Abstract
Trypanosoma cruzi, the causative agent of Chagas' disease, preferentially infects cardiac and digestive tissues. Baboons living in Texas (Papio hamadryas) and cynomolgus monkeys (Macaca fascicularis) have been reported to be infected naturally with T. cruzi. In this study, we retrospectively reviewed cases of animals that were diagnosed with lymphocytic myocarditis and used a polymerase chain reaction (PCR)-based method (S36/S35 primer set) to amplify T. cruzi DNA from archived frozen and formalin-fixed paraffin-embedded (FFPE) cardiac tissues. We show that the PCR method is applicable in archived frozen and FFPE tissues and the sensitivity is in the femtogram range. A positive correlation between PCR positivity and lymphocytic myocarditis in both baboons and cynomolgus monkeys is shown. We also show epicarditis as a common finding in animals infected with T. cruzi.
Introduction
Chagas' disease is an illness caused by infection with the kinetoplastid protozoan parasite Trypanosoma cruzi. The parasite is typically transmitted to humans by sanguivorous insects belonging to the genus Triatoma (Order Hemiptera, Family Reduviidae) and commonly known as “cone-nosed” bugs. Non-vector-borne routes of transmission include blood transfusion, organ or tissue transplantation, congenital, laboratory exposure, and consumption of contaminated foods.1–4 The disease is widespread in Central and South America, where currently about 8 million people are estimated to be infected and 109 million are at risk of infection.3 In 2006, an estimated 12,500 people died caused by complications arising from chronic, symptomatic Chagas' disease in Latin America, primarily from heart pathologies.3 Approximately two-thirds of deaths of patients with Chagasic heart disease were caused by sudden cardiac arrest, 25–30% were caused by refractory congestive heart failure, and 10–15% were caused by thromboembolic events.3 It is clear that this organism preferentially infects cardiac tissue, although it can infect other organs. South of the Amazon basin, 10–15% of the chronically infected develop chronic megacolon or megaesophagus, indicating tropism of this organism for the gastrointestinal system.3
The parasites and reduviid insect vectors are also present in the southern United States.5–7 In the state of Texas, the distribution of triatome bugs is widespread encompassing all major geographic regions.8
In addition to humans, T. cruzi naturally infects a wide variety of mammalian species including opossums, armadillos, and nonhuman primates.9–11 Chagas' disease is increasingly diagnosed in domestic pets in the southern United States. Dogs are generally regarded as important in the transmission of Chagas' disease around human settlements as they maintain blood parasitemia long after infection and are a preferred source of blood meals for Triatoma infestans, which is the most important vector of T. cruzi.12–15
Trypanosoma cruzi is a kinetoplasid protozoa and contains a complex network of catenated circular DNA molecules within its single mitochondrion.16,17 Each network of circular DNA (also called kinetoplast DNA or kDNA) consists of 5–20 × 103 minicircles and 20–50 maxicircles.16,17 Kinetoplast DNA, especially the minicircles, are good candidates for the development of detection and classification methods of T. cruzi.
Polymerase chain reaction (PCR) is currently used as an adjunct to immunologic diagnosis of Chagas' disease.4,18 The S35/S36 primer set used for PCR amplification of T. cruzi is based on the amplification of kinetoplast DNA.19
The Southwest National Primate Research Center (SNPRC) located in San Antonio, TX, is home to ∼2,500 nonhuman primates, including baboons, chimpanzees, and two species of macaque, most of which are housed in large, open, outdoor cages. In 1984, it was discovered that some primates at the SNPRC were infected with T. cruzi11; the animals are possibly bitten by the insect vector while resting or sleeping, but the primary route of infection is believed to be opportunistic eating of the insect vectors.20 At the SNPRC, complete necropsies are carried out on all animals that die. Over 1,000 necropsies are carried out every year.
The aims of this study were 3-fold: 1) to explore the use of a PCR-based method that uses the S35/S36 primer set to amplify T. cruzi DNA from baboon and cynomolgus monkey heart tissues, 2) to determine if PCR positivity is correlated with lymphocytic myocarditis, and 3) to determine the use of frozen and formalin-fixed paraffin-embedded (FFPE) tissue samples for detecting presence of T. cruzi by PCR.
Materials and Methods
Study animals.
Baboons.
Archived cardiac tissue samples from 26 baboons with a diagnosis of myocarditis were used in this study. Both frozen and FFPE samples were available for 13; only FFPE samples were available for the other 13. Archived cardiac tissue samples from 20 baboons with no diagnosis of myocarditis were used as controls. The FFPE samples were available from 12 and frozen from eight of these baboons. The demographics of the baboons are shown in Table 1.
Table 1.
Demographic information for baboons used in this study, and data obtained
| Animal no. | Age (years) | Sex | Myocarditis | PCR result (frozen) | PCR result (FFPE) | Amastigotes | Type of inflammation | Location of inflammation | Serology |
|---|---|---|---|---|---|---|---|---|---|
| 13545 | 16 | F | + | + | + | − | L, P, H, E | Epicardium/Myocardium | + |
| 14735 | 6.4 | F | + | + | + | − | L, P | Epicardium/Myocardium | + |
| 15238 | 14 | F | + | + | + | − | L, P, H, E | Epicardium/Myocardium | + |
| 15330 | 13.6 | F | + | + | + | − | L, P, H | Myocardium | + |
| 15564 | 13.2 | F | + | + | + | − | L, P | Epicardium/Myocardium | + |
| 16219 | 12.6 | F | + | + | + | − | L, P, H, E | Epicardium/Myocardium | + |
| 28146 | 6.0 | F | + | + | + | − | L, P, H, E | Myocardium | + |
| 10793 | 15 | M | + | + | − | − | L | Myocardium | + |
| 16191 | 12.6 | F | + | + | − | − | L, P, H, E | Myocardium | + |
| 19124 | 9.8 | F | + | + | − | − | L, P, H | Myocardium | + |
| 15796 | 7.6 | F | + | − | + | − | L, P, H, E | Epicardium/Myocardium | + |
| 27284 | 7.0 | F | + | − | + | − | L, P, H | Myocardium | + |
| 27292 | 7.0 | F | + | − | + | − | L, P | Epicardium/Myocardium | + |
| 11716 | 9.0 | M | + | ND | + | + | L, P, H, E, N | Epicardium/Myocardium | + |
| 1X3641 | 25.0 | F | + | ND | + | − | L, P | Epicardium/Myocardium | + |
| 8251 | 17.0 | M | + | ND | − | − | L, P, H, E, N | Epicardium/Myocardium | − |
| 8369 | 17.0 | M | + | ND | − | − | L, P | Epicardium/Myocardium | + |
| 10520 | 15.9 | F | + | ND | − | − | L, P, H | Epicardium/Myocardium | + |
| 14905 | 9.6 | F | + | ND | − | − | L, P, H | Epicardium/Myocardium | − |
| 15615 | 11.4 | F | + | ND | − | − | L | Epicardium/Myocardium | − |
| 17759 | 6.0 | F | + | ND | − | − | L, P | Myocardium | ID |
| 1X2001 | 27.4 | F | + | ND | − | − | L, P, H, E | Epicardium/Myocardium | + |
| 1X2176 | 30.5 | F | + | ND | − | − | L, P | Epicardium/Myocardium | + |
| 1X3731 | 24.6 | F | + | ND | − | − | L, P, H, E | Epicardium/Myocardium | + |
| 1X3846 | 24.8 | F | + | ND | − | − | L | Myocardium | − |
| 1X3873 | 24.7 | F | + | ND | − | − | L, P | Epicardium/Myocardium | + |
| 12098 | 8.3 | M | − | ND | − | − | ID | ||
| 12667 | 12.8 | F | − | ND | − | − | − | ||
| 13001 | 12.2 | F | − | ND | − | − | − | ||
| 13202 | 12.2 | F | − | ND | − | − | − | ||
| 13352 | 11.5 | F | − | ND | − | − | − | ||
| 14131 | 10.6 | F | − | ND | − | − | − | ||
| 14624 | 10.4 | F | − | ND | − | − | − | ||
| 14629 | 10.4 | F | − | ND | − | − | − | ||
| 15983 | 8.0 | F | − | ND | − | − | − | ||
| 18571 | 6.0 | M | − | ND | − | − | ID | ||
| 18623 | 6.1 | M | − | ND | − | − | ID | ||
| 28023 | 2.3 | M | − | ND | − | − | − | ||
| 7223 | 22.6 | M | − | − | ND | − | − | ||
| 8274 | 21.6 | F | − | − | ND | − | − | ||
| 8624 | 21.2 | F | − | − | ND | − | − | ||
| 9129 | 20.4 | F | − | − | ND | − | − | ||
| 15654 | 7.6 | M | − | − | ND | − | − | ||
| 16560 | 6.0 | M | − | − | ND | − | − | ||
| 26988 | 7.2 | F | − | − | ND | − | − | ||
| 27998 | 7.2 | M | − | − | ND | − | − |
PCR = polymerase chain reaction; FFPE = formalin-fixed paraffin-embedded; L = lymphocytes, P = plasma cells, H = histiocytes/macrophages, E = eosinophils, N = neutrophils; ND = not done; ID = indeterminate.
Cynomolgus monkeys.
Archived cardiac tissue samples from 19 cynomolgus monkeys with a diagnosis of myocarditis were used in this study. Both frozen and FFPE samples were available for 11; only FFPE samples were available for the other eight. Archived cardiac tissue samples from 12 cynomolgus monkeys with no diagnosis of myocarditis were used as controls. Both frozen and FFPE tissue samples were available for three of the control monkeys; only FFPE samples were available for the other nine. The demographics of the cynomolgus monkeys are provided in Table 2.
Table 2.
Demographic information for cynomolgus monkeys used in study and data obtained
| Animal no. | Age (years) | Sex | Myocarditis | PCR result (frozen) | PCR result (FFPE) | Amastigotes | Type of inflammation | Location of inflammation | Serology |
|---|---|---|---|---|---|---|---|---|---|
| 20443 | 2.8 | F | + | + | + | − | L | Epicardium/Myocardium | + |
| 20567 | 14.7 | F | + | + | + | − | L, P, H | Epicardium/Myocardium | + |
| 20575 | 14.6 | M | + | + | + | − | L, P, H | Epicardium/Myocardium | + |
| 20657 | 10.2 | F | + | + | + | − | L, P | Epicardium/Myocardium | ID |
| 20787 | 17.7 | F | + | + | + | − | L, P, H | Epicardium/Myocardium | + |
| 20805 | 7.5 | F | + | + | + | − | L, P, H | Myocardium | + |
| 20857 | 20.6 | F | + | + | + | − | L,P, H, E | Epicardium/Myocardium | ID |
| 21268 | 14.5 | F | + | + | + | − | L, P | Epicardium/Myocardium | + |
| 20281 | 3.5 | M | + | + | − | − | L, P | Epicardium/Myocardium | + |
| 20445 | 15.8 | F | + | + | − | − | L, P | Epicardium/Myocardium | + |
| 20476 | 3.1 | M | + | + | − | − | L, P, H, E | Epicardium/Myocardium | + |
| 20883 | 1.7 | M | + | ND | + | + | L, P, H | Epicardium/Myocardium | − |
| 22345 | 20.6 | F | + | ND | + | + | L, P, H | Epicardium/Myocardium | ND |
| 23463 | 3.0 | F | + | ND | + | + | L, P, H, N | Epicardium/Myocardium | ND |
| 23605 | 1.6 | F | + | ND | + | + | L, P | Epicardium/Myocardium | ND |
| 24478 | 2.2 | F | + | ND | + | + | L, P, H | Epicardium/Myocardium | ND |
| 24776 | 0.5 | F | + | ND | + | − | L, P, H, N | Epicardium/Myocardium | ND |
| 20161 | 17.7 | F | + | ND | − | − | L | Myocardium | ND |
| 21277 | 15.3 | F | + | ND | − | − | L | Myocardium | − |
| 20564 | 12.0 | F | − | − | − | − | + | ||
| 21020 | 3.3 | F | − | − | − | − | ND | ||
| 21395 | 14.7 | F | − | − | − | − | + | ||
| 20446 | 13.7 | M | − | ND | − | − | − | ||
| 20455 | 9.9 | F | − | ND | − | − | − | ||
| 20656 | 10.2 | F | − | ND | − | − | ID | ||
| 20852 | 20.6 | F | − | ND | − | − | − | ||
| 20895 | 3.2 | M | − | ND | − | − | ID | ||
| 20991 | 13.7 | F | − | ND | − | − | − | ||
| 21203 | 6.0 | F | − | ND | − | − | − | ||
| 21383 | 14.7 | F | − | ND | − | − | ID | ||
| 26662 | 1.8 | F | − | ND | − | − | − |
PCR = polymerase chain reaction; FFPE = formalin-fixed paraffin-embedded; L = lymphocytes, P = plasma cells, H = histiocytes/macrophages, E = eosinophils, N = neutrophils; ND = not done; ID = indeterminate.
Pathology examination.
Complete necropsies were performed on all animals. Samples of the left ventricle were fixed in 10% neutral-buffered formalin, embedded in paraffin, and archived at room temperature.
Myocarditis was diagnosed by a veterinary pathologist if, on microscopic observation, animals exhibited focal to diffuse primarily lymphocytic myocardial infiltrates, with minimum admixture of other inflammatory cells. Clusters of organisms morphologically consistent with T. cruzi amastigotes were seen in some tissue sections.
All animals were provided care in compliance with the Guide for the Care and Use of Laboratory Animals. All procedures were approved by the Institutional Animal Care and Use Committee. Texas Biomedical Research Institute is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International.
PCR method.
DNA extraction.
The DNA was obtained from the frozen left ventricle and from FFPE tissue blocks. For FFPE samples, a 3 mm tissue punch (Acuderm Inc., Fort Lauderdale, FL) was used to acquire samples from the wax blocks; these samples were then de-paraffinized overnight in xylene. For frozen tissues and de-paraffinized FFPE tissues, DNA was extracted using the DNeasy kit (Qiagen, Valencia, CA), according to the manufacturer's specifications and quantified by a spectrophotometer (NanoDrop Technologies, Wilmington, DE).
PCR procedure.
Standard techniques of conventional PCR were used to test for the presence of parasite and host DNA from all animals. Primers are listed in Table 3. Reference DNA for T. cruzi (Tulahuen strain) was purchased from American Type Culture Collection (ATCC, Manassas, VA) and was used as the positive control.
Table 3.
Primers used for the detection of Trypanosoma cruzi in cardiac tissue from baboons and cynomolgus monkeys
A 330-base pair (bp) kinetoplast minicircle DNA sequence specific to T. cruzi was amplified using primers S35 and S36, and Platinum Taq DNA polymerase (Life Technologies, Carlsbad CA) under “touchdown” conditions.19 Amplification reactions were performed in a final volume of 25 μL containing 0.5 U of Platinum Taq DNA polymerase high fidelity (Life Technologies), 0.2 mM of each of the dNTPs, 2 mM MgSO4, 0.4 μM of each primer, 1× high fidelity buffer, and 1 μL of template DNA. The thermal cycling conditions were as follows: Briefly, following an initial denaturation step at 94°C for 2 minutes, the annealing temperature was set to 59°C for two cycles, and thereafter decreased by 1° for every 2 cycles until the temperature reached 52°C. At those conditions the samples were run for an additional 20 cycles. The PCR product was loaded onto 1% agarose gels and electrophoresis was performed. The gels were then stained with ethidium bromide, visualized under UV illumination, and photo-documented. The presence of a 330-bp band indicated positivity of T. cruzi.
In separate reactions, a 244-bp sequence of the 16S ribosomal RNA (rRNA) gene was amplified from the samples as a control for failure to obtain good quality DNA. It was done using the primer set L2513 and H271421; we also investigated the suitability of AmpliTaq Gold 360 DNA polymerase (Life Technologies) as a substitute for Platinum Taq DNA polymerase high fidelity for our PCR method.
Serological analysis.
Conventional enzyme-linked immunosorbent assay (ELISA), and immunochromatographic serodiagnostic kits are used to screen a subset of colony animals for antibodies to T. cruzi. For the purposes of colony management, a positive outcome in both of these assay methods is accepted as indicating that the animal is infected with T. cruzi.
Statistical analysis.
We determined the correlation between T. cruzi PCR results and myocarditis in two species. For each species we had data from frozen and FFPE samples. Statistical comparisons were made using two-sided Fisher's exact test of independence to show the relationship between myocarditis and PCR positivity in both frozen and FFPE samples. Statistical analysis was performed using the R system for statistical computing.22
Results
Histopathology.
Histopathological evaluation indicated that in baboons and cynomolgus monkeys with myocarditis, the heart contained focal to multifocal collections of lymphocytes and lesser plasma cells within the myocardium and along the epicardial surface. In all cases the vast majority of inflammatory cells were lymphocytes, often with plasma cells, and frequently some histiocytic infiltration. In a few cases, there was rare infiltration by eosinophils and neutrophils, with the latter occurring in cases with active necrosis. Eosinophilic infiltration was more common in baboons when compared with cynomolgus monkeys. However, in one cynomolgus monkey, eosinophilic infiltration was much more intense. Epicardial inflammation was present in 18 of 23 baboons (Table 1). Three other baboon slides did not have the epicardium on the sections examined and therefore we do not know if there was epicardial inflammation in those baboons. In cynomolgus monkeys, epicardial inflammation was present in 16 of 19 (Table 2). Occasionally scattered foci of shrunken and pale staining myocardial fibers were observed. Clusters of organisms morphologically consistent with T. cruzi amastigotes were seen in some tissue sections. Trypansoma cruzi amastigotes were seen in one of the 26 (3.8%) baboons and five of the 19 (26%) cynomolgus monkeys with myocarditis (Tables 1 and 2).
PCR analysis.
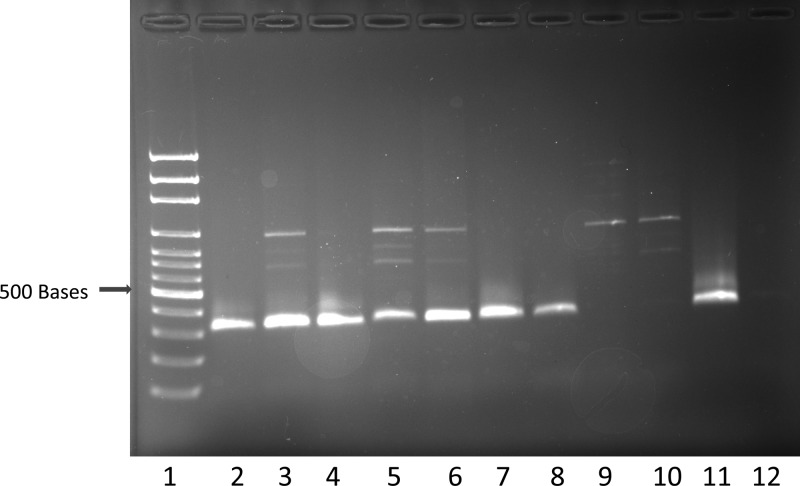
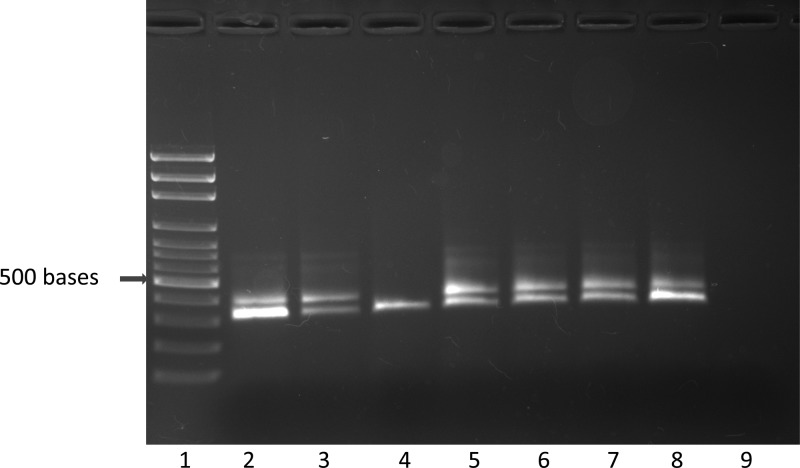
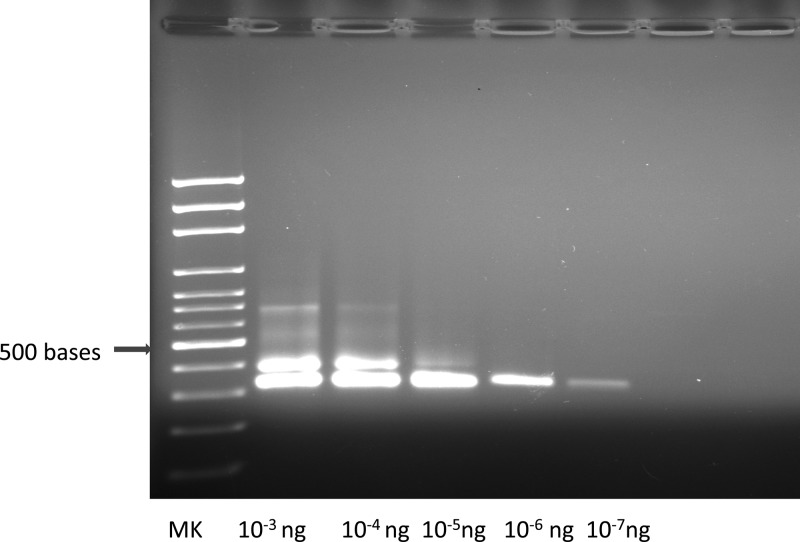
Using the Platinum Taq DNA polymerase (Life Technologies), the S35/S36 primer set successfully amplified T. cruzi DNA from baboon and cynomolgus monkey frozen tissues and also from archived FFPE tissue blocks (Figures 1 and 2 ). Successful PCR amplification with primer set S35/S36 resulted in a single 330-bp amplicon. However, in samples with high DNA concentrations, and also when FFPE-derived DNA was used as a template for PCR, another band of higher molecular weight was observed in addition to the 330-bp amplicon (Figures 1–3). We do not know the origin of this band, but it does not interfere with the scoring of the 330-bp amplicon. The minimal amount of pure T. cruzi DNA that was detected with the PCR method was 2.39 femtograms (2.39 × 10−15 g) (Figure 3). The total DNA (host plus parasite) concentrations of the samples used in this study ranged from 6–1,474 ng/μL.
Figure 1.
Polymerase chain reaction (PCR) amplification of Trypanosoma cruzi from frozen baboon left ventricle using the S35/S36 primer pair. Lane 1 = size marker, lanes 2–8 = positive results, lanes 9–10 = negative results, lane 11 = T. cruzi positive control (Tulahuen strain), lane 12 = water control.
Figure 2.
Polymerase chain reaction (PCR) amplification of Trypanosoma cruzi from cynomolgus monkey formalin-fixed paraffin-embedded left ventricle using the S35/S36 primer set. Lane 1 = size marker, lanes 2–7 = positive results, lane 8 = T. cruzi positive control (Tulahuen strain), lane 9 = water control.
Figure 3.
Polymerase chain reaction (PCR) amplification of serially diluted Trypanosoma cruzi (Tulahuen strain) DNA using the S35/S36 primer set. MK = size marker; ng = nanograms.
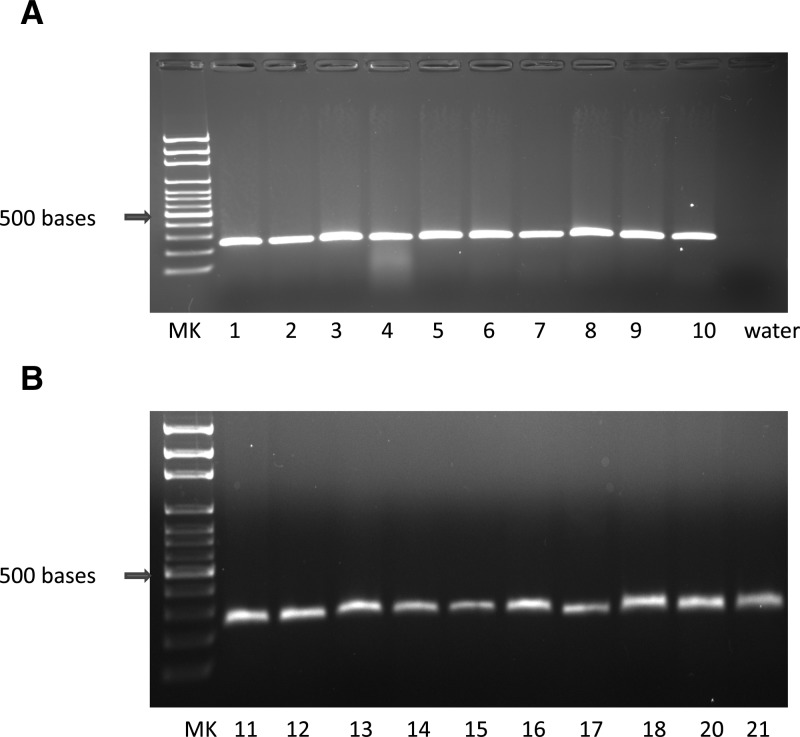
The 16S rRNA primer set (host DNA control) successfully amplified DNA from all baboon and cynomolgus monkey samples that were tested. Therefore, it can be used in future studies as a positive control for failure to obtain good quality DNA (Figure 4). Although the AmpliTaq Gold 360 DNA polymerase amplified T. cruzi DNA from frozen tissues, it failed to amplify T. cruzi DNA from FFPE tissues.
Figure 4.
Polymerase chain reaction (PCR) amplification of host DNA from baboon ventricles (panel A) and cynomolgus monkey ventricles (panel B) using the L2513/H2714 (rRNA) primer set. MK = size marker.
Tables 1 and 2 show the age, sex, myocarditis status, and PCR results from frozen and FFPE samples using the S35/S36 primer set, presence or absence of amastigotes, and serologic results for the baboons and cynomolgus monkeys. Baboons ranged in age from 2.3 to 30.5 years. Cynomolgus monkeys ranged in age from 0.5 to 20.6 years.
Overall, 15 of the 26 baboons with myocarditis had a positive PCR result (58%) using frozen or FFPE samples (Table 1). Twelve of 26 FFPE samples that exhibited myocarditis had positive PCR results (46%). Using frozen samples, 10 of 13 baboons that had myocarditis had positive PCR results (77%). Three of the animals with positive PCR results from frozen tissue were negative from FFPE samples, and three animals with positive PCR results from FFPE samples were negative for frozen samples. The one baboon with amastigotes had a positive PCR result on the FFPE sample; a frozen sample was not available from this animal. None of the animals without myocarditis had a positive PCR result.
Overall, 17 of the 19 cynomolgus monkeys with myocarditis had a positive PCR result (89%) from frozen or FFPE tissue samples (Table 2). From FFPE samples, 14 of 19 cynomolgus monkeys that had myocarditis had positive PCR results (74%). All 11 frozen samples from cynomolgus monkeys that had myocarditis had positive PCR results (100%). Three animals with positive PCR results from frozen samples were negative from FFPE samples. The five cynomolgus monkeys from which amastigotes were detected had positive PCR results on FFPE samples; frozen samples were not available from these animals. None of the animals without myocarditis had a positive PCR result.
Statistical analyses are summarized in Table 4; the results indicate an association between nonspecific lymphocytic myocarditis and T. cruzi PCR positivity in both baboons and cynomolgus monkeys in our colony.
Table 4.
Two-sided Fisher's exact test of independence for the relationship between myocarditis and PCR positivity for Trypanosoma cruzi in frozen and formalin-fixed paraffin-embedded (FFPE) tissues from baboons and cynomolgus monkeys
| Myocarditis/PCR | –/– | +/– | –/+ | +/+ | P |
|---|---|---|---|---|---|
| Baboon-frozen | 8 | 3 | 0 | 10 | 0.001 |
| Baboon-FFPE | 12 | 14 | 0 | 12 | 0.0065 |
| Cynomolgus monkey-frozen | 3 | 0 | 0 | 11 | 0.0027 |
| Cynomolgus monkey-FFPE | 12 | 5 | 0 | 14 | 8.8 × 10−5 |
PCR = polymerase chain reaction; FFPE = formalin-fixed paraffin-embedded.
Serology.
All baboons that did not have myocarditis had negative serology (17 of 20) or were indeterminate (3 of 20). Of the 26 animals with myocarditis, 21 were seropositive, four were seronegative, and one was indeterminate (Table 1). The serology data for cynomolgus monkeys was incomplete and therefore no serological correlations were made.
Discussion
The S35/S36 primer has been used in the diagnosis of T. cruzi in humans, and domestic and wild animals before.23–25 The sensitivity of our assay (femtogram range) is similar to what others have reported when using the same primers.24 Fernandes and others24 reported that as little as 0.1 fg of pure T. cruzi DNA could be detected by PCR using the S35/S36 primer set. The robustness of our PCR method was shown by the fact that T. cruzi DNA could be amplified from both frozen tissue samples and archived FFPE tissue blocks; excess host DNA did not interfere with the PCR reactions. Another added advantage of our current PCR method is that it is easy to determine whether an animal is positive or not based on a clear PCR band of 330 bp. Previously, we used the TCZ1/TCZ2 primer set in a PCR method to detect T. cruzi in nonhuman primate samples, however, it was not always possible to determine positivity of a sample because the amplicon bands occasionally appeared as a smear.26,27 In both our previous and current studies, it was determined that the type of Taq DNA polymerase used in the PCR amplification is important for successful amplification to occur, especially in FFPE samples.27
The detection rates were lower for FFPE samples as compared with frozen samples in both cynomolgus monkeys and baboons, indicating that frozen samples are preferred for detecting the presence of T. cruzi by our method. We expect that frozen samples will give higher sensitivity than FFPE samples regardless of the PCR method that is used.
However, despite the lower sensitivity of PCR when applied to FFPE samples, the successful use of PCR on DNA derived from FFPE tissue is significant. We emphasize the importance of being able to use FFPE samples in the diagnosis of Chagas disease and also other diseases in nonhuman primates. The FFPE blocks are usually collected at necropsy by pathologists and can be stored indefinitely at room temperature; furthermore, PCR quality DNA can be recovered from them decades after fixation; they therefore represent a precious resource that can be used retrospectively to study many disease conditions in addition to Chagas disease. For example, we have recently performed studies of orthoreovirus infection in our baboon colony using PCR on DNA from FFPE samples archived for up to 24 years.28
Three baboons gave positive results from PCR of frozen samples and negative results from PCR of FFPE samples, and conversely three were positive from PCR of FFPE samples and negative from PCR of frozen samples. The former type of discrepancy is to be expected because PCR should logically yield more sensitive results from frozen samples. However, the latter type of discrepancy illustrates that FFPE samples can yield positive results even when frozen samples from the same animal do not. We know from histological results that the parasites are not uniformly distributed in the tissue, and we speculate that some FFPE sections were cut through a focus of parasites, whereas the frozen sample from the same animal was devoid of parasites.
We also observed one discrepant result between serology (negative for cynomolgus monkey 20883) and PCR (positive) and histology (presence of amastigotes). It is clear from the histological results that the monkey was infected, yet it was negative on both tests by serology. The only serum sample available from that monkey was drawn on July 22, 2005, and the monkey died on August 21, 2005. We presume the monkey became infected sometime between the date of the blood draw and its death.
In this study, we report extensive epicardial inflammation in both baboons and cynomolgus monkeys; this type of inflammation has not been reported in nonhuman primates and not well studied in humans. It is important that this type of inflammation is studied so that its role in the pathogenesis of Chagas disease is clarified.
A previous study from our institute reported an association between nonspecific lymphocytic myocarditis and T. cruzi serology positivity in baboons29; our results further confirm that association and extend it to our cynomolgus monkey colony.
ACKNOWLEDGMENTS
We are grateful for the technical assistance of Allen Ford, Sam Galindo, Janice MacRossin, and Michael Torres, in collecting, processing, and assaying samples; and Michaelle Hohmann, Jesse Martinez, and Renee Escalona, for histopathology support.
Footnotes
Financial support: This research was funded in part by National Institutes of Health grant P51 OD013986, which supports the Southwest National Primate Research Center. Dr. Mubiru was supported by the National Institutes of Health under award K01 OD010973. Nonhuman primates were housed in facilities constructed with support from Research Facilities Improvement Programs grants C06 RR015456 and C06 RR014578 from the National Institutes of Health.
Authors' addresses: James N. Mubiru, Edward J. Dick Jr., Michael Owston, R. Mark Sharp, Jane F. VandeBerg, Robert E. Shade, and John L. VandeBerg, Southwest National Primate Center, Texas Biomedical Research Institute, San Antonio, TX, E-mails: jmubiru@txbiomed.org, edick@txbiomed.org, mowston@txbiomed.org, msharp@txbiomed.org, janev@txbiomedgenetics.org, bshade@txbiomed.org, and jlv@txbiomedgenetics.org. Alice Yang, Department of Biology, St. Mary's University, San Antonio, TX, E-mail: ayang@mail.stmarytx.edu.
Reprint requests: James N. Mubiru, Southwest National Primate Center, Texas Biomedical Research Institute, P.O. Box 760549, San Antonio, TX 78245-0549, E-mail: jmubiru@txbiomed.org.
References
- 1.Bittencourt AL. Congenital Chagas disease. Am J Dis Child. 1976;130:97–103. doi: 10.1001/archpedi.1976.02120020099020. [DOI] [PubMed] [Google Scholar]
- 2.Bern C, Montgomery SP, Herwaldt BL, Rassi A, Jr, Marin-Neto JA, Dantas RO, Maguire JH, Acquatella H, Morillo C, Kirchhoff LV, Gilman RH, Reyes PA, Salvatella R, Moore AC. Evaluation and treatment of Chagas disease in the United States: a systematic review. JAMA. 2007;298:2171–2181. doi: 10.1001/jama.298.18.2171. [DOI] [PubMed] [Google Scholar]
- 3.Rassi A, Jr, Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375:1388–1402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- 4.Noya BA, Díaz-Bello Z, Colmenares C, Zavala-Jaspe R, Abate T, Contreras R, Losada S, Artigas D, Mauriello L, Ruiz-Guevara R, Noya O. The performance of laboratory tests in the management of a large outbreak of orally transmitted Chagas disease. Mem Inst Oswaldo Cruz. 2012;107:893–898. doi: 10.1590/s0074-02762012000700009. [DOI] [PubMed] [Google Scholar]
- 5.Grant IH, Gold JW, Wittner M, Tanowitz HB, Nathan C, Mayer K, Reich L, Wollner N, Steinherz L, Ghavimi F, O'Reilly RJ, Armstrong D. Transfusion-associated acute Chagas disease acquired in the United States. Ann Intern Med. 1989;111:849–851. doi: 10.7326/0003-4819-111-10-849. [DOI] [PubMed] [Google Scholar]
- 6.Cimo PL, Luper WE, Scouros MA. Transfusion-associated Chagas' disease in Texas: report of a case. Tex Med. 1993;89:48–50. [PubMed] [Google Scholar]
- 7.Beard CB, Pye G, Steurer FJ, Rodriguez R, Campman R, Peterson AT, Ramsey J, Wirtz RA, Robinson LE. Chagas disease in a domestic transmission cycle, southern Texas, USA. Emerg Infect Dis. 2003;9:103–105. doi: 10.3201/eid0901.020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjos SA, Snowden KF, Craig TM, Lewis B, Ronald N, Olson JK. Distribution and characterization of canine Chagas disease in Texas. Vet Parasitol. 2008;152:249–256. doi: 10.1016/j.vetpar.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Gleiser CA, Yaeger RG, Ghidoni JJ. Trypanosoma cruzi infection in a colony-born baboon. J Am Vet Med Assoc. 1986;189:1225–1256. [PubMed] [Google Scholar]
- 10.Yeo M, Acosta N, Llewellyn M, Sánchez H, Adamson S, Miles GA, López E, González N, Patterson JS, Gaunt MW, de Arias AR, Miles MA. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasitol. 2005;35:225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Grieves JL, Hubbard GB, Williams JT, VandeBerg JL, Dick EJ, Jr, López-Alvarenga JC, Schlabritz-Loutsevitch NE. Trypanosoma cruzi in non-human primates with a history of stillbirths: a retrospective study (Papio hamadryas spp.) and case report (Macaca fascicularis) J Med Primatol. 2008;37:318–328. doi: 10.1111/j.1600-0684.2008.00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gürtler RE, Solard ND, Lauricela MA, Haedo AS, Pietrokovski SM, Alberti AA, Wisnivesky-Colli C. Dynamics of transmission of Trypanosoma cruzi in a rural area of Argentina. III. Persistence of T. cruzi parasitemia among canine reservoirs in a two-year follow-up. Rev Inst Med Trop Sao Paulo. 1986;28:213–219. doi: 10.1590/s0036-46651986000400002. [DOI] [PubMed] [Google Scholar]
- 13.Gürtler RE, Cécere MC, Petersen RM, Rubel DN, Schweigmann NJ. Chagas disease in north-west Argentina: association between Trypanosoma cruzi parasitemia in dogs and cats and infection rates in domestic Triatoma infestans. Trans R Soc Trop Med Hyg. 1993;87:12–45. doi: 10.1016/0035-9203(93)90400-k. [DOI] [PubMed] [Google Scholar]
- 14.Gurtler RE, Cohen JE, Cecere MC, Chuit R. Shifting host choices of the vector of Chagas disease Triatoma infestans and the availability of hosts in houses in north-west Argentina. J Appl Ecol. 1997;34:699–715. [Google Scholar]
- 15.Umezawa ES, Souza AI, Pinedo-Cancino V, Marcondes M, Marcili A, Camargo LM, Camacho AA, Stolf AM, Teixeira MM. TESA-blot for the diagnosis of Chagas disease in dogs from co-endemic regions for Trypanosoma cruzi, Trypanosoma evansi and Leishmania chagasi. Acta Trop. 2009;111:15–20. doi: 10.1016/j.actatropica.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Simpson L. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Annu Rev Microbiol. 1987;41:363–382. doi: 10.1146/annurev.mi.41.100187.002051. [DOI] [PubMed] [Google Scholar]
- 17.Degrave W, Fragoso SP, Britto C, van Heuverswyn H, Kidane GZ, Cardoso MA, Mueller RU, Simpson L, Morel CM. Peculiar sequence organization of kinetoplast DNA minicircles from Trypanosoma cruzi. Mol Biochem Parasitol. 1988;27:63–70. doi: 10.1016/0166-6851(88)90025-4. [DOI] [PubMed] [Google Scholar]
- 18.Degrave W, Fernandes O, Thiemann O, Wincker P, Britto C, Cardoso A, Pereira JB, Bozza M, Lopes U, Morel C. Detection of Trypanosoma cruzi and Leishmania using the polymerase chain reaction. Mem Inst Oswaldo Cruz. 1994;89:367–368. doi: 10.1590/s0074-02761994000300013. [DOI] [PubMed] [Google Scholar]
- 19.Sturm NR, Degrave W, Morel C, Simpson L. Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas' disease. Mol Biochem Parasitol. 1989;33:205–214. doi: 10.1016/0166-6851(89)90082-0. [DOI] [PubMed] [Google Scholar]
- 20.Argañaraz ER, Hubbard GB, Ramos LA, Ford AL, Nitz N, Leland MM, VandeBerg JL, Teixeira AR. Blood-sucking lice may disseminate Trypanosoma cruzi infection in baboons. Rev Inst Med Trop Sao Paulo. 2001;43:271–276. doi: 10.1590/s0036-46652001000500007. [DOI] [PubMed] [Google Scholar]
- 21.Kitano T, Umetsu K, Tian W, Osawa M. Two universal primer sets for species identification among vertebrates. Int J Legal Med. 2007;121:423–427. doi: 10.1007/s00414-006-0113-y. [DOI] [PubMed] [Google Scholar]
- 22.R Core Team R: a language and environment for statistical computing. 2013. http://www.R-project.org/ R Foundation for Statistical Computing, Vienna, Austria. Foundation for Statistical Computing, Vienna, Austria. Available at.
- 23.Wincker P, Britto C, Pereira JB, Cardoso MA, Oelemann W, Morel CM. Use of a simplified polymerase chain reaction procedure to detect Trypanosoma cruzi in blood samples from chronic chagasic patients in a rural endemic area. Am J Trop Med Hyg. 1994;51:771–777. doi: 10.4269/ajtmh.1994.51.771. [DOI] [PubMed] [Google Scholar]
- 24.Fernandes CD, Tiecher FM, Balbinot MM, Liarte DB, Scholl D, Steindel M, Romanha A. Efficacy of benznidazol treatment for asymptomatic chagasic patients from state of Rio Grande do Sul evaluated during a three years follow-up. Mem Inst Oswaldo Cruz. 2009;104:27–32. doi: 10.1590/s0074-02762009000100004. [DOI] [PubMed] [Google Scholar]
- 25.Cominetti MC, Andreotti R, Oshiro ET, Dorval ME. Epidemiological factors related to the transmission risk of Trypanosoma cruzi in a Quilombola community, State of Mato Grosso do Sul, Brazil. Rev Soc Bras Med Trop. 2011;44:576–581. doi: 10.1590/s0037-86822011000500009. [DOI] [PubMed] [Google Scholar]
- 26.Moser DR, Kirchhoff LV, Donelson JE. Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J Clin Microbiol. 1989;27:1477–1482. doi: 10.1128/jcm.27.7.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams JT, Mubiru JN, Schlabritz-Loutsevitch NE, Rubicz RC, VandeBerg JL, Dick EJ, Jr, Hubbard GB. Polymerase chain reaction detection of Trypanosoma cruzi in Macaca fascicularis using archived tissues. Am J Trop Med Hyg. 2009;81:228–234. [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Dick EJ, Jr, Reddy BY, Yang A, Mubiru J, Hubbard GB, Owston MA. Reovirus-associated meningoencephalomyelitis in baboons. Vet Pathol. 2013 doi: 10.1177/0300985813497487. July 26, 2013 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrade MC, Dick EJ, Jr, Guardado-Mendoza R, Hohmann ML, Mejido DC, VandeBerg JL, DiCarlo CD, Hubbard GB. Nonspecific lymphocytic myocarditis in baboons is associated with Trypanosoma cruzi infection. Am J Trop Med Hyg. 2009;81:235–239. [PMC free article] [PubMed] [Google Scholar]