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 2002;532:147–152. doi: 10.1016/s0014-5793(02)03660-8. [DOI] [PubMed] [Google Scholar]
- 37.Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun. 2003;303:91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- 38.Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, Warscheid B, Sa-Miranda C, Azevedo JE. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J Biol Chem. 2008;283:14190–14197. doi: 10.1074/jbc.M800402200. [DOI] [PubMed] [Google Scholar]
- 39.Scheffner M, Huibregtse JM, Howley PM. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci U S A. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorick KL, Jensen JP, Weissman AM. Expression, purification, and properties of the Ubc4/5 family of E2 enzymes. Methods Enzymol. 2005;398:54–68. doi: 10.1016/S0076-6879(05)98006-3. [DOI] [PubMed] [Google Scholar]
- 41.Herr RA, Harris J, Fang S, Wang X, Hansen TH. Role of the RING-CH domain of viral ligase mK3 in ubiquitination of non-lysine and lysine MHC I residues. Traffic. 2009;10:1301–1317. doi: 10.1111/j.1600-0854.2009.00946.x. [DOI] [PubMed] [Google Scholar]
- 42.Cadwell K, Coscoy L. The specificities of Kaposi’s sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets. J Virol. 2008;82:4184–4189. doi: 10.1128/JVI.02264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 44.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 45.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Paveltich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 46.Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Nacusi L, Sheaff RJ, Liu X. Ubiquitination of p21Cip1/WAF1 by SCFSkp2: substrate requirement and ubiquitination site selection. Biochemistry. 2005;44:14553–14564. doi: 10.1021/bi051071j. [DOI] [PubMed] [Google Scholar]
- 49.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 51.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 52.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 53.Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, Luzio J, Lehner PJ. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 58.Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hazes B, Read RJ. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 1997;36:11051–11054. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- 60.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng JM, Schwartz D, Elias JE, Thoreen CC, Cheng DM, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nature Biotechnology. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 62.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]


