Abstract
Thirty pools from 900 (540 females and 360 males) Phlebotomus perfiliewi sandflies collected during the summer of 2012 in the Fermo area (Marche Region, central Italy) were tested for the presence of Phleboviruses. A nested polymerase chain reaction was performed using degenerated primers amplifying a fragment of the polymerase gene (large segment) and a fragment of the nucleoprotein gene (small segment) of the genus Phlebovirus. One pool was positive for Toscana virus, as expected from results of studies in the area, and six pools were positive for a putative novel Phlebovirus. Virus isolation in Vero cells was performed. Minimum field infection rates/1,000 insects processed for the novel and Toscana viruses were 6.7 and 1.0, respectively. Phylogenetic analysis of the novel Phlebovirus, tentatively named Fermo virus, placed it in the Sandfly Fever Naples virus serocomplex.
Phlebotomine sandflies are vectors of zoonotic pathogens (e.g., Leishmania spp. and Bartonella spp.) and arboviruses (e.g., Phlebovirus [family Bunyaviridae], Vesiculovirus [family Rhabdoviridae], and Orbivirus [family Reoviridae]) that affect human health.1
The genus Phlebovirus is divided into nine antigenic complexes that includes 37 of the 53 recognized viruses.2 Recent studies on phlebotomine sandfly surveillance in Europe highlighted the presence of new Phleboviruses (e.g., Punique, Adria, Massilia and reassortant Granada viruses), suggesting that viruses in this group need to be identified.3 Five Phlebotomus-borne (PhB) viruses have been isolated in Italy. Four viruses belong to the genus Phlebovirus: Sandfly fever Naples virus (SFNV) and Sandfly fever Sicilian virus (SFSV), which were isolated from soldiers during World War II; and Toscana virus (TOSV) and Arbia virus (ARBV), which have been isolated from Phlebotomus pernicious and Phlebotomus perfiliewi sandflies.3 Radi virus is the only PhB virus (family Rhabdoviridae, genus Vesiculovirus) that has been isolated in Italy.4 SFSV and SFNV are the causative agents of transient febrile illnesses in humans, and TOSV is neurovirulent and a major cause of summer aseptic meningitis in countries where it circulates.5,6 The purpose of our study was to monitor the infection rate and the viral stability of PhB viruses in a well-known focus of TOSV in Italy in the framework of the European FP7 EDENext project of surveillance of sandfly-borne diseases.
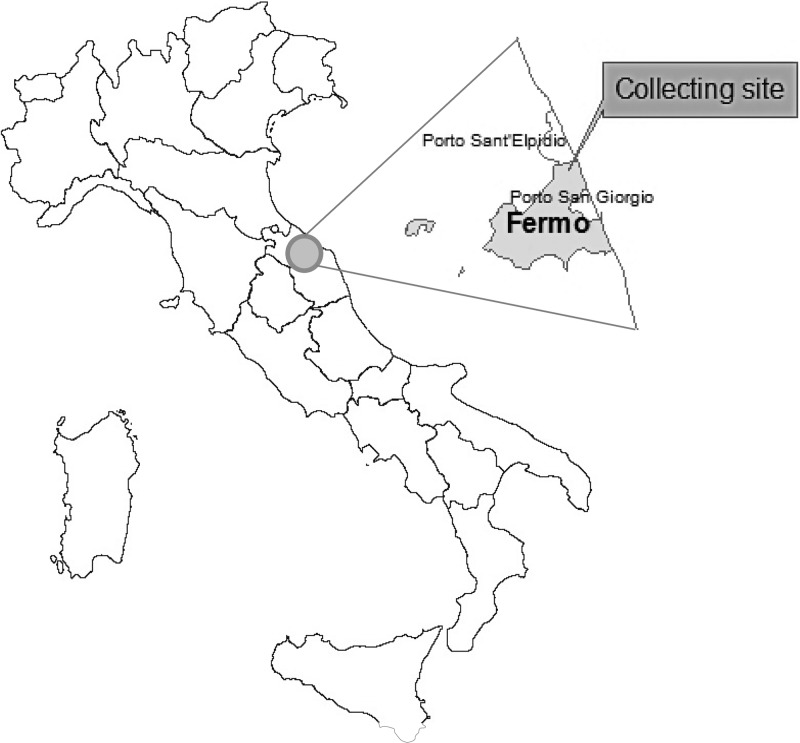
Sandfly and arbovirus surveillance was conducted during the second half of August 2012 in a rural area of Fermo commune (Marche region, central Italy), located 7 km from the Adriatic coast (43°11′55.84″N, 13°44′12.08″E, Torre Mattiucci) (Figure 1). The selected site was restricted to an area that had been surveyed and found to be monospecific for the phlebotomine species P. perfiliewi.4 In previous studies, no differences were found in monthly virus isolation rates during the activity season of this species (June–September). Thus, the sandfly collection period was selected according to the highest historical density peaks.
Figure 1.
Marche Region of central Italy showing the study site (Fermo commune) where Fermo virus was isolated during the summer of 2012.
A total of 2,980 sandflies (2,268 females and 712 males) were collected by using standard CDC miniature light traps (Hausherr Machine Works, Toms River, NJ) and transported alive to the laboratory, where they were pooled separately by sex (30 specimens/pool) and then stored at −80°C pending examination. Before analysis of PhB viruses, sandfly monospecificity of the collected samples was re-confirmed for a large representative subsample of total catches (250 males and 250 females). Identification used standard morphologic procedures and taxonomic keys,7,8 which required clarification and permanent mounting of specimens. All sandflies were identified as P. perfiliewi.
A total of 30 pools from a total of 900 sandflies (540 females) were assayed for PhB viruses. The pools were homogenized and supernatants were used for virus isolation in Vero cells and viral RNA extraction by using the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer's protocol. Two nested polymerase chain reactions (PCRs) were performed by using degenerated primers specific for regions of the polymerase (large [L]) and nucleoprotein (small [S]) genes.9,10
Phlebovirus RNA was detected by nested PCR in 7 of 30 pools (6 pools of females and 1 pool of males) and in Vero cells, which showed a cytopathic effect. The overall minimum field infection rate/1,000 insects processed was 7.7 (Table 1).
Table 1.
Phleboviruses isolated from Phlebotomus perfiliewi sandflies collected in Fermo commune, Marche Region, Italy*
| Sex | No. sandflies/no. pools | No. (%) Phlebovirus isolates (MFIR) | |
|---|---|---|---|
| RT-PCR | Vero cells | ||
| F | 540/18 | 6 (11.1) | 6 (11.1) |
| M | 360/12 | 1 (2.7) | 1 (2.7) |
| Total | 900/30 | 7 (7.7) | 7 (7.7) |
MFIR = minimum field infection rate/1,000 sandflies; RT-PCR = reverse transcription polymerase chain reaction.
The PCR products were purified by electrophoresis and gel extraction (QIAGEN) and were sequenced on both strands by using forward and reverse primers for the L and S genes. Sequences obtained were aligned with other Phlebovirus sequences available in the GenBank database. Analysis was performed by using CLUSTAL in DAMBE software (http://dambe.bio.uottawa.ca/dambe.asp), and sequences were analyzed in MEGA version 5.2 (www.megasoftware.net/).
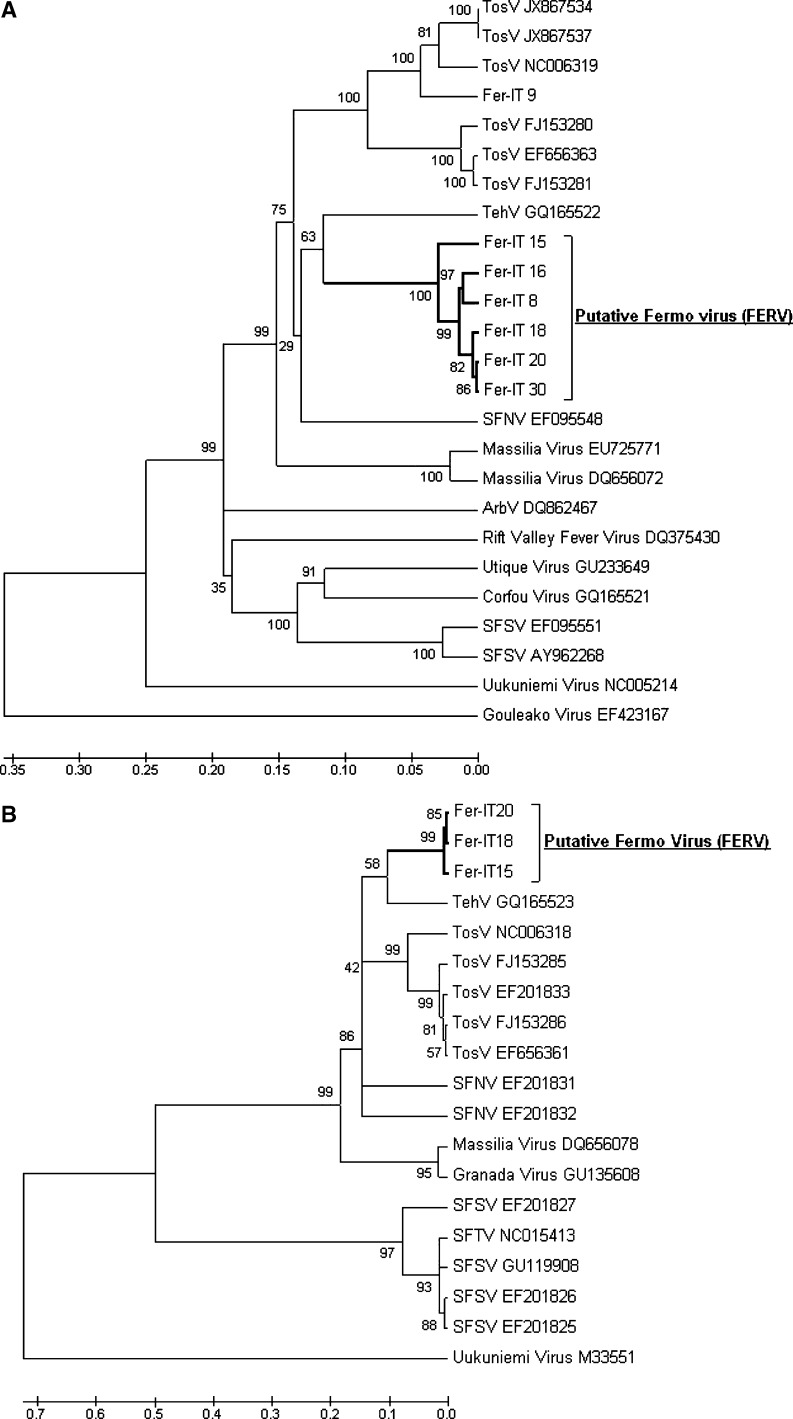
Results of phylogenetic analysis of the L and S genes are shown in Figure 2. Analysis of polymerase gene sequences (521 nucleotides) of the isolates obtained from five female pools and one male pool (Fer-IT8, Fer-IT15, Fer-IT16, Fer-IT18, Fer-IT20, and Fer-IT30) placed these viruses in a branch distinct from other Phleboviruses analyzed. Intra-genetic analysis of this isolate group showed minimal variation among nucleotide identities and amino acid similarities, ranging from 0.2% to 7.8% and 0% to 10.2%, respectively (Figure 2A and Table 2). Specific primers for Phleboviruses within the SFNV complex were used to confirm the Fermo virus isolates belonging to Naples serocomplex.9 Analysis of the variation of nucleotide identities within members of the SFNV complex (e.g., TOSV A and B genotypes, SFNV, and Tehran virus) showed genetic distances ranging from 22.7% to 30.0% (8.4% to 21.0% amino acid similarities) (Table 2).
Figure 2.
Phylogenetic trees of partial sequences of A, polimerase segment (L) and B, nucleoprotein segment (S) of Marche Region isolates and other selected phleboviruses. Trees were constructed by using the neighbor-joining method and 1,000 bootstrap values. Analysis was performed in Mega version 5.2 (www.megasoftware.net/). Sequences were aligned by using Clustal W (www.clustal.org/). Prototype sequences with accession numbers are indicated. Accession numbers of Fermo virus (FERV) sequences are Fer-IT8 HG321367, Fer-IT9 HG793786, Fer-IT15 HG793787, Fer-IT16 HG793791, FerIT18 HG793788, Fer-IT20 HG793789, and Fer-IT30 HG793790. Strains of the novel virus isolated in this study are indicated in bold. Scale bars indicate percentage diversity. TOSV = Toscana virus; SFSV = sandfly fever Sicilian virus; SFTV = sandfly fever Turkey virus; TehV = Tehran virus; SFNV = sandfly fever Naples virus; ArbV = Arbia virus.
Table 2.
Nucleotide and amino acid (in parentheses) diversity (%) of sequences of polymerase (L) and nucleoprotein (S) genes of FERV, TOSV genotypes A and B, SFNV, and TEHV*
| Nucleotide and amino acid diversity of large segment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| FERV isolates | TOSV A | TOSV B | SFNV | TEHV | |||||
| Fer-IT8 | 28.6 (18.6) | 28.8 (21.0) | 27.0 (22.2) | 23.7 (11.4) | |||||
| Fer-IT16 | 29.6 (18.0) | 30.0 (17,4) | 27.4 (22.2) | 25.0 (11.4) | 2.2 (8.4) | ||||
| Fer-IT15 | 27.4 (21.6) | 28.0 (24,0) | 27.4 (24.6) | 22.7 (14.4) | 6.8 (5.4) | 7.8 (10.2) | |||
| Fer-IT18 | 27.8 (16.2) | 28.0 (19.2) | 26.4 (19.8) | 22.9 (9.0) | 2.4 (6.0) | 3.4 (3.6) | 4.8 (7.8) | ||
| Fer-IT20 | 27.6 (15.6) | 27.8 (18.6) | 26.0 (19.2) | 22.7 (8.4) | 2.0 (4.2) | 3.8 (4.2) | 5.2 (8.4) | 0.8 (1.8) | |
| Fer-IT30 | 27.6 (15.6) | 27.8 (18.6) | 26.0 (19.2) | 22.7 (8.4) | 1.8 (4.2) | 3.6 (4.2) | 5.0 (8.4) | 0.6 (1.8) | 0.2 (0.0) |
| Nucleotide and amino acid diversity of small segment | |||||||||
| FERV isolates | TOSV A | TOSV B | SFNV | TEHV | |||||
| Fer-IT15 | 21.6 (11.8) | 21.6 (11.8) | 21.2 (7.1) | 16.9 (7.1) | |||||
| Fer-IT18 | 21.6 (11.8) | 21.6 (11.8) | 22.0 (7.1) | 17.3 (7.1) | 1.6 (0.0) | ||||
| Fer-IT20 | 21.6 (11.8) | 20.8 (11.8) | 22.0 (7.1) | 16.9 (7.1) | 1.2 (0.0) | 0.04 (0.0) | |||
FERV = Fermo virus; TOSV = Toscana virus; SFNV = sandfly fever Naples virus; TEHV = Tehran virus.
In addition, analysis of nucleoprotein gene sequences (258 nucleotide) from three of six isolates (Fer-IT15, Fer-IT18, and Fer-IT20) showed that they were closely related to each other in nucleotide composition (0.4–1.6%) and identical in amino acid composition (Figure 2B and Table 2). The analysis of the S sequences of these three isolates showed that they were distinct from the SFNV group members (19.6–22.0% in nucleotide composition) and distantly related to SFSV and Uukuniemi virus (44.3–46.7% and 49.8–51.0% in nucleotide composition, respectively) (Figure 2B). These results on the partial sequences of the L and S genes suggested the presence in these isolates of a novel virus belonging to the genus Phlebovirus, which we tentatively name Fermo virus (FERV).
One of the seven isolates (Fer-IT9) clustered in the TOSV subgroup. Its genetic distance with known TOSV was 10.2% (Figure 2A), thus confirming the persistence of TOSV in populations of P. perfiliewi in central Italy. The overall minimum field infection rate was higher (1.0) than that previously recorded in Italy,4,11 indicating possible annual variations in infection rates.
The isolation of FERV from one pool of male sandflies strongly suggests that this putative new virus, similar to TOSV, ARBV, and Radi virus, is transmitted transovarially.11 Analysis of our isolates did not identify ARBV. Furthermore, four isolates that were negative by nested PCR amplification for members of the genus Phlebovirus showed a cytopathic effect in Vero cell cultures and their identification is in progress. Previous studies of viral ecology showed the simultaneous presence of TOSV and ARBV in the same population of a sandfly species,4 as well as mechanisms for potential virus maintenance and amplification by transovarial11 and venereal transmission.12 These ecologic niche characteristics, associated with possible co-infections in the same vector, could result in the emergence of reassortants of viruses with different degrees of pathogenecity.
Further studies of full-genome sequencing and serologic characterization are in progress to confirm the taxonomic status of the novel FERV. Moreover, investigations are needed to monitor seasonal and annual distribution of local viruses, as well as human seroprevalence, and to determine the pathogenicity of FERV by analyzing patient samples with summer fever or neurologic disease.
ACKNOWLEDGMENTS
This study was catalogued by the EDEnext Steering Committee as EDENext195 (http://www.edenext.eu).
Disclaimer: The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission.
Footnotes
Financial support: This study was supported by European Union grant FP7-261504 EDENext.
Authors' addresses: Maria Elena Remoli, Claudia Fortuna, Antonella Marchi, Paola Bucci, and Maria Grazia Ciufolini, Unit of Viral Diseases and Attenuated Vaccines, Department of Infectious, Parasitic and Immunomediated Disease, Istituto Superiore Di Sanità, Rome, Italy, E-mails: mariaelena.remoli@iss.it, claudia.fortuna@iss.it, antonella.marchi@iss.it, paola.bucci@iss.it, and mariagrazia.ciufolini@iss.it. Claudio Argentini, Global Health Unit, Department of Therapeutic Research and Medicines Evaluation, Istituto Superiore di Sanità, Rome, Italy, E-mail: claudio.argentini@iss.it. Gioia Bongiorno, Michele Maroli, Luigi Gradoni, and Marina Gramiccia, Unit of Vector-Borne Diseases and International Health, Department of Infectious, Parasitic and Immunomediated Disease, Istituto Superiore di Sanità, Rome, Italy, E-mails: gioia.bongiorno@iss.it, michele.maroli@gmail.com, luigi.gradoni@iss.it, and marina.gramiccia@iss.it.
References
- 1.Maroli M, Feliciangeli D, Bichaud L, Charrel RN, Gradoni L. Phlebotomine sandflies and spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol. 2013;27:123–147. doi: 10.1111/j.1365-2915.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- 2.King AM, Adams MJ, Caestens EB, Lefkowitz EJ. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier; 2012. [Google Scholar]
- 3.Depaquit J, Grandadam M, Fouque F, Andry PE, Peyrefitte C. Arthropod-borne viruses transmitted by Phletomine sandflies in Europe: a review. Euro Surveill. 2010;15:19507. [PubMed] [Google Scholar]
- 4.Ciufolini MG, Maroli M, Miceli N, Nicoletti L, Cecchetti S, Amore R, Percopo S, Verani P. Monitoring of Phlebotominae sandflies transmitted viruses activity in Marche region (Italy) Arbo Info Exch. 1990:58–59. [Google Scholar]
- 5.Charrel RN, Gallian P, Navarro-Mari J-M, Nicoletti L, Papa A, Sánchez-Seco MP, Tenorio A, de Lamballerie X. Emergence of Toscana virus in Europe. Emerg Infect Dis. 2005;1:1657–1663. doi: 10.3201/eid1111.050869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venturi G, Madeddu G, Rezza G, Ciccozzi M, Pettinato ML, Cilliano M, Fiorentini C, Mura MS, Ciufolini MG. Detection of Toscana virus central nervous system infections in Sardinia Island, Italy. J Clin Virol. 2007;40:90–91. doi: 10.1016/j.jcv.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Theodor O. Psychodidae-Phlebotomine. In: Lindner, E, editor. Die Fliegen der Palaearktischen Region, 9c. Stuttgart, Germany: Schweizerbart'sche Verlagsbuchhandlung; 1958. pp. 1–55. [Google Scholar]
- 8.Léger N, Pesson B, Madulo-Leblond G, Abonnenc E. Differentiation of females of the subgenus Larroussius Nitzulescu 1931 (Diptera-Phlebotomidae) of the Mediterranean region. Ann Parasitol Hum Comp. 1983;58:611–623. [PubMed] [Google Scholar]
- 9.Charrel RN, Izri A, Temmam S, Delaunay P, Toga I, Dumon H, Marty P, de Lamballerie X, Parola P. Cocirculation of 2 genotypes of Toscana virus, southeastern France. Emerg Infect Dis. 2007;13:465–468. doi: 10.3201/eid1303.061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sánchez-Seco MP, Echevarría JM, Hernández L, Estévez D, Navarro-Marí JM, Tenorio AJ. Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. Med Virol. 2003;71:140–149. doi: 10.1002/jmv.10465. [DOI] [PubMed] [Google Scholar]
- 11.Verani P, Ciufolini MG, Caciolli S, Renzi A, Nicoletti L, Sabatinelli G, Bartolozzi D, Volpi G, Amaducci L, Coluzzi M. Ecology of viruses isolated from sand flies in Italy and characterized of a new Phlebovirus (Arbia virus) Am J Trop Med Hyg. 1988;38:433–439. doi: 10.4269/ajtmh.1988.38.433. [DOI] [PubMed] [Google Scholar]
- 12.Ciufolini MG, Maroli M, Guandalini E, Marchi A, Verani P. Experimental studies on the maintenance of Toscana and Arbia viruses (Bunyaviridae: Phlebovirus) Am J Trop Med Hyg. 1989;40:669–675. doi: 10.4269/ajtmh.1989.40.669. [DOI] [PubMed] [Google Scholar]