Abstract
Reliable laboratory testing is of great importance to detect Bartonella bacilliformis infection. We evaluated the sensitivity and specificity of the enzyme-linked immunosorbent assay (ELISA) using recombinant protein Pap31 (rPap31) for the detection of antibodies against B. bacilliformis as compared with immunofluorescent assay (IFA). Of the 302 sera collected between 1997 and 2000 among an at-risk Peruvian population, 103 and 34 samples tested positive for IFA-immunoglobulin G (IgG) and IFA-IgM, respectively. By using Youden's index, the cutoff values of ELISA-IgG at 0.915 gave a sensitivity of 84.5% and specificity of 94%. The cutoff values of ELISA-IgM at 0.634 gave a sensitivity of 88.2% and specificity of 85.1%. Using latent class analysis, estimates of sensitivity and specificity of almost all the assays were slightly higher than those of a conventional method of calculation. The test is proved beneficial for discriminating between infected and non-infected individuals with the advantage of low-cost and high-throughput capability.
Introduction
Tropical bartonellosis caused by Bartonella bacilliformis remains a major health threat to populations living in endemic areas and travelers visiting such regions. As a result of favorable ecological conditions for the principal suspected sand fly vector, Lutzomyia verrucarum,1 the disease is historically confined to remote valleys located between altitudes of 800 and 3,000 m above sea level on the western slopes of the Andes Mountains in Peru, Columbia, and Ecuador. However, the disease has recently expanded over a broader geographical range including lower elevations, high forest regions, and valleys located in the eastern portions of the Andes.2–4 The bacteria is known to invade and replicate inside human erythrocytes and endothelial cells causing the disease, which is classically manifested in one of two distinct ways, either as acute onset of fever with hemolytic anemia (Oroya fever) or with angiogenic skin lesions called verruga peruana.1 Although the first manifestation is life-threatening with case fatality rates that can reach as high as 88% in untreated patients, verruga peruana is benign and self-limiting.5 Without treatment, these skin lesions can persist for up to 6 months.1
The diagnosis of B. bacilliformis infection remains problematic as the spectrum of clinical manifestations is more highly variable than previously described, resulting in misdiagnosis and delay of appropriate treatment.6 Although the disease is typically biphasic: acute anemia, followed some months later by the chronic dermal phase, Oroya fever is rarely seen in endemic regions, whereas verruga peruana is common. Conversely, Oroya fever appears to be more common in areas of non-endemicity.7 Furthermore, one-third of patients have skin lesions without a history of fever and nearly one-fourth of patients are asymptomatic.8 The reservoir of infection remains unknown. In endemic areas, bacteremia was found in 0.5% of healthy individuals and in nearly half of the patients with verruga peruana at the time of diagnosis, suggesting that humans may serve as the reservoir for infection.8 Prompt diagnosis with rapid and reliable diagnostic tests would be of great clinical value to reduce suffering and death from the disease, and it may have an added benefit of helping to control disease transmission.
The two main types of assays used for diagnosing the disease are pathogen or antigen detection methods and serological or antibody detection methods. Techniques for pathogen detection, which include thin blood smear, culture, and polymerase chain reaction (PCR) are not always reliable for detecting the pathogen. The Giemsa or Wright staining of the blood smear to detect intraerythrocytic bacilli may be the only test available for diagnosis of acute bartonellosis in endemic areas. The specificity of the test is very high (96%) but the sensitivity remains fairly low (36%) for detection of the organism.9,10 In addition, B. bacilliformis is difficult to isolate in laboratory cultures, as it requires special media and a long incubation time of up to 8 weeks. The PCR assay requires special equipment, dedicated laboratory space, and highly skilled personnel. Serological testing, in several formats, is now increasingly used to detect the antibody for diagnosing the disease. Currently, the indirect immunofluorescent assay (IFA) using irradiated whole cell antigen preparations from co-cultivated Vero cells is considered the most sensitive serological test for diagnosing human bartonellosis.10 In a previous study, a titer of 1:40 or greater was considered positive for IFA-immunoglobulin G (IgG) and a titer of 1:5 or greater was considered positive for IFA-IgM for detection of antibodies against B. bacilliformis.11 In a study done by Chamberlin and others in Peru, the cutoff titers were adjusted to account for the high background antibody levels in the endemic population. Therefore, a cutoff titer of 1:256 or greater or a 4-fold rise of antibody in paired sera was selected for diagnosing human bartonellosis, resulting in a sensitivity of 82%, and specificity of 92%.12 Despite its advantages, IFA is costly, time consuming, requires a fluorescent microscope, and relies on highly trained personnel to interpret the results.
The enzyme-linked immunosorbent assay (ELISA) is considered more desirable because it is more objective, relatively inexpensive, and has the capability of being high throughput especially when automated. Previously, ELISA was done using soluble whole cell B. bacilliformis antigen but it was limited by its low specificity.13,14
Recently, Pap31 (GenBank accession no. ABA60112.1), also known as hemin-binding protein A (HbpA) in Bartonella spp., was identified in the virulent Peruvian strain of B. bacilliformis. The protein was found to be highly expressed in cultures of B. bacilliformis and immunologically dominant; thus, it is a good candidate to be used in ELISA.15 Furthermore, as a homologue of the bacteriophage-associated protein, it was recognized by the host's immune response during Bartonella henselae infections.16 Recombinant Pap31 (rPap31) can be produced in bulk, is easily purified, and remains antigenic even after several freeze–thaw cycles. During the initial assay development by Taye and others,15 a total of 136 samples from 29 IFA positive and 107 IFA negative sera were tested by ELISA using the rPap31 antigen. The results showed that the 95% confidence interval (CI) of the optical density (OD) values for the IFA negative samples did not overlap with the 95% CI of the OD values for the IFA positive samples. However, an adequate sample size is needed to ensure that the assay will yield results with the desired precision.
The purpose of this study was to determine the sensitivity and specificity of the ELISA assay using the recombinant protein antigen, rPap31, for detection of antibody against B. bacilliformis compared with IFA as the reference standard, using a larger sample size.
Material and Methods
Serum samples.
The samples used in our study were collected among Peruvian populations at risk of B. bacilliformis infection in the Caraz and Cusco regions of Peru between 1997 and 2000.8,12 All samples were anonymized before use in the current study and have remained in storage at −70°C since that time. The samples used in our study were collected under two previous projects at the same study site in a village near Caraz city, ∼475 km northeast of Lima. Officials from the Ministry of Health selected this area as being representative of an area with long established bartonellosis endemicity. When samples were collected they were all tested by IFA for detection of IgG and IgM, as previously described.11 The only information provided for this study was the IFA results. Therefore, we were not able to indicate the number of samples taken from each study. To reduce the bias from the selection process, we examined all the samples that had adequate volume for testing both ELISA IgG and IgM without knowing the IFA results. An IFA titer of 1:40 or greater was defined as positive for IgG17 and a titer of 1:5 or greater was defined as positive for IgM antibody. All samples were tested anonymously by two persons who were trained to perform an ELISA assay. The ELISA results were evaluated independently without knowing the IFA results.
To test possible cross-reaction to pap31-ELISA, 10 sera containing antibodies to bacteria other than B. bacilliformis were assessed as follows: five serum samples containing antibodies to Coxiella burnetii, four archived sera from patients with brucellosis, one serum sample from a patient with B. henselae endocarditis.
Ethical considerations.
The samples used in this study were obtained under a previous study protocol (PMB-208710) that received approval from the institutional review board of the Uniformed Services University of the Health Sciences (USUHS) and adhered to all guidelines of the U.S. Department of Health and Human Services. This study was approved by the Office of Research at the Uniformed Services University of Health Sciences in 2012 (T08732-02) and was determined to be non-human research and exempt from human use regulations.
Laboratory techniques.
The recombinant Pap31 antigen was prepared as described previously.15 The ELISA was carried out in 96-well ELISA microplates (Polystyrene; Microlon 200, U-bottom, Greiner Bio-one, Monroe, NC). The wells were coated with 0.3 μg/well of recombinant Pap31 antigen prepared in phosphate buffered saline (PBS) and bovine serum albumin (BSA) as the negative control for nonspecific binding of the serum components. The plates were coated and kept at 4°C for 48 hr. After incubation, the plates were rinsed twice with 200 μL of 1× PBS, blocked with 10% skim milk at room temperature (RT) for 1 hr, and again rinsed twice with PBS.
Serum samples, diluted at 1:100 in 5% skim milk, were added at a volume of 100 μL/ well in duplicates and were incubated at RT for 1 hr. The plates were then washed 3 times with 200 μL of 0.1% Tween 20 in 1× PBS (1× PBST) to remove unbound primary antibody. For each wash step, the plates were incubated on a rocker for 5 min before the washing solution was discarded. After the three wash steps, the plates were subjected to a final rinse with PBS. The IgG and IgM were detected with biotinylated goat anti-human IgG (Santa Cruz Biotechnology, CA) or IgM (Sigma, MS) diluted 1:1,000 and 1:500, respectively, in 5% BSA. The secondary antibody was added at a volume of 100 μL/ well, incubated at RT for 1 hr, and the plates were washed 3 times, as described previously, and rinsed once with PBS. Bound antibody was detected with streptavidine-horseradish peroxidase (HRP) conjugate diluted to 1:8000 in 5% BSA and added to the wells at 100 μL/ well. After incubating for 1 hr at RT, the plates were washed 3 times with PBST and once with PBS. The ABTS peroxidase substrate and peroxidase substrate solution B (KPL, Inc., Gaithersburg, MD) were mixed at a ratio of 1:1, 100 μL/ well was added to the plate and plates were incubated in the dark at RT for 30 min. The OD was measured using an ELISA plate reader (SpectraMax 190 (BC-MD SMX190), Molecular Devices, Inc., Long Beach, CA) using dual wavelengths (405 nm and 650 nm).
Data analysis and outcome measures.
The data were analyzed using SPSS v.16.0 (SPSS, Inc., Chicago, IL) for Windows (Microsoft Corp., Redmond, WA). The duplicate of each sample for rPap31 and BSA was averaged. The corrected OD of each sample was calculated by subtracting the average background OD of the BSA from the average, corresponding, rPap 31 OD. The OD of all the samples was expressed as a mean with standard deviation. A student's t test for independent samples was used to compare the average OD values of positive and negative groups, defined by IFA. The IFA results were expressed as titer values. The correlation between IFA titer and the OD were assessed by Spearman's rank correlation. To determine the optimal cutoff values for the ELISA assay, a receiver operating characteristics (ROC) curve was constructed and a table that displayed every possible cutoff value for a positive OD was generated. The area under the ROC curve (AUC) was also calculated. The cutoff values of the IgG and IgM ELISA assays were selected as the ones that maximized the sum of sensitivity and specificity by using Youden's index (J = sensitivity + specificity − 1).18 We also calculated the likelihood ratio for a positive test (LR+) and the likelihood ratio for a negative test (LR−). The LR is the probability of a specific test result in people who have disease divided by the probability of the same finding in people who do not have disease. The likelihood ratio was determined by using the following formulas: LR+ = sensitivity/1 − specificity, LR− = 1-sensitivity/specificity.19
In the absence of a consensus reference standard, we used latent class analysis (LCA) to estimate the sensitivity and specificity of IFA and pap-31 ELISA simultaneously. The LCA uses a non-linear statistical model to classify groups of subject who have shared characteristics into latent class. The LCA can achieve this by maximizing a mathematical likelihood function based on the patients' distributional pattern of response of input variables. In this study, B. bacilliformis infection (having positive antibody against B. bacilliformis) was defined as the latent variable, and the result of serostatus obtained with each assay (IFA and ELISA) were the manifest variables or input items. Log-likelihood, Akaike information criteria (AIC) and Bayesian information criteria (BIC) were used to assess goodness of fit of each model. In addition to a traditional LCA, which requires assumption of conditional independence of tests, a random effect model was also run to account for possible conditional dependence between the tests. Preferred models are those that minimize values of the BIC, AIC, and log-likelihood. The R program (http://www.r-project.org) was used to calculate LCA with the package poLCA (version 1.4) and random LCA (version 0.8-6).20,21
A sample size of 100 positive sera confirmed by IFA will have 80% power to detect a significant difference in sensitivity between IFA and ELISA tests if the sensitivity is 80% and 70%, respectively; and the correlation between IFA and ELISA is 0.7, based on McNemar's test with a 5%, two-sided significance level. A sample size of 100 negative sera will have 85% power to detect a significant difference in specificity between IFA and ELISA tests if the specificity is 80% and 90%, respectively. In addition, a sample size of 100 positive and 100 negative sera is sufficient to estimate sensitivity or specificity with a margin of error of 10% points based on a 95% two-sided confidence interval and a “worst-case” sensitivity or specificity of 50%.22
Results
A total of 307 serum samples were available for testing. Of these, five were excluded because of an inadequate amount and were subsequently known to be negative by IFA. Of 302 sera that were examined in the study, 103 samples tested positive for IFA-IgG and 34 samples tested positive for IFA-IgM. There were 22 samples that tested positive for both IgG and IgM. Of 103 sera with positive IFA-IgG, 85 had a titer of 1:64, 13 had a titer of 1:128, and 5 had a titer of 1:512. Of 34 sera with positive IFA-IgM, 3 had a titer of 1:20, 26 had a titer of 1:64, and 5 had a titer of 1:128.
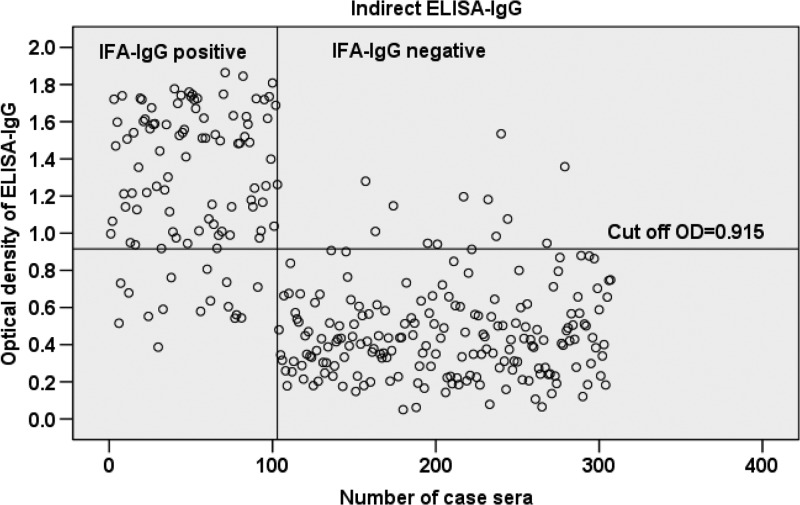
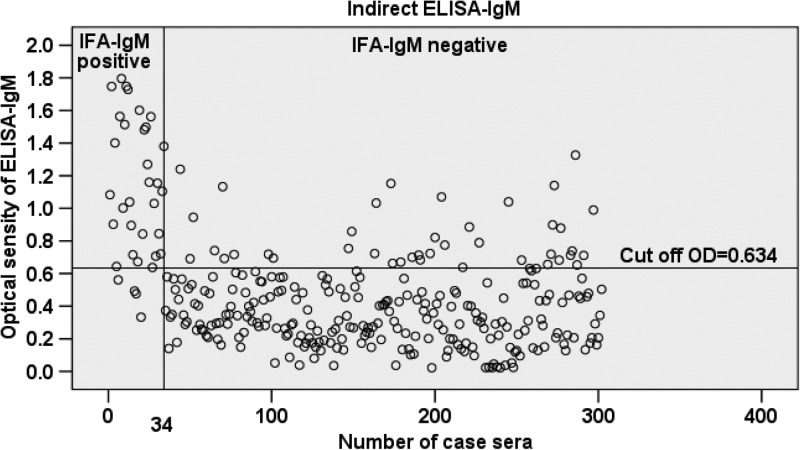
The mean OD value was significantly higher among sera that were IFA positive for either IgG or IgM compared with those that were IFA negative for either IgG or IgM (P < 0.001) (Table 1). The 95% CI of the mean OD of IFA negative and positive samples for both IgG and IgM did not overlap. A scatterplot analysis of the 302 sera tested by the ELISA-IgG and IgM based on rPap31 antigen is shown in Figures 1 and 2. A significant positive correlation was found between the IFA titer and OD values for both IgG and IgM (P < 0.001), but it was stronger for IgG (rs = 0.71) than that for IgM (rs = 0.47).
Table 1.
Mean optical density (OD) and 95% confidence interval (CI) of the recombinant Pap31-based enzyme-linked immunosorbent assay (ELISA) in relation to immunofluorescent assay (IFA)
| IFA | ELISA-OD | P value | |
|---|---|---|---|
| Mean (SD) | 95% CI | ||
| IFA-IgG | |||
| Negative (N = 199) | 0.467 (0.26) | 0.43–0.50 | |
| Positive (N = 103) | 1.286 (0.39) | 1.21–1.36 | < 0.001 |
| IFA-IgM | |||
| Negative (N = 268) | 0.367 (0.25) | 0.36–0.42 | |
| Positive (N = 34) | 1.100 (0.43) | 0.95–1.25 | < 0.001 |
Figure 1.
Scatterplot analysis of 302 sera measured by enzyme-linked immunosorbent assay (ELISA)-immunoglobulin G (IgG) using rPap31 antigen. The vertical line separates immunofluorescent assay (IFA)-IgG-positive sera (sera nos. 1 to 103) from negative sera (sera nos. 104 to 302). The horizontal line indicates the cutoff value of the ELISA-IgG assay.
Figure 2.
Scatterplot analysis of 302 sera measured by enzyme-linked immunosorbent assay (ELISA)-immunoglobulin M (IgM) using rPap31 antigen. The vertical line separates immunofluorescent assay (IFA)-IgM-positive sera (sera nos. 1 to 34) from negative sera (sera nos. 35 to 302). The horizontal line indicates the cutoff value of the ELISA-IgM assay.
The performance of the rPap31-based ELISA compared with IFA for detection of antibody against B. bacilliformis is shown in Table 2. The cutoff value of ELISA-IgG at 0.915 and ELISA-IgM at 0.634 was selected to maximize the sensitivity and specificity of the test. Overall, the ELISA-IgG assay showed a high sensitivity (84.5%) and specificity (94%). The ELISA-IgM had a higher sensitivity (88.2%) but a lower specificity (85.1%) than ELISA-IgG. A number of false positives were found for the ELISA-IgM, resulting in a lower specificity than IgG. In addition, no cross-reactivity was observed when testing 10 serum samples containing antibodies against C. burnetii, Brucella spp., and Bartonella henselae.
Table 2.
Performance of the recombinant Pap31-based enzyme-linked immunosorbent assay (ELISA) compared with immunofluorescent assay (IFA) for detection of antibody against Bartonella bacilliformis
| No. ELISA-IgG | No. IFA positive (n) | No. IFA negative (n) | Total |
|---|---|---|---|
| Positive | 87 | 12 | 99 |
| Negative | 16 | 187 | 203 |
| Total | 103 | 199 | 302 |
| Performance of ELISA-IgG test at cutoff value* 0.915 (J = 0.79) | |||
| Sensitivity* 84.5% -Specificity* 94% | |||
| Positive likelihood ratio 14.1 -Negative likelihood ratio 0.16 | |||
| No. ELISA-IgM | No. IFA positive (n) | No. IFA negative (n) | Total |
| Positive | 30 | 40 | 70 |
| Negative | 4 | 228 | 232 |
| Total | 34 | 268 | 302 |
| Performance of ELISA-IgM test at cutoff value* 0.634 (J = 0.73) | |||
| -Sensitivity* 88.2% -Specificity* 85.1% | |||
| -Positive likelihood ratio 5.9 -Negative likelihood ratio 0.14 | |||
Using Youden index (J) = Max [sensitivity + specificity − 1] can range from 0 to 1.
Goodness of fit for one-class and two-class traditional LCA is shown in Table 3. Only minor differences of BIC and estimates were found when analyzing with traditional LCA or a random effect model. Based on a goodness of fit measure, the two-class model with traditional LCA was the best fitting. The estimates of sensitivity and specificity with the best model are shown in Table 4. The estimates indicated that ELISA-IgG was comparable to IFA-IgG in terms of sensitivity (94% versus 96%, respectively) and specificity (96% versus 95%, respectively), although ELISA-IgM was much more sensitive (93% versus 66%, respectively) but slightly less specific (90% versus 99%, respectively) than IFA-IgM.
Table 3.
Goodness of fit for one-class and two-class traditional latent class analysis
| Statistics | IFA-IgG and ELISA-IgG | IFA-IgM and ELISA-IgM | ||
|---|---|---|---|---|
| 1 Class | 2 Class | 1 Class | 2 Class | |
| AIC | 773.71 | 577.23 | 543.56 | 473.04 |
| BIC | 781.13 | 595.79* | 550.98 | 491.59† |
| Loglik | −384.86 | −283.62 | −269.78 | −231.52 |
BIC of two-class with random effect model for IFA-IgG & ELISA-IgG = 601.49.
BIC of two-class with random effect model for IFA-IgM & ELISA-IgM = 497.31.
IFA = immunofluorescent assay; IgG = immunoglobulin G; IgM = immunoglobulin M; ELISA = enzyme-linked immunosorbent assay; AIC = Akaike information criteria; BIC = Bayesian information criteria.
Table 4.
Sensitivity and specificity of IFA and ELISA assay based on two-class model with traditional latent class analysis*
| Assay | % Sensitivity (95% CI) | % Specificity (95% CI) |
|---|---|---|
| IFA-IgG | 96 (95,97) | 95 (92, 97) |
| ELISA-IgG | 94 (92, 96) | 96 (92, 98) |
| IFA-IgM | 66 (55, 76) | 99 (98, 100) |
| ELISA-IgM | 93 (87, 100) | 90 (87, 93) |
IFA = immunofluorescent assay; ELISA = enzyme-linked immunosorbent assay; IgG = immunoglobulin G; CI = confidence interval; IgM = immunoglobulin M.
Considering LR+, people infected with B. bacilliformis were about 14 times and 5.9 times more likely to have a positive ELISA-IgG and IgM result, respectively, than are those who had never been infected. Whereas the probability of the infected individuals having a negative ELISA-IgG and IgM were about a sixth (0.16) and a seventh times (0.14) that of those who have never been infected.
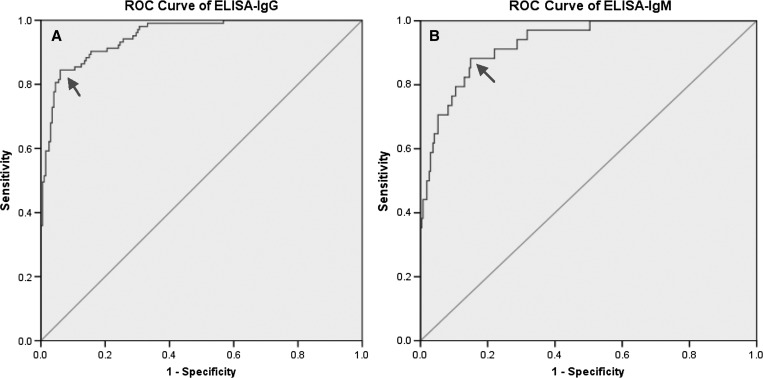
Further performance comparison of the IFA and the ELISA was conducted by plotting the ROC curve (Figure 3). The ROC curve almost reached an ideal slope, with an AUC of 0.95 (95% CI = 0.93 0.97) for the ELISA-IgG (Figure 3A) and AUC of 0.93 (95% CI = 0.89–0.97) for the ELISA-IgM (Figure 3B).
Figure 3.
Receiver operating characteristics (ROC) curves of rPap31-based enzyme-linked immunosorbent assay (ELISA) for detection of antibody against Bartonella bacilliformis. (A) Shows the ROC curve of ELISA-immunoglobulin G (IgG) with an area under the ROC curve (AUC) of 0.95 (95% confidence interval [CI] = 0.93–0.97). (B) Shows the ROC curve of ELISA-IgM with an AUC of 0.93 (95% CI = 0.89–0.97).
Discussion
The results of this study explicitly show that the use of rPap31 antigen in the ELISA format is suitable for detecting serum antibody to B. bacilliformis. With a clearly distinguished mean OD and 95% CI, the method is able to differentiate B. bacilliformis seropositive sera from seronegative sera. Using the cutoff value at 0.915, rPap31-based ELISA is 84.5% sensitive and 94% specific for detecting IgG antibody based on the reference test, IFA. With the cutoff value at 0.634, the ELISA-IgM resulted in a sensitivity of 88.2% and a specificity of 85.1%. With a high LR+ for ELISA-IgG, a positive test is good at ruling in those previously infected with B. bacilliformis.
As anticipated, all observed sensitivity and specificity estimates were greater than 80%. For estimating the performance of the ELISA-IgG test at a cutoff value of 0.915, the observed sample size was 103 positive and 199 negative sera. Based on a sensitivity and specificity of 80%, the observed sample size was sufficient to estimate sensitivity with a margin of error of 7.8% points and specificity with a margin of error of 5.6% points. For estimating the performance of the ELISA-IgM test at a cutoff value of 0.634, the sample size was 34 positive and 268 negative sera. Assuming a proportion of 80% for sensitivity and specificity, the observed sample size was sufficient to estimate sensitivity with a margin of error of 13.7% points and specificity with a margin of error of 4.8% points. Calculations are based on a 95% two-sided CI for a single proportion.
In a previously developed assay by Knobloch,13 crude cell antigen was used in ELISA to detect antibodies against B. bacilliformis. The assay was sensitive but suffered from cross-reactivity with anti-Chlamydia psittaci antibody. Another antigen that was identified as a major protein of B. bacilliformis was the recombinant 65 kDa protein (Bb65). The protein is predominantly located within the bacterial cytoplasm without any exposed epitopes on the cell wall surface. When applied to the ELISA format, Bb65 was 90% sensitive for detection of persisting IgG antibody from sera collected 1 to 3 years after onset of Oroya fever. However, this assay failed to detect antibodies during the first 2 weeks of fever.23 Unlike Bb65, the rPap 31-based ELISA showed good sensitivity and specificity for detection of IgM antibodies. In addition, the rPap31-based ELISA is comparable with the immunoblot assay using sonicated whole cell antigens, which was 70% sensitive in diagnosing acute bartonellosis and 94% sensitive in diagnosis of chronic bartonellosis with 100% specificity, using healthy individuals as the control group.24 A significant positive correlation between IFA titer and ELISA OD for detection of antibody against B. bacilliformis was also observed. Such a correlation is found to be stronger for IgG (rs = 0.71) than IgM (rs = 0.47). This has been reported in other studies, where varying correlations have been observed among different serological tests and among the same serological tests from different sources.25 For example, the correlation of the recombinant line immunoblot to ELISA and to IFA for the detection of Lyme disease was found to be varied from rs 0.673 to 0.905, respectively.26
A significant number of false positive results were observed for the ELISA-IgM as compared with IFA, resulting in a lower specificity for IgM than IgG. It is important to realize that sensitivity and specificity may change depending on the reference test used as the gold standard, which is not necessarily the true representation of disease status. Although there is no widely accepted suitable standard, we selected IFA as our reference test because it showed good performance when being compared against a composite reference standard including blood smear, culture, and PCR in a previous study.12 However, the IFA test is not 100% accurate. Under the conditions where we are dealing with probabilities of diagnosis rather than true knowledge of the disease, LCA is another approach that often used to estimate the diagnostic characteristics of an existing and new test. Overall, the two-class model of traditional LCA provided a slightly higher sensitivity and specificity of almost all the assays than the conventional method. The IFA-IgG was the most sensitive, whereas IFA-IgM was the most specific but was the least sensitive for detecting antibodies against B. bacilliformis. With the two-class model of traditional LCA, both ELISA-IgG and IgM had a sensitivity and specificity > 90%.
The IFA test had been shown to exhibit considerable cross-reactivity with other pathogens such as Brucella spp., Chalmydia spp., Coxiella burnetii, or other Bartonella species.14 In contrast, we did not observe any cross-reactivity of rPap31 to antibodies against antigenically similar bacteria including C. burnetii, Brucella spp., and B. henselae. It must be noted that using specific immunoreactive protein Pap31 as the antigen in an ELISA format is likely to be more specific than using whole cell antigen in an IFA format for detecting antibodies because the numerous antigens presented in the whole cell can result in cross-reaction of antibodies to other microorganisms or tissue components.27
In addition, comparison of the Pap 31-like protein of B. bacilliformis using the FASTA similarity search (www.ebi.ac.uk/Tools/sss/fasta/), showed that the amino acid sequence had only 39–51% similarity to those of other Bartonella species such as B. henselae, B. clarridgeiae, or B. rochalimae (which was first identified in patients having fever and bacteremia after visiting Peru). Furthermore, with the sequence identity well below 40% with those from organisms like Brucella spp., Chalmydia spp., and C. burnetii, Pap31 is unlikely to show cross-reactivity with antibodies against those organisms.28 In addition, our test is more likely to have a higher sensitivity than the IFA for detecting antibodies. While the ELISA plates were coated with a higher amount of a single specific immunoreactive antigen, the IFA was coated with the whole cell antigen, which also contained non-immunogenic proteins. The reduced amount of immunoreactive proteins may lead to less sensitive detection of the disease-specific antibodies.25,29 Previous evaluation of the IFA, ELISA, and indirect hemagglutination among 187 sera from Peruvian residents in a bartonellosis-endemic area by Knobloch and others reported that the ELISA test using soluble B. bacilliformis antigen was more sensitive for detecting IgG antibody than IFA. Of the 102 ELISA-positive samples, only 51% were positive by IFA-IgG.11
The rPap31-based ELISA is comparable to IFA for the detection of antibody against B. bacilliformis. Furthermore, the ELISA using recombinant Pap31 protein as the antigen has many distinct advantages over the IFA using whole cell antigen. Although the IFA assay is based on subjective determinations of a positive result, the ELISA results, based on OD values, are more objective. The use of a recombinant antigen, instead of whole cell antigen, in the ELISA provides high quality antigen, which can greatly improve the sensitivity and specificity of the test.
The population in the endemic area may have antibodies without necessarily having symptoms. Nearly one-fourth of infections were asymptomatic and asymptomatic bacteremia was found in 0.5% of the population living in the endemic area.8 Even though using the test to diagnose acute or chronic disease is of paramount importance to reduce mortality and morbidity in the population, detection of antibodies against B. bacilliformis to evaluate exposure and infection shall be helpful to better understand the epidemiology of the disease, both clinically and subclinically. The likelihood ratio is useful across the array of disease frequencies. A LR+ above 10 means that a positive test is good at ruling in a diagnosis, whereas a LR− below 0.1 is good at ruling out a diagnosis.19 Considering a big and modest increase of the LR+ of ELISA-IgG and IgM and their moderately low LR−, the test could be helpful to determine the probabilities of having infection across different populations with varying prevalence.
The main limitation of this study is the lack of clinical data including status or stage of disease, duration of the disease, or the date that serum was collected in relation to symptoms. Therefore, claims regarding the value of the rPap31 ELISA test for diagnosis of disease cannot be overstated. Furthermore, using stored sera (frozen since 1997) was another limitation of our study because the antibody may have degraded after repeated freeze–thaw cycles. The test may perform differently if using recently collected samples. Future prospective studies will address these issues.
Our study was designed to distinguish between infected and uninfected individuals by detection of antibody against B. bacilliformis. Considering its good performance, the ELISA test is still useful for screening of infection, which is beneficial for the assessment of actual disease burdens, particularly in endemic area. Whether the rPap31 ELISA is of value for diagnosis of acute or chronic disease requires further evaluation. Furthermore, the test provides a high-throughput format and requires minimal laboratory equipment, making it a suitable alternative to the IFA, with the advantage of low cost. The reliable performance of rPap31-ELISA also suggests its potential to be developed as a “point of care” rapid test for use in rural endemic areas with limited resources.
ACKNOWLEDGMENTS
We thank John Antony J Prakash for critical comments and Cara Olsen for statistical support. We are also thankful to Sasima Tongsai for assistance with the latent class analysis.
Disclaimer: The opinions and assertions contained herein are the private ones of the authors and are not to be construed as official or as reflecting the views of the Uniformed Services University of the Health Sciences (USUHS), the Department of the Navy, the Department of Defense (DOD), or the U. S. Government.
Footnotes
Financial support: This work was supported by Work Unit No. (WUN) 6000.RAD1.J.A0310 (to Naval Medical Research Center) and USUHS grant G187WI.
Disclosure: CCC and WMC are employees of the U. S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by an employee of the U.S. Government as part of that person's official duties, and “copyright protection” under this title is not available for any work of the United States Government.
Authors' addresses: Nasikarn Angkasekwinai and John Grieco, Department of Preventive Medicine and Biometrics, Uniformed Services University of the Health Sciences, Bethesda, MD, E-mails: nasikarn@gmail.com and jgrieco@usuhs.edu. Erin H. Atkins, Chien Chung Chao, and Wei Mei Ching, Viral and Rickettsial Diseases Department, Infectious Disease Directorate, Naval Medical Research Center, Silver Spring, MD, E-mails: eshuber1@gmail.com, Chien-Chung.Chao@med.navy.mil, and WeiMei.Ching@med.navy.mil. Sofia Romero, Naval Medical Research Institute Detachment, Lima, Peru, E-mail: sofiaromero_m@yahoo.com.
Reprint requests: Wei Mei Ching, Viral and Rickettsial Diseases Department, Infectious Disease Directorate, Naval Medical Research Center, 503 Robert Grant Avenue, RM3N71 Silver Spring, MD 20910.
References
- 1.Maguina C, Gotuzzo E. Bartonellosis. New and old. Infect Dis Clin North Am. 2000;14:1–22. doi: 10.1016/s0891-5520(05)70215-4. vii. [DOI] [PubMed] [Google Scholar]
- 2.Lydy SL, Eremeeva ME, Asnis D, Paddock CD, Nicholson WL, Silverman DJ, Dasch GA. Isolation and characterization of Bartonella bacilliformis from an expatriate Ecuadorian. J Clin Microbiol. 2008;46:627–637. doi: 10.1128/JCM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huarcaya E, Maguina C, Torres R, Rupay J, Fuentes L. Bartonelosis (Carrion's Disease) in the pediatric population of Peru: an overview and update. Braz J Infect Dis. 2004;8:331–339. doi: 10.1590/s1413-86702004000500001. [DOI] [PubMed] [Google Scholar]
- 4.Kosek M, Lavarello R, Gilman RH, Delgado J, Maguina C, Verastegui M, Lescano AG, Mallqui V, Kosek JC, Recavarren S, Cabrera L. Natural history of infection with Bartonella bacilliformis in a nonendemic population. J Infect Dis. 2000;182:865–872. doi: 10.1086/315797. [DOI] [PubMed] [Google Scholar]
- 5.Gray GC, Johnson AA, Thornton SA, Smith WA, Knobloch J, Kelley PW, Obregon Escudero L, Arones Huayda M, Wignall FS. An epidemic of Oroya fever in the Peruvian Andes. Am J Trop Med Hyg. 1990;42:215–221. doi: 10.4269/ajtmh.1990.42.215. [DOI] [PubMed] [Google Scholar]
- 6.Maguina C, Garcia PJ, Gotuzzo E, Cordero L, Spach DH. Bartonellosis (Carrion's disease) in the modern era. Clin Infect Dis. 2001;33:772–779. doi: 10.1086/322614. [DOI] [PubMed] [Google Scholar]
- 7.Birtles RJ, Fry NK, Ventosilla P, Caceres AG, Sanchez E, Vizcarra H, Raoult D. Identification of Bartonella bacilliformis genotypes and their relevance to epidemiological investigations of human bartonellosis. J Clin Microbiol. 2002;40:3606–3612. doi: 10.1128/JCM.40.10.3606-3612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlin J, Laughlin LW, Romero S, Solorzano N, Gordon S, Andre RG, Pachas P, Friedman H, Ponce C, Watts D. Epidemiology of endemic Bartonella bacilliformis: a prospective cohort study in a Peruvian mountain valley community. J Infect Dis. 2002;186:983–990. doi: 10.1086/344054. [DOI] [PubMed] [Google Scholar]
- 9.Ellis BA, Rotz LD, Leake JA, Samalvides F, Bernable J, Ventura G, Padilla C, Villaseca P, Beati L, Regnery R, Childs JE, Olson JG, Carrillo CP. An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am J Trop Med Hyg. 1999;61:344–349. doi: 10.4269/ajtmh.1999.61.344. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez Clemente N, Ugarte-Gil CA, Solorzano N, Maguina C, Pachas P, Blazes D, Bailey R, Mabey D, Moore D. Bartonella bacilliformis: a systematic review of the literature to guide the research agenda for elimination. PLoS Negl Trop Dis. 2012;6:e1819. doi: 10.1371/journal.pntd.0001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knobloch J, Solano L, Alvarez O, Delgado E. Antibodies to Bartonella bacilliformis as determined by fluorescence antibody test, indirect hemagglutination and ELISA. Trop Med Parasitol. 1985;36:183–185. [PubMed] [Google Scholar]
- 12.Chamberlin J, Laughlin L, Gordon S, Romero S, Solorzano N, Regnery RL. Serodiagnosis of Bartonella bacilliformis infection by indirect fluorescence antibody assay: test development and application to a population in an area of bartonellosis endemicity. J Clin Microbiol. 2000;38:4269–4271. doi: 10.1128/jcm.38.11.4269-4271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knobloch J. Analysis and preparation of Bartonella bacilliformis antigens. Am J Trop Med Hyg. 1988;39:173–178. doi: 10.4269/ajtmh.1988.39.173. [DOI] [PubMed] [Google Scholar]
- 14.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–2274. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taye A, Chen H, Duncan K, Zhang Z, Hendrix L, Gonzalez J, Ching W. Production of recombinant protein Pap31 and its application for the diagnosis of Bartonella bacilliformis infection. Ann N Y Acad Sci. 2005;1063:280–285. doi: 10.1196/annals.1355.045. [DOI] [PubMed] [Google Scholar]
- 16.Dabo SM, Confer AW, Anderson BE, Gupta S. Bartonella henselae Pap31, an extracellular matrix adhesin, binds the fibronectin repeat III13 module. Infect Immun. 2006;74:2513–2521. doi: 10.1128/IAI.74.5.2513-2521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amano Y, Rumbea J, Knobloch J, Olson J, Kron M. Bartonellosis in Ecuador: serosurvey and current status of cutaneous verrucous disease. Am J Trop Med Hyg. 1997;57:174–179. doi: 10.4269/ajtmh.1997.57.174. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R, Indrayan A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr. 2011;48:277–287. doi: 10.1007/s13312-011-0055-4. [DOI] [PubMed] [Google Scholar]
- 19.Grimes DA, Schulz KF. Refining clinical diagnosis with likelihood ratios. Lancet. 2005;365:1500–1505. doi: 10.1016/S0140-6736(05)66422-7. [DOI] [PubMed] [Google Scholar]
- 20.Linzer D, Lewis J. poLCA: Polytomous Variable Latent Class Analysis. 2013. R package version 1.4. [Google Scholar]
- 21.Beath K. randomLCA: Random effects latent class analysis. 2013. R package version 0.8–6. [Google Scholar]
- 22.Sim J, Wright CC. The kappa statistic in reliability studies: use, interpretation, and sample size requirements. Phys Ther. 2005;85:257–268. [PubMed] [Google Scholar]
- 23.Knobloch J, Schreiber M. Bb65, a major immunoreactive protein of Bartonella bacilliformis. Am J Trop Med Hyg. 1990;43:373–379. doi: 10.4269/ajtmh.1990.43.373. [DOI] [PubMed] [Google Scholar]
- 24.Mallqui V, Speelmon EC, Verastegui M, Maguina-Vargas C, Pinell-Salles P, Lavarello R, Delgado J, Kosek M, Romero S, Arana Y, Gilman RH. Sonicated diagnostic immunoblot for bartonellosis. Clin Diagn Lab Immunol. 2000;7:1–5. doi: 10.1128/cdli.7.1.1-5.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander A, Berner R, Ruess M. Serodiagnosis of cat scratch disease: response to Bartonella henselae in children and a review of diagnostic methods. Eur J Clin Microbiol Infect Dis. 2001;20:392–401. doi: 10.1007/pl00011280. [DOI] [PubMed] [Google Scholar]
- 26.Lencakova D, Fingerle V, Stefancikova A, Schulte-Spechtel U, Petko B, Schreter I, Wilske B. Evaluation of recombinant line immunoblot for detection of Lyme disease in Slovakia: comparison with two other immunoassays. Vector Borne Zoonotic Dis. 2008;8:381–390. doi: 10.1089/vbz.2007.0216.A. [DOI] [PubMed] [Google Scholar]
- 27.Reed KD. Laboratory testing for Lyme disease: possibilities and practicalities. J Clin Microbiol. 2002;40:319–324. doi: 10.1128/JCM.40.2.319-324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edkins TJ, Koller-Eichhorn R, Alhadeff JA, Mayer U, Faust H, Del Tito BJ. Assessment of potential cross-reactivity of human endogenous matrix metalloproteinases with collagenase Clostridium histolyticum antibodies in human sera obtained from patients with Dupuytren's contracture. Clin Vaccine Immunol. 2012;19:562–569. doi: 10.1128/CVI.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson B, Lu E, Jones D, Regnery R. Characterization of a 17-kilodalton antigen of Bartonella henselae reactive with sera from patients with cat scratch disease. J Clin Microbiol. 1995;33:2358–2365. doi: 10.1128/jcm.33.9.2358-2365.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]