Abstract
The natural extracellular matrix (ECM), with its multitude of evolved cell-instructive and cell-responsive properties, provides inspiration and guidelines for the design of engineered biomaterials. One strategy to create ECM-mimetic materials is the modular design of protein-based engineered ECM (eECM) scaffolds. This modular design strategy involves combining multiple protein domains with different functionalities into a single, modular polymer sequence, resulting in a multifunctional matrix with independent tunability of the individual domain functions. These eECMs often enable decoupled control over multiple material properties for fundamental studies of cell-matrix interactions. In addition, since the eECMs are frequently composed entirely of bioresorbable amino acids, these matrices have immense clinical potential for a variety of regenerative medicine applications. This brief review demonstrates how fundamental knowledge gained from structure-function studies of native proteins can be exploited in the design of novel protein-engineered biomaterials. While the field of protein-engineered biomaterials has existed for over 20 years, the community is only now beginning to fully explore the diversity of functional peptide modules that can be incorporated into these materials. We have chosen to highlight recent examples that either (1) demonstrate exemplary use as matrices with cell-instructive and cell-responsive properties or (2) demonstrate outstanding creativity in terms of novel molecular-level design and macro-level functionality.
Keywords: Protein engineering, Extracellular matrix, Modular design strategy, Tissue engineering, Recombinant protein synthesis, Three-dimensional hydrogels
1. Introduction: The rationale for creating engineered extracellular matrices (eECM)
The native extracellular matrix (ECM) is a complex and heterogeneous material containing numerous multifunctional proteins that provide cells with structural support and biochemical signals to facilitate a series of important cellular processes [1]. First, the ECM presents integrin-specific ligands and binding affinity to other cell-surface receptors that combine to initiate cell adhesion. Second, through complex and dynamic interactions with cells, the matrix provides a three-dimensional (3D) mechanical framework. These biochemical and biomechanical cues activate highly regulated signaling pathways that allow for ensuing cellular responses such as spreading, migration, proliferation, and differentiation.
In order to mimic these important functions of native ECM in the design of biomaterial scaffolds, the strategy of using isolated ECM components (collagen, fibronectin, etc) or their mixtures harvested from tissues has been widely adopted [2]. Although often highly cell adhesive, these biomaterial scaffolds have proved challenging to standardize and use for in vivo applications due to poorly defined chemical structure, inconsistent batch-to-batch reproducibility, and risk of immunogenicity. In addition, it is extremely difficult to manipulate and customize the ECM scaffolds for a specific cellular microenvironment or to study fundamental aspects of cell-material interactions, because all material factors are intertwined and coupled together, resulting in largely observation-based outcomes.
Motivated to design tunable biomaterials that emulate the native ECM, researchers have been developing engineered ECM (eECM) that combines multiple structural and biofunctional features [3, 4]. Using recombinant protein technologies, eECM offers enormous possibilities in the design of reproducible, highly tunable, and modular protein scaffolds [5–9]. The four major advantages of creating eECM using protein engineering are: 1) to gain better control over decoupled material variables for mechanistic studies of cell-matrix interactions, 2) to achieve more physiologically relevant in vitro cultures, 3) to create more reproducible materials for clinical therapies, and 4) to create more complex and dynamic materials with multi-functionality, responsiveness, and bioactivity. These four advantages are discussed in more detail below.
Towards goal 1, eECM can be customized to have consistent material properties with only one variable factor of interest, such as cell-adhesive ligand density, matrix compliance, structural formation, and cell-instructive biochemical signals. For example, elastin-like protein (ELP) hydrogels have been designed with either a cell-adhesive arginine-glycine-aspartic acid (RGD) ligand or non-adhesive, sequence-scrambled RDG in their otherwise identical primary amino acid sequences [10]. Thus, blending these two engineered proteins together prior to crosslinking into a bulk hydrogel affords a direct control over the bioactive ligand density. Independently, the matrix stiffness of these hydrogels can be tuned by altering the density of crosslinks [11]. This system has been used to evaluate the independent effects of RGD ligand density and matrix stiffness on neurite outgrowth from three-dimensional cultures of dorsal root ganglia [12].
Towards goal 2, once synthesized, eECM proteins can be fabricated through a variety of techniques to create matrices that mimic certain structural features of the native ECM. These material structures include 2D surface patterning at the micro- and nanoscale [13], 3D hydrogels [12, 14], porous scaffolds [15], and fibrous structures [16]. The eECM can then be seeded with cells to create either 2D or 3D cultures that recapitulate aspects of the cell niche and produce cell responses distinct from standard 2D tissue culture polystyrene with ECM coatings. These in vitro cultures may result in cell morphologies and levels of gene expression that are more reminiscent of in vivo tissue.
Towards the creation of consistent materials for clinical therapies, protein engineering offers a highly reproducible synthetic strategy. Because of the high fidelity of protein translation, recombinant proteins present precisely controlled, monodispersed sequences and biochemical compositions at the molecular level, a feature that is normally improbable in natural or synthetic materials [17]. In addition to reproducibility and customizability, eECM is also biodegradable and yields non-toxic degradation products, which is desirable for clinical usage.
Towards goal 4, the modular design strategy of eECM enables direct incorporation of diverse peptide building blocks into the backbone of a single protein sequence. This modular approach results in the synthesis of multi-functional materials that combine the functionality of each individual peptide domain. For example, novel, protein-engineered, cell-delivery vehicles have been developed using several peptide-based gelation mechanisms including leucine-zipper self-assembly [18–20], enzyme-triggered self-assembly [21, 22], chemical crosslinking [11, 12, 14, 23], and hetero-assembly of molecular recognition peptides [24–27]. In addition to these domains that enable structural gelation, ECM-mimetic domains that are either cell-instructive or cell-responsive can be included. Examples include cell-adhesive [28–32], growth factor mimetic [33–38], or enzyme-degradable domains [11, 39–41]. Finally, more complex designs can be achieved by adding functional domains that interact with inorganic materials [42–45] or respond to dynamic environmental stimuli [46–48].
In this review, we describe the toolbox that is currently used to generate protein-engineered eECM biomaterials in Section 2. We focus our attention on eECM fabricated purely from protein-engineered materials using canonical amino acids. In Section 3, we describe the wide selection of peptide building blocks and domains available for the modular design of eECM, with an emphasis on interactions with mammalian cells. Finally, we discuss emerging new functionalities and peptide modules for eECM design in Section 4, including binding domains with inorganic materials, anti-microbial peptides, immuno-modulatory peptides, and dynamic peptides triggered by environmental stimuli.
2. Toolbox to design and synthesize eECM
2.1. Direct peptide synthesis versus recombinant protein synthesis
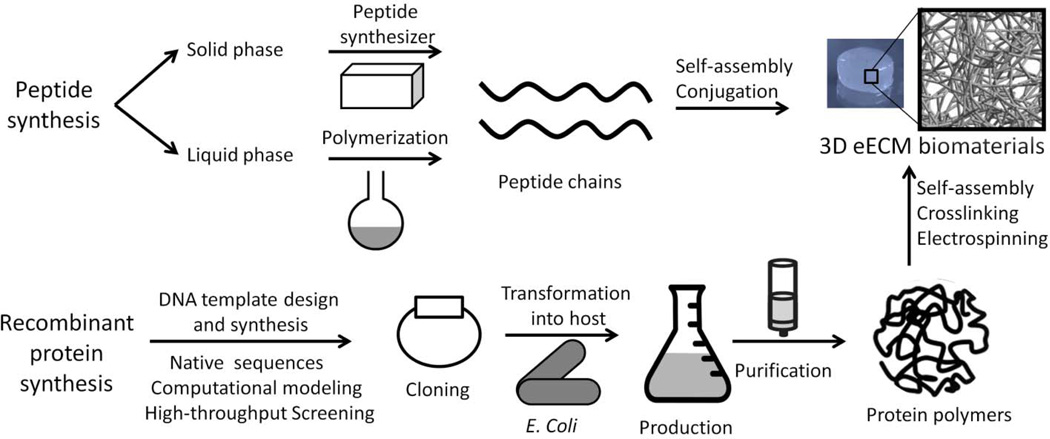
As protein technologies continue to evolve, there are now several toolkits available to achieve reproducible and tunable eECM with virtually limitless possibilities. To design a multi-functional eECM, the first step is the selection of primary amino acid sequences with desirable structural, gelation, degradation, or bioactive roles. Once the desired amino acid sequence has been designed, two common techniques can be used to synthesize the eECM: direct chemical synthesis of small peptide building blocks or recombinant biochemical synthesis of large proteins (Figure 1). Direct peptide synthesis is usually realized through solid phase peptide synthesis (SPPS) using a peptide synthesizer. The advent of peptide synthesizers allows for automatic and efficient production of up to 30–50 amino acid residues in sub-gram quantities at reasonable cost [49]. These small peptides can be ligated together to create much longer sequences of up to 150 amino acids [50]. SPPS ensures precise chemical structures and enables the addition of modifications to the functional groups of individual amino acids during synthesis. Liquid phase peptide synthesis (LPPS) is a classical method that requires sequential incorporation of amino acids followed by removal of protecting groups [51]. LPPS is still commonly used for large-scale production of gram-scale quantities of a given peptide, although this synthetic route is much slower and more labor intensive than SPPS.
Figure 1.
Schematic of peptide and recombinant protein synthesis to construct eECM biomaterials. Peptide synthesis is conducted by either solid phase or liquid phase synthesis that results in short peptide chains for self-assembly or conjugation into eECMs. Recombinant protein synthesis involves the modular design of peptide modules derived from native sequences, predicted through computational modeling, or identified by high throughput combinatorial screening. These modules are concatenated and translated into a DNA template, which is then cloned into a genetic vector and transformed into the host cell of choice, where production of the engineered protein occurs. After purification, the target protein is further processed via self-assembly, crosslinking, or electrospining into 3D eECM biomaterials.
Recombinant protein synthesis can produce much larger proteins with more complex structural features, resulting in more complex functionalities. This biochemical synthesis strategy requires several genetic engineering steps, each of which may require optimization to achieve efficient yields of functional protein. First, a DNA template that encodes the target amino acid sequence is designed and chemically synthesized. This engineered gene is then inserted into a plasmid vector that enables gene replication and transcription. The vector is transformed into a host organism that expresses the target protein. The target protein is then harvested and purified from other endogenous proteins and contaminants. This templated synthesis offers precise control of long protein sequences with multiple functional modules each presented at a specific location, facilitating independent tuning of mechanical properties and bioactivity within the final biomaterial.
2.2. Protein engineering strategies for designing eECM
The most common strategy for designing engineered protein biomaterials with multi-functionality is to first design individual peptide domains and then to mix and match these peptide domains to create a versatile family of ECM-mimetic materials. This modular design strategy can use peptide domains derived from native sequences, predicted through computational modeling, or identified by high throughput combinatorial screening. Common peptide modules that are rationally derived from native sequences include silk-like [52], elastin-like [53], and resilin-like [54] domains. Protein sequences based on these peptide modules often display properties reminiscent of their native proteins. Rational modification of the native sequences is often performed using site-specific amino acid mutation, which requires knowledge of the underlying protein structure-function relationship and identification of appropriate target sites for mutation [55].
To better choose sites for mutation and to predict the resultant properties, many computational algorithms have been developed that employ physics-based modeling to estimate energetic protein interactions. As an example, the WW domain, a small protein interaction module that undergoes heteroassembly with a proline-rich peptide sequence, has been engineered using a computer-based model to construct an artificial sequence that functions like its natural counterpart [56, 57]. This molecular recognition has been further utilized to create physical hydrogels that encapsulate cells without environmental triggers [24]. A new computational method has recently been used to design self-assembling proteins with high accuracy. This method includes a symmetrical docking of peptides in a target with symmetric architecture and a subsequent design of low-energy, protein-protein interfaces to drive self-assembly [58]. Besides rational or computational engineering of native sequences, combinatorial engineering using high throughput screening methods provides a powerful alternative to generating proteins with complex properties. Also known as directed evolution, this technology involves first creating a diverse gene and corresponding protein library and then screening and identifying mutants with the desired properties [11, 42, 59–61]. For example, combinatorial engineering has been used to identify clones with high affinity to specific integrin receptors [60]. It is also a widely used method to identify peptides or proteins capable of binding to target inorganic material surfaces, such as hydroxyapatite [42].
3. Protein modules to create eECMs
3.1. Structural domains for cell-compatible encapsulation
The formation of 3D ECM-mimetic matrices entrapping viable cells is a crucial step for functional tissue engineering [62]. To construct a 3D structure for cell encapsulation, a crosslinking mechanism is often needed for eECM materials. Current state-of-the-art techniques can be classified into two main categories: physical crosslinking, which includes self-assembly and molecular recognition, and chemical crosslinking, which typically utilizes primary amine and thiol groups to form covalent bonds between protein polymer chains. A combination of both crosslinking mechanisms has also been used to design hydrogels for potential cell encapsulation [63]. Physical hydrogels are generally endowed with shear-thinning and self-healing properties via reversible sol-gel transitions, but they are subject to quick erosion and low mechanical moduli. In comparison, chemical hydrogels are typically much more stable in situ and possess higher mechanical rigidity, although special attention must be made to developing a cell-compatible crosslinking chemistry.
One classic example of a self-assembly module is the leucine zipper domain, which enables reversible self-assembly through coiled-coil associations [64]. This motif contains hydrophobic leucine residues and charged residues that form amphiphilic α-helical structures that multimerize into coiled-coils as junction points in the engineered hydrogel network. The gelation process can be reversibly triggered by external stimuli such as temperature, pH, and ionic strength [19]. These injectable hydrogels display rapid recovery after injection, and more than 95% of seeded cells survive the injection process [65]. These cyto-compatible, leucine zipper hydrogels were modified with RGD cell-binding ligands to promote adhesion, spreading, and polarization of human fibroblast cells, which remained rounded on unmodified hydrogels [20]. When presented as two-dimensional substrates, these materials promote the viability and proliferation of human fibroblasts, human umbilical vein endothelial cells, and rat neural stem cells [66]. One potential drawback to the use of leucine zipper hydrogels is their stability in biological environments, as they erode when placed in contact with cell culture media. However, leucine zipper domains can be stabilized to inhibit dissolution by subsequent chemical crosslinking using end-linked cysteine residues that form disulfide covalent bonds [18].
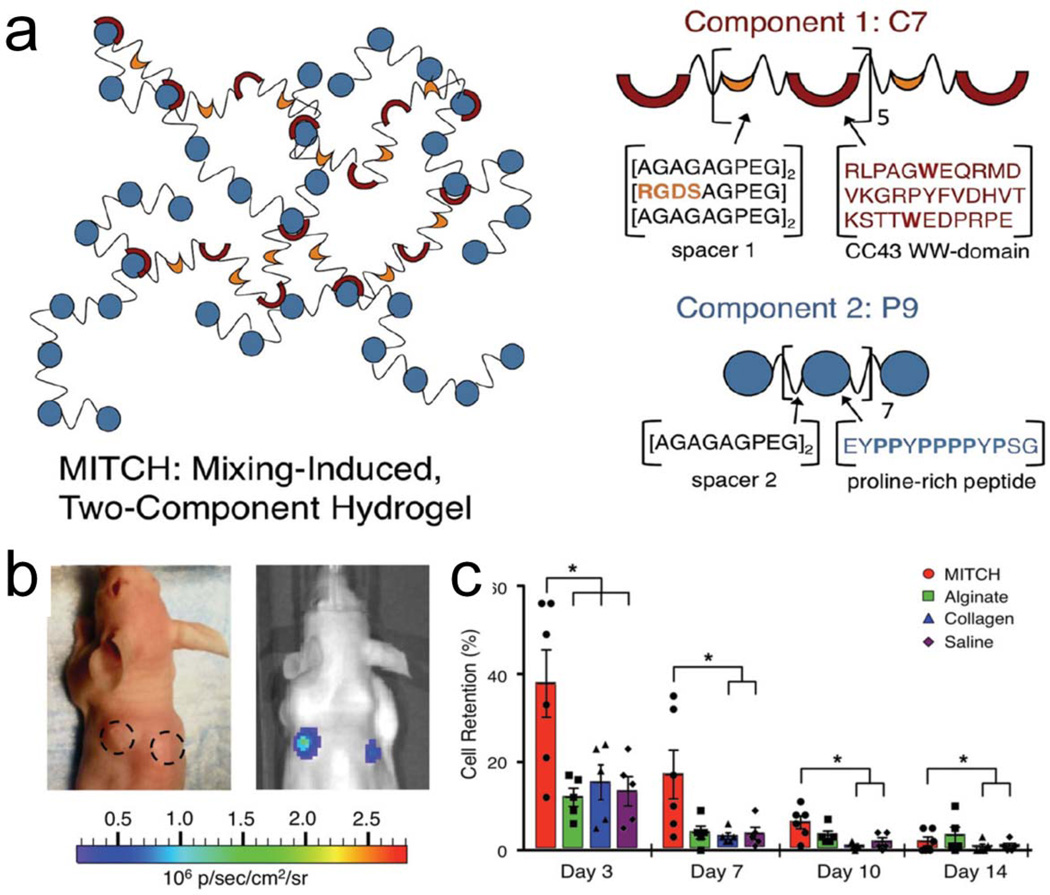
Molecular recognition is another important motif for creating physical hydrogels. In one example, a mixing-induced two-component hydrogel (MITCH) system, where one component is a block copolymer containing multiple repeats of the WW domain that specifically recognize proline-rich peptide domains encoded in a second component. The two components undergo a sol-gel transition by forming physical associations upon mixing without the need for any external environmental triggers [24]. [67]. The resulting hydrogel is shear-thinning, injectable, and able to self-heal, facilitating the encapsulation of neural stem cells and supporting their growth and differentiation [24]. In addition, MITCH has been used as a stem cell-delivery vehicle for adipose-derived stem cells in vivo in a subcutaneous mouse model. MITCH-delivery resulted in higher levels of cell survival and retention compared to other common hydrogels such as collagen and alginate [68] (Figure 2).
Figure 2.
A physically crosslinked hydrogel formed upon mixing of two modularly designed polypeptides for the encapsulation of mouse adipose-derived stem cells (mASCs). (a) Schematic of Mixing-Induced, Two-Component Hydrogel (MITCH) network (left) formed after mixing of individual components (right). Protein sequences shown using single letter amino acid abbreviations. (b) Images of mASC (Fluc+) transplant sites in nude mice; sites demarcated with dotted lines (day 0, left) and with bioluminescence (BLI) total flux overlay (day 3, right). (c) BLI measurements of cell retention. Data normalized to day 1 and reported as mean ± SEM; n = 5 or 6; * p < 0.0001. Adapted from reference [100], copyright 2012, reprinted with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
The design of reversible hydrogels was also demonstrated using molecular recognition between a protein and a peptide conjugated to a multi-arm polyethylene glycol (PEG) spacer. For example, a recombinant tax-interactive protein-1 (TIP1) with a PDZ domain was fused to each end of the triangular trimeric CutA protein. Upon mixing with PDZ-binding peptides conjugated to PEG, a 3D physical gel was formed at physiological pH and temperature, entrapping chondrocytes with high viability [69]. TIP-1 was further modified into a fusion protein with four binding sites and mixed with self-assembled nanofibers formed by TIP-1 binding polypeptides, leading to the formation of molecular hydrogels [27]. Similarly, a physical hydrogel design was created based on a structural tetratricopeptide repeat (TPR) that recognizes TPR-binding peptide modules conjugated to PEG. These two components form self-supporting hydrogels upon mixing at room temperature [70]. As a further example, a self-assembling dock-and-lock system was used to fabricate hydrogels through molecular recognition between an A-kinase anchoring protein domain and a docking and dimerization domain [71]. This specific, dynamic, and rapidly associating protein-ligand interaction enabled two components to form shear-thinning and rapidly self-healing hydrogels for cytocompatible encapsulation of human mesenchymal stem cells (hMSCs) [71].
As an alternative to the above-described physical hydrogels that rely on transient crosslinking, permanent chemical crosslinking mechanisms have been widely used to create 3D protein hydrogels for cell encapsulation. In one strategy, enzymatic reactions enable mild and biocompatible crosslinking reactions to occur between protein polymers that act as enzyme substrates. For example, tissue transglutaminase (tTG), a multifunctional enzymatic crosslinker that stabilizes tissues [72], has been used to catalyze covalent bonding between lysine and glutamine amino acids. In one demonstration, 3D hydrogels were created using tTG to form crosslinks between two classes of protein polymers containing either lysine or glutamine reactive sites that were evenly spaced along the protein backbone. Under physiological conditions, complete crosslinking through tTG occurred within 2 min. Mouse 3T3 and primary human fibroblasts encapsulated in this 3D hydrogel showed high cell viability and displayed spreading on 2D gel surfaces [21]. In another study, tTG was used to crosslink elastin-like polypeptides (ELPs) designed for potential cartilage repair therapies. Cells maintained their chondrocytic phenotype in the ELP hydrogels in vitro and restructured the ELP matrix to deposit cartilage ECM components [73].
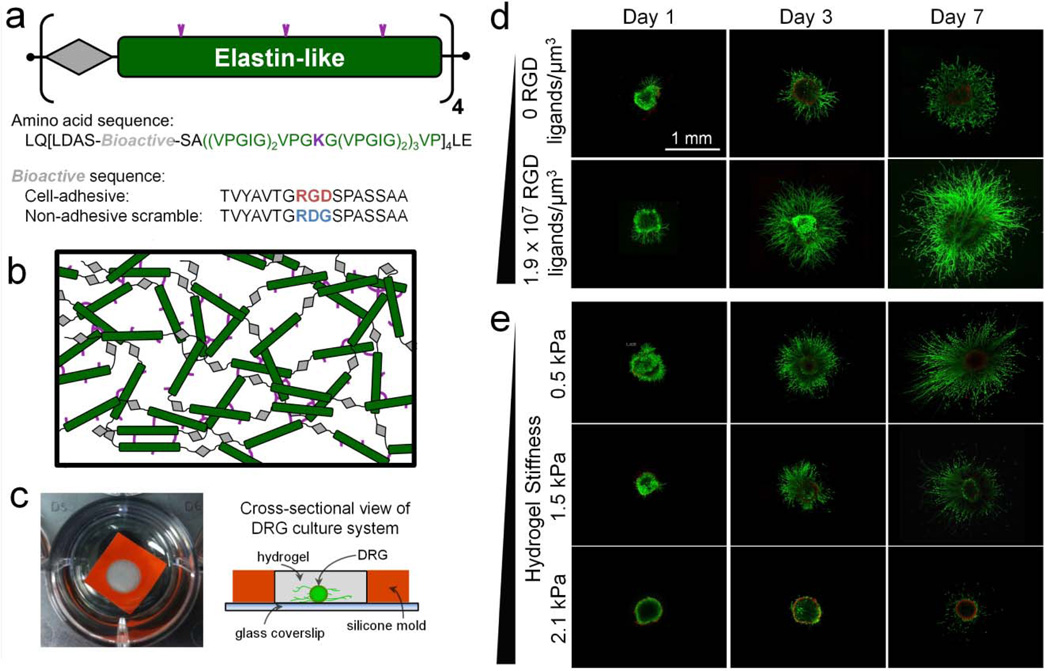
In addition to enzymatic crosslinking, chemical hydrogels can also be formed using small molecule crosslinkers. As an example, ELP materials are frequently modified to include lysine residues, which enable primary amine reactive crosslinking. These ELP hydrogels have been created using various chemical crosslinkers, including trissuccinimidyl aminotriacetate [74], disuccinimidyl suberate [11], and phosphine-based catalysts [12, 14, 23]. Similarly, resilin-like polypeptides (RLPs), which are based on the elastomeric structural protein resilin, have been crosslinked by phosphine-based catalysts to form ECM-mimetic hydrogels [39, 75, 76]. Recently, another novel family of recombinant elastomeric proteins have been reported based on abductin, a protein found in the inner hinge of bivalves [82]. Numerous types of cells have been encapsulated in these eECM hydrogels to probe cell-material interactions as well as to build up tissue-engineered constructs. For example, by encapsulating dorsal root ganglia within a tunable family of ELP matrices, the effects of integrin ligand density and matrix stiffness on neurite outgrowth was analyzed in a decoupled manner, showing longer neurite outgrowth in gels with higher ligand density and lower stiffness [12] (Figure 3). As a further example, an integrin-binding, RLP hydrogel with stiffness close to human cartilage tissue supported the adhesion, spreading, and 3D encapsulation of hMSCs [39, 75].
Figure 3.
A chemically crosslinked ELP hydrogel for the encapsulation of dorsal root ganglia (DRG). (a) ELPs were designed with modular repeats of bioactive (grey) and elastin-like (green) sequences. The bioactive domains were either an extended RGD sequence or a non-adhesive, scrambled RDG sequence. (b) The elastin-like structural domains included lysines (purple) for site-specific crosslinking with amine-reactive crosslinkers yielding a 3D hydrogel network. (c) Photograph (left) and schematic (right) of ELP hydrogel within a 5-mm silicone mold (orange) to encapsulate a single DRG cluster for culture within a 24-well plate. (d) Neurites extended from the initial DRG explant in hydrogels with 0 and 1.9 × 107 cell-adhesive RGD ligands µm−3 over the course of 7 days culture. (e) Neurites extended by day 1 in 0.5 kPa hydrogels, by day 3 in 1.5 kPa hydrogels, and not until day 7 in 2.1 kPa hydrogels. Adapted from reference [12], copyright 2012, reprinted with permission from Elsevier.
3.2. Cell-instructive domains
While the structural and crosslinking domains described in the previous section provide a scaffolding interactions to build up 3D matrices, a host of other peptide modules can be introduced into the eECMs to impart additional biofunctionality. To promote cell adhesion and spreading, well-known cell-adhesive domains, such as RGD, IKVAV, and YIGSR amino acid sequences, naturally found in various ECM proteins, are frequently integrated into the material [29–32]. In addition to the naturally evolved cell-adhesive sequences, domains identified through protein engineering strategies may be particularly useful in eECM development. For example, the RGD sequence has been engineered into a cystine-knot peptide motif to modulate its integrin-binding affinity and specificity [60, 77].
RGD ligands have been encoded into ELP, RLP, and silk-like proteins to promote interactions with a variety cell types [12, 14, 78]. In a particularly elegant example, RGD accessibility was tuned by fusion to a coiled-coil domain, which was functionalized on gold nanoparticles and subsequently immobilized on substrates. RGD accessibility and thereby cell adhesion was reversibly controlled via co-immobilization and removal of leucine-zipper coiled coils through heterodimerization [79]. An alternative cell-binding domain, CS5, has been added into ELPs to illustrate the effects of distal amino acid selection on the resultant accessibility and affinity of the cell-binding domain [28]. In another study, N-cadherin, a key cell-cell adhesion protein in neural development, was fused to the Fc region of the IgG antibody to create an artificial ECM surface that enables neural stem cells to maintain their undifferentiated state and preserve their differentiation potential [80]. In another novel design, a fibronectin type III domain from human tenascin-C was used in the creation of ECM-mimetic hydrogels that encourage the spreading of human lung fibroblast cells [81].
In addition to cell-adhesion domains, other bioinstructive moieties such as growth factors and cell signaling components have been added to the protein-engineered biomaterials toolbox. For example, fibroblast growth factor-2 (FGF-2) fused to a fibronectin fragment presented a synergistic effect on osteoblast cell adhesion and proliferation [33]. However, the direct encoding of longer bioactive sequences into the protein backbone can lead to decreased functionality, since conjugation at the N- and/or C-termini may hinder the complex folding that is typical of these factors. Therefore as an alternative strategy, fully folded soluble factors have been immobilized to the scaffold, either by covalent conjugation or affinity binding. As an example, vascular endothelial growth factor (VEGF), a heparin-binding growth factor, served as elastic crosslinks in a noncovalently assembled hydrogel network. In the presence of VEGF receptors, the hydrogel is selectively eroded, and the VEGF released from these hydrogels increased proliferation of VEGF-responsive cell lines [34].
An alternative method for growth factor immobilization within eECMs is to conjugate the bioactive signals to an ECM-binding domain, which binds to ECM matrices via affinity-based interactions. For example, transforming growth factor-beta (TGF-β) has been recombinantly engineered to contain a collagen-binding domain [35]. Similarly, genetically engineered human bone morphogenetic protein-2 (BMP-2) was fused to a collagen-binding domain for potential bone repair. A collagen matrix loaded with the engineered BMP-2 induced better bone formation in a rabbit mandible defect model [36, 37]. Additionally, platelet-derived growth factor BB (PDGF-BB) has been immobilized to a collagen-binding domain. The engineered PDGF-loaded collagen scaffolds encouraged cell proliferation in vitro and were uniformly cellularized and vascularized in vivo [38].
3.3. Cell-responsive domains
The native ECM is able to respond to changes in the local microenvironment, as cells survive, migrate, and secrete their own ECM. To mimic this functionality, amino acid sequences that can be cleaved at specific sites by cell-secreted proteases have been introduced into eECM biomaterials. One common motif is domains degradable by matrix metalloproteinases (MMPs). As an example of eECM multifunctionality, a single RLP has been engineered with a cell-adhesive RGD sequence, a heparin-binding domain for non-covalent immobilization and release of growth factors, and an MMP-cleavable site to enable proteolytic degradation [83]. The MMP-sensitive domains are susceptible to MMP-1 enzymatic degradation within two days. hMSCs encapsulated in these RLP hydrogels with various degradation rates displayed high viability [39]. Similarly, silk-elastin-like protein polymers (SELPs) have been modified to include a sequence that is sensitive to MMP, which yielded complete cleavage of all full-length polymers in two days [41].
As an alternative approach, peptide sequences that are sensitive to degradation by the proteases tissue plasminogen activator (tPA) and urokinase plasminogen activator (uPA) have been encoded into ELP hydrogels. These domains were designed to be responsive to neuronal growth cones, which locally secrete tPA and uPA, in order to clear a path for elongating neurites. Minor amino acid sequence mutations yielded hydrogel variants with identical initial mechanical properties but with degradation kinetics that spanned two orders of magnitude [40]. Neuronal cell adhesion, neuronal differentiation, and neurite outgrowth were all supported by these matrices [11].
4. Emerging functionalities for eECMs
While the majority of domains used in eECMs to date have been inspired by commonly occurring motifs in the native ECM, functional domains from non-matrix proteins can be repurposed to create more complex eECM designs. These emerging functionalities include linkage with inorganic materials, antimicrobial activities, immune modulators, and dynamically responsive modules. These new functionalities represent the versatility of protein sequences and expand the potential of eECM biomaterials for a plethora of tissue engineering and biotechnology applications.
4.1. Linkage with inorganic materials
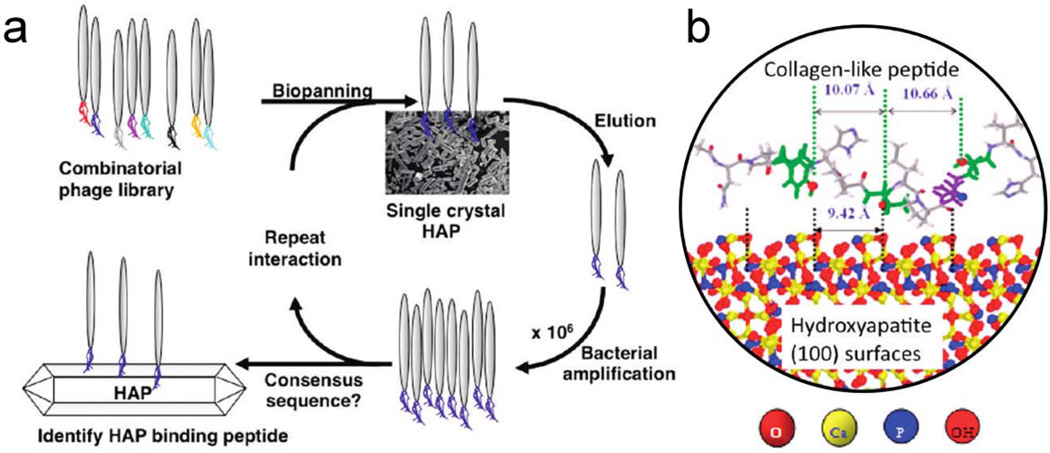
Many peptides are endowed with the capability to specifically bind to or control the synthesis of diverse inorganic materials [84]. These inorganic-binding peptide modules offer new possibilities to create composite eECMs that include both organic and inorganic components. The biogenesis of hybrid inorganic/organic composites, such as bone, often includes the process of templated mineralization [42]. Thus, biomimetic synthesis of bone-like structures requires novel organic scaffolds that mediate the mineralization of hydroxyapatite (HAP), the major inorganic component of bone. Using phage display, a 12-residue peptide has been identified that binds to single-crystal HAP. This peptide, mimicking the tripeptide repeat of type I collagen, was found to template the nucleation and growth of crystalline HAP [42] (Figure 4). Similarly, a silk/silica fusion protein was designed by fusing the self-assembling domains of spider dragline silk with the silaffin-derived R5 peptide that initiates silica mineralization [43–45]. This engineered silk protein served as the template for the formation of 3D, composite, porous networks with tunable silica morphologies and distributions. hMSCs were found to attach, proliferate, and differentiate toward osteogenic lineages with early bone formation on these composite structures [44]. Besides bone-related applications, cell-material interactions have been probed using a series of peptides bound to gold, platinum, glass, and titanium with RGD sequences. These inorganic-binding peptides control the adhesion and spreading of fibroblast cells through the immobilization of RGD ligands on solid surfaces [85]. In a highly creative application, the rational design of thermostable vaccines was achieved by engineering biomimetic nucleating peptides to induce virus self-biomineralization under physiological conditions. The engineered, self-biomineralized vaccine was found to have better thermostability for long-term storage at ambient temperature [86].
Figure 4.
Design of short hydroxyapatite (HAP)-binding peptides. (a) Schematic diagram of the phage display process for single-crystalline HAP whiskers. (b) A model of the proposed interaction of the collagen-like peptide CLP12 with the single-crystalline HAP surface, showing closely matched distances between adjacent hydroxyl residues and the HAP crystal lattice. Adapted from reference [42], copyright 2011, reprinted with permission from American Chemical Society.
4.2. Antimicrobial activity
Multidrug-resistant bacteria are a severe threat to public health. Conventional antibiotics are becoming increasingly ineffective as a result of evolving drug resistance, and therefore it is imperative to find new antibacterial strategies. Natural antimicrobial peptides (AMPs) are innate immune system effectors to defend host organisms against microbes, but most of them have relatively modest antibiotic activity. Enhanced variants have been developed using rational design, optimization strategies, and computer-assisted design strategies [87]. Induced amphipathic α-helical conformations were found to play an important role in the antimicrobial activity of these peptides [88]. Additionally, most of the AMPs tend to form amyloid-like structures to destabilize phospholipid bilayers, suggesting that these aggregation-prone structures may have served as templates from which AMPs were evolutionarily derived [89]. Based on these structural guidelines and computer-assisted technologies that relate primary sequence to peptide structure, more potent, cost-effective, broad-spectrum peptides are being identified as potential next-generation, anti-infective peptides [87]. Some AMPs have already been included as eECM modules to create multifunctional biomaterials for medical use. In one study, three new fusion proteins were designed, cloned, and evaluated for function by fusing the sequence of dragline spider silk with three different antimicrobial peptides. These engineered proteins demonstrated compatibility with mammalian cells while offering resistance to Gram negative Escherichia coli and Gram positive Staphylococcus aureus [90].
4.3. Immune modulatory peptides
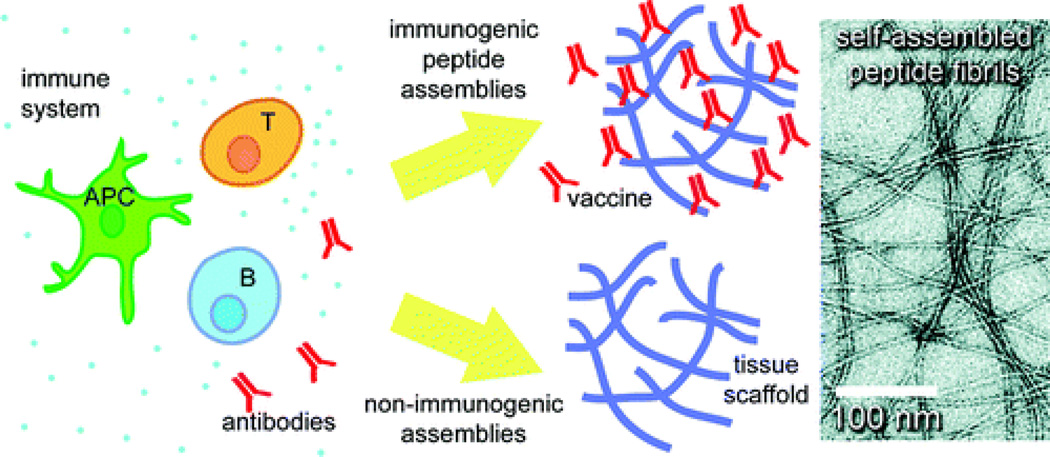
Understanding the immune responses against eECMs is of significant importance in their ultimate clinical use. Peptides are generally poor immunogens and typically require coadministration with adjuvants to elicit antibody responses; however, their immunogenicity can enhance significantly upon assembly and multimerization of supramolecular stuctures [91]. Self-assembling peptides have shown significant variability in immunogenicity, with many peptides inducing minimal antibody responses, but others inducing strong responses in the absence of any supplemental adjuvants [91–94]. Collier et al. have exploited these interactions by designing self-assembling peptides for use as chemically defined immune adjuvants [92]. Peptide epitopes, assembled into nanofibers via a short synthetic fibrillization domain, elicited high antibody titers without any adjuvant. This strategy represents a simple, chemically defined method to dramatically elevate antibody responses to peptide epitopes [93]. These same ideas can also be used to create eECMs with minimal immunogenicity. For example, the self-assembling peptide, OVA-Q11, which elicits a strong, T cell-dependent antibody response in mice, was modified by deleting the amino acid regions in the peptide that are recognized by T cells, thereby greatly diminishing immune responses [94] (Figure 5). Taken together, this work demonstrates that eECMs can be modulated either to raise a strong antibody response, and hence find potential use in immunotherapies, or to avoid such a response, and hence find use as scaffolds for regenerative medicine applications.
Figure 5.
Schematic of a self-assembling peptide system that can be modulated to either raise a strong antibody response or to avoid such a response. The immune system (left) consists of antigen-presenting cells (APC), T cells, and B cells. Peptide assemblies containing the OVA323–339 antigen (top middle) elicit strong antibody responses, while assemblies without the antigen (bottom middle) elicit no significant responses. The engineered peptide system underwent self-assembly to form nanofibrils (right). Adapted from reference [94], copyright 2012, reprinted with permission from American Chemical Society.
4.4 Dynamically responsive peptides
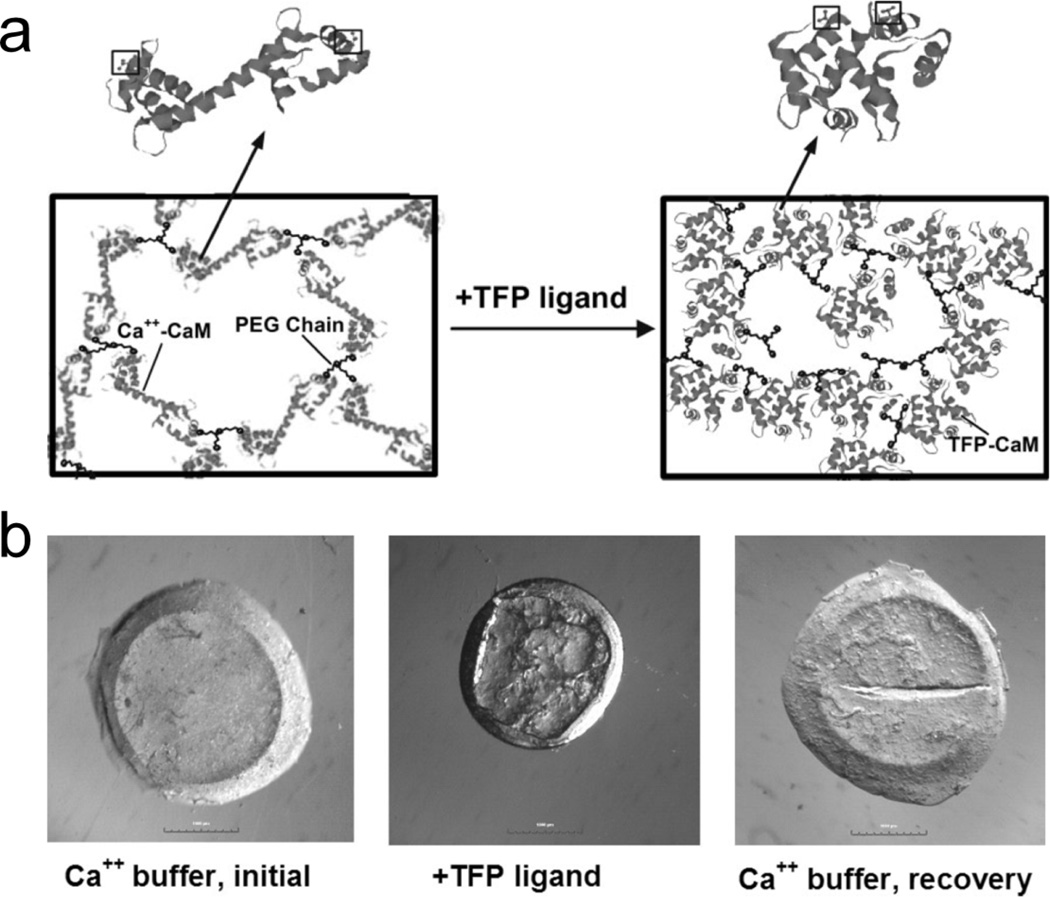
Hydrogels that harness protein motion to achieve dynamic responsiveness have great potential in biotechnology applications. Through careful design, protein motion at the molecular level can be translated into macroscopic changes in hydrogel properties. For example, calmodulin, a hinge motion protein, has been designed into hydrogels that collapse upon binding of calcium ions or other specific biochemical ligands, leading to significant decreases in hydrogel volume [95]. In one example, a mutant version of calmodulin was used as a crosslinker to form a poly(ethylene glycol)-based hydrogel network [46]. In another design, poly(ethylene glycol)-calmodulin conjugates were photocrosslinked to form dynamic hydrogels that could undergo tunable volume change (up to 80%) based on the gelation conditions [47] (Figure 6).
Figure 6.
A dynamic, protein-based hydrogel using a hinge-motion protein, calmodulin (CaM). (a) Schematic representation of hydrogel network structure with CaM in the extended (left) and collapsed (right) conformations. Cysteine residues engineered into the protein are delineated by boxes. (b) CaM-based hydrogels undergo substantial volume changes as a result of trifluoperazine (TFP) ligand binding. Photomicrographs showing CaM-based hydrogels with CaM in extended conformation (left) and collapsed conformation (middle). The volume decrease was recovered when gels were returned to an environment favoring the extended CaM conformation (right). Adapted from reference [47], copyright 2007, reprinted with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
In addition to calmodulin, catalytic enzymes have been used as modules in the design of dynamic biomaterials. One study employed a protein that underwent a nanoscale conformational change upon binding to a substrate, which was translated into macroscale mechanical motion and control of hydrogel swelling [48]. As another example, a glucose-responsive hydrogel was fabricated by conjugating a glucose/galactose-binding protein within an acrylamide hydrogel network, resulting in a quantitative, accordion-like dynamic response upon addition of glucose [26]. Dynamic hydrogels can also be generated by metallothionein conformational changes that respond to heavy metal ions. Upon binding of various toxic heavy metal ions, such as mercury and cadmium, metallothionein undergoes collapse from an extended coil to a more compact, globular morphology. When incorporated into poly(acrylamide) hydrogels, metallothionein’s conformational shift was translated into an up to 80% decrease in hydrogel swelling [96].
Photo-switchable self-assembly is another emerging strategy in the fabrication of smart, functional eECMs. Reversible optical control of protein structure and function offers the possibility of probing and manipulating complex cell microenvironments [97]. Photocontrollable peptides that are reversible are often based on α helix-coil transitions. For example, a simple light-activated system employed a peptide designed to self-assemble into hydrogels depending on its intramolecular folded conformational state [98]. This system utilized a photocaged peptide that remains unfolded in aqueous medium and a freely, soluble unfolded peptide that is stable to ambient light. Irradiation of the solution released the photocage and triggered peptide folding to produce amphiphilic α-hairpins that self-assembled into viscoelastic hydrogels. Fibroblasts seeding indicated that the gel surface supported cell adhesion, proliferation, and migration [98]. Azobenzene is another widely used, light-responsive molecule. In one example, an azobenzene-linked, symmetrical gemini α-helical peptide was designed to enable light-switched, self-assembly due to reversible, molecular transitions between Z- and U-structures. The self-assembled morphology was observed to reversibly change between nanofibers and nanospheres in acidic medium, and between nanospheres and vesicles in basic medium [99].
5. Conclusions
In summary, protein eECMs have been designed with molecular precision to fabricate 3D biomaterial scaffolds that mimic many of the features of native ECMs. With the advancement of recombinant protein engineering technology and various crosslinking mechanisms, eECMs have been designed with independently controlled functional modules that encompass fibril structural domains, highly tunable mechanical compliance, cell-instructive biochemical functionalities, and tailored cell-responsive degradation profiles. The repertoire of peptide modules that can be successfully designed into protein-based eECMs is fast developing, which enables the virtually limitless design of multifunctional materials. Furthermore, additional functionality using synthetic organic materials or inorganic components can be combined and sequestered in the eECM matrix with ease. This emerging class of biomaterials offers a versatile platform with decoupled material parameters for the development of physiologically relevant in vitro cultures. These scaffolds are well suited for fundamental biological studies of cell-matrix interactions as well as for translational applications in clinical therapies.
Acknowledgements
The authors acknowledge support from NSF (DMR-0846363), NIH (R01-DK085720, DP2-OD006477, R21-AR062359) and the California Institute for Regenerative Medicine (RT2-01938).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature Biotechnology. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Barker TH. The role of ECM proteins and protein fragments in guiding cell behavior in regenerative medicine. Biomaterials. 2011;32:4211–4214. doi: 10.1016/j.biomaterials.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Collier JH, Segura T. Evolving the use of peptides as components of biomaterials. Biomaterials. 2011;32:4198–4204. doi: 10.1016/j.biomaterials.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maskarinec SA, Tirrell DA. Protein engineering approaches to biomaterials design. Curr Opin Biotech. 2005;16:422–426. doi: 10.1016/j.copbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Romano NH, Sengupta D, Chung C, Heilshorn SC. Protein-engineered biomaterials: Nanoscale mimics of the extracellular matrix. Bba-Gen Subjects. 2011;1810:339–349. doi: 10.1016/j.bbagen.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sengupta D, Heilshorn SC. Protein-Engineered Biomaterials: Highly Tunable Tissue Engineering Scaffolds. Tissue Eng Part B-Re. 2010;16:285–293. doi: 10.1089/ten.teb.2009.0591. [DOI] [PubMed] [Google Scholar]
- 8.Kiick KL. Biosynthetic methods for the production of advanced protein-based materials. Polym Rev. 2007;47:1–7. [Google Scholar]
- 9.DiMarco RL, Heilshorn SC. Multifunctional Materials through Modular Protein Engineering. Advanced Materials. 2012;24:3923–3940. doi: 10.1002/adma.201200051. [DOI] [PubMed] [Google Scholar]
- 10.Liu JC, Tirrell DA. Cell Response to RGD Density in Cross-Linked Artificial Extracellular Matrix Protein Films. Biomacromolecules. 2008;9:2984–2988. doi: 10.1021/bm800469j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straley KS, Heilshorn SC. Independent tuning of multiple biomaterial properties using protein engineering. Soft Matter. 2009;5:114–124. [Google Scholar]
- 12.Lampe KJ, Antaris AL, Heilshorn SC. Design of three-dimensional engineered protein hydrogels for tailored control of neurite growth. Acta Biomaterialia. 2013;9:5590–5599. doi: 10.1016/j.actbio.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta D, Gilbert PM, Johnson KJ, Blau HM, Heilshorn SC. Protein-Engineered Biomaterials to Generate Human Skeletal Muscle Mimics. Advanced Healthcare Materials. 2012;1:785–789. doi: 10.1002/adhm.201200195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung C, Lampe KJ, Heilshorn SC. Tetrakis(hydroxymethyl) Phosphonium Chloride as a Covalent Cross-Linking Agent for Cell Encapsulation within Protein-Based Hydrogels. Biomacromolecules. 2012;13:3912–3916. doi: 10.1021/bm3015279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werkmeister JA, Ramshaw JAM. Recombinant protein scaffolds for tissue engineering. Biomed Mater. 2012;7 doi: 10.1088/1748-6041/7/1/012002. [DOI] [PubMed] [Google Scholar]
- 16.Benitez PL, Sweet JA, Fink H, Chennazhi KP, Nair SV, Enejder A, et al. Sequence-Specific Crosslinking of Electrospun, Elastin-Like Protein Preserves Bioactivity and Native-Like Mechanics. Advanced Healthcare Materials. 2013;2:114–118. doi: 10.1002/adhm.201200115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes S, Leonor IB, Mano JF, Reis RL, Kaplan DL. Natural and genetically engineered proteins for tissue engineering. Prog Polym Sci. 2012;37:1–17. doi: 10.1016/j.progpolymsci.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen W, Lammertink RGH, Sakata JK, Kornfield JA, Tirrell DA. Assembly of an artificial protein hydrogel through leucine zipper aggregation and disulfide bond formation. Macromolecules. 2005;38:3909–3916. [Google Scholar]
- 19.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Reversible hydrogels from self-assembling artificial proteins. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 20.Mi L, Fischer S, Chung B, Sundelacruz S, Harden JL. Self-Assembling Protein Hydrogels with Modular Integrin Binding Domains. Biomacromolecules. 2005;7:38–47. doi: 10.1021/bm050157p. [DOI] [PubMed] [Google Scholar]
- 21.Davis NE, Ding S, Forster RE, Pinkas DM, Barron AE. Modular enzymatically crosslinked protein polymer hydrogels for in situ gelation. Biomaterials. 2010;31:7288–7297. doi: 10.1016/j.biomaterials.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toledano S, Williams RJ, Jayawarna V, Ulijn RV. Enzyme-Triggered Self-Assembly of Peptide Hydrogels via Reversed Hydrolysis. Journal of the American Chemical Society. 2006;128:1070–1071. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]
- 23.Nettles DL, Kitaoka K, Hanson NA, Flahiff CM, Mata BA, Hsu EW, et al. In situ crosslinking elastin-like polypeptide gels for application to articular cartilage repair in a goat osteochondral defect model. Tissue Eng Pt A. 2008;14:1133–1140. doi: 10.1089/ten.tea.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foo CTSWP, Lee JS, Mulyasasmita W, Parisi-Amon A, Heilshorn SC. Two-component proteinengineered physical hydrogels for cell encapsulation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22067–2272. doi: 10.1073/pnas.0904851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan W, Yang J, Kopeckova P, Kopecek J. Smart Hydrogels Containing Adenylate Kinase: Translating Substrate Recognition into Macroscopic Motion. Journal of the American Chemical Society. 2008;130:15760-+. doi: 10.1021/ja805634x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrick JD, Luckett MR, Khatwani S, Wei Y, Deo SK, Bachas LG, et al. Glucose Responsive Hydrogel Networks Based on Protein Recognition. Macromolecular Bioscience. 2009;9:864–868. doi: 10.1002/mabi.200800337. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Chu X, Wang L, Wang H, Liang G, Zhang J, et al. Rational Design of a Tetrameric Protein to Enhance Interactions between Self-Assembled Fibers Gives Molecular Hydrogels. Angewandte Chemie International Edition. 2012;51:4388–4392. doi: 10.1002/anie.201108612. [DOI] [PubMed] [Google Scholar]
- 28.Heilshorn SC, Liu JC, Tirrell DA. Cell-binding domain context affects cell behavior on engineered proteins. Biomacromolecules. 2005;6:318–323. doi: 10.1021/bm049627q. [DOI] [PubMed] [Google Scholar]
- 29.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5:497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura M, Mie M, Mihara H, Nakamura M, Kobatake E. Construction of a multi-functional extracellular matrix protein that increases number of N1E-115 neuroblast cells having neurites. Journal of biomedical materials research Part B, Applied biomaterials. 2009;91:425–432. doi: 10.1002/jbm.b.31418. [DOI] [PubMed] [Google Scholar]
- 31.Elloumi I, Kobayashi R, Funabashi H, Mie M, Kobatake E. Construction of epidermal growth factor fusion protein with cell adhesive activity. Biomaterials. 2006;27:3451–3458. doi: 10.1016/j.biomaterials.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Jung JP, Moyano JV, Collier JH. Multifactorial optimization of endothelial cell growth using modular synthetic extracellular matrices. Integrative biology : quantitative biosciences from nano to macro. 2011;3:185–196. doi: 10.1039/c0ib00112k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang J-H, Chung C-P. Engineering and expression of a recombinant fusion protein possessing fibroblast growth factor-2 and fibronectin fragment. Biotechnol Lett. 2004;26:1837–1840. doi: 10.1007/s10529-004-5278-1. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi N, Zhang L, Chae BS, Palla CS, Furst EM, Kiick KL. Growth factor mediated assembly of cell receptor-responsive hydrogels. Journal of the American Chemical Society. 2007;129:3040-+. doi: 10.1021/ja0680358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrades JA, Wu LT, Hall FL, Nimni ME, Becerra J. Engineering, expression, and renaturation of a collagen-targeted human bFGF fusion protein. Growth Factors. 2001;18:261–275. doi: 10.3109/08977190109029115. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Lin H, Zhao Y, Wang B, Zhao Y, Liu Y, et al. Activation of demineralized bone matrix by genetically engineered human bone morphogenetic protein-2 with a collagen binding domain derived from von Willebrand factor propolypeptide. Journal of Biomedical Materials Research - Part A. 2007;80:428–434. doi: 10.1002/jbm.a.30900. [DOI] [PubMed] [Google Scholar]
- 37.Chen B, Lin H, Wang J, Zhao Y, Wang B, Zhao W, et al. Homogeneous osteogenesis and bone regeneration by demineralized bone matrix loading with collagen-targeting bone morphogenetic protein-2. Biomaterials. 2007;28:1027–1035. doi: 10.1016/j.biomaterials.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Lin H, Chen B, Sun W, Zhao W, Zhao Y, Dai J. The effect of collagen-targeting platelet-derived growth factor on cellularization and vascularization of collagen scaffolds. Biomaterials. 2006;27:5708–5714. doi: 10.1016/j.biomaterials.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Tong Z, Jia X, Kiick KL. Resilin-like polypeptide hydrogels engineered for versatile biological function. Soft Matter. 2013;9:665–673. doi: 10.1039/C2SM26812D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straley KS, Heilshorn SC. Dynamic, 3D-Pattern Formation Within Enzyme-Responsive Hydrogels. Advanced Materials. 2009;21:4148-+. [Google Scholar]
- 41.Gustafson JA, Price RA, Frandsen J, Henak CR, Cappello J, Ghandehari H. Synthesis and characterization of a matrix-metalloproteinase responsive silk-elastinlike protein polymer. Biomacromolecules. 2013;14:618–625. doi: 10.1021/bm3013692. [DOI] [PubMed] [Google Scholar]
- 42.Chung WJ, Kwon KY, Song J, Lee SW. Evolutionary screening of collagen-like peptides that nucleate hydroxyapatite crystals. Langmuir. 2011;27:7620–7628. doi: 10.1021/la104757g. [DOI] [PubMed] [Google Scholar]
- 43.Foo CWP, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, et al. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9428–9433. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mieszawska AJ, Nadkarni LD, Perry CC, Kaplan DL. Nanoscale Control of Silica Particle Formation via Silk-Silica Fusion Proteins for Bone Regeneration. Chemistry of Materials. 2010;22:5780–5785. doi: 10.1021/cm101940u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J, Wong C, George A, Kaplan DL. The effect of genetically engineered spider silk-dentin matrix protein 1 chimeric protein on hydroxyapatite nucleation. Biomaterials. 2007;28:2358–2367. doi: 10.1016/j.biomaterials.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 46.Murphy WL, Dillmore WS, Modica J, Mrksich M. Dynamic hydrogels: translating a protein conformational change into macroscopic motion. Angewandte Chemie. 2007;46:3066–3069. doi: 10.1002/anie.200604808. [DOI] [PubMed] [Google Scholar]
- 47.Sui Z, King WJ, Murphy WL. Dynamic materials based on a protein conformational change. Advanced Materials. 2007;19:3377-+. [Google Scholar]
- 48.Yuan W, Yang J, Kopeckova P, Kopecek J. Smart hydrogels containing adenylate kinase: translating substrate recognition into macroscopic motion. J Am Chem Soc. 2008;130:15760–15761. doi: 10.1021/ja805634x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amblard M, Fehrentz JA, Martinez J, Subra G. Methods and protocols of modern solid phase Peptide synthesis. Molecular biotechnology. 2006;33:239–254. doi: 10.1385/MB:33:3:239. [DOI] [PubMed] [Google Scholar]
- 50.Raibaut L, Ollivier N, Melnyk O. Sequential native peptide ligation strategies for total chemical protein synthesis. Chem Soc Rev. 2012;41:7001–7015. doi: 10.1039/c2cs35147a. [DOI] [PubMed] [Google Scholar]
- 51.Bayer E, Mutter M. LIQUID-PHASE SYNTHESIS OF PEPTIDES. Nature. 1972;237:512-&. doi: 10.1038/237512a0. [DOI] [PubMed] [Google Scholar]
- 52.Megeed Z, Cappello J, Ghandehari H. Genetically engineered silk-elastinlike protein polymers for controlled drug delivery. Adv Drug Deliver Rev. 2002;54:1075–1091. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 53.Nettles DL, Chilkoti A, Setton LA. Applications of elastin-like polypeptides in tissue engineering. Adv Drug Deliver Rev. 2010;62:1479–1485. doi: 10.1016/j.addr.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, et al. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 55.Urry DW, Luan CH, Parker TM, Gowda DC, Prasad KU, Reid MC, et al. TEMPERATURE OF POLYPEPTIDE INVERSE TEMPERATURE TRANSITION DEPENDS ON MEAN RESIDUE HYDROPHOBICITY. Journal of the American Chemical Society. 1991;113:4346–4348. [Google Scholar]
- 56.Macias MJ, Gervais V, Civera C, Oschkinat H. Structural analysis of WW domains and design of a WW prototype. Nature Structural Biology. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 57.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 58.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman AP, Levin AM, Lahti JL, Cochran JR. Engineered Cystine-Knot Peptides that Bind αvβ3 Integrin with Antibody-Like Affinities. Journal of Molecular Biology. 2009;385:1064–1075. doi: 10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sidhu SS, Bader GD, Boone C. Functional genomics of intracellular peptide recognition domains with combinatorial biology methods. Current opinion in chemical biology. 2003;7:97–102. doi: 10.1016/s1367-5931(02)00011-x. [DOI] [PubMed] [Google Scholar]
- 62.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 63.Sallach RE, Cui W, Wen J, Martinez A, Conticello VP, Chaikof EL. Elastin-mimetic protein polymers capable of physical and chemical crosslinking. Biomaterials. 2009;30:409–422. doi: 10.1016/j.biomaterials.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 65.Olsen BD, Kornfield JA, Tirrell DA. Yielding Behavior in Injectable Hydrogels from Telechelic Proteins. Macromolecules. 2010;43:9094–9099. doi: 10.1021/ma101434a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fischer SE, Liu X, Mao H-Q, Harden JL. Controlling cell adhesion to surfaces via associating bioactive triblock proteins. Biomaterials. 2007;28:3325–3337. doi: 10.1016/j.biomaterials.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 67.Mulyasasmita W, Lee JS, Heilshorn SC. Molecular-level engineering of protein physical hydrogels for predictive sol-gel phase behavior. Biomacromolecules. 2011;12:3406–3411. doi: 10.1021/bm200959e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parisi-Amon A, Mulyasasmita W, Chung C, Heilshorn SC. Protein-engineered injectable hydrogel to improve retention of transplanted adipose-derived stem cells. Adv Healthc Mater. 2013;2:428–432. doi: 10.1002/adhm.201200293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ito F, Usui K, Kawahara D, Suenaga A, Maki T, Kidoaki S, et al. Reversible hydrogel formation driven by protein–peptide–specific interaction and chondrocyte entrapment. Biomaterials. 2010;31:58–66. doi: 10.1016/j.biomaterials.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 70.Grove TZ, Osuji CO, Forster JD, Dufresne ER, Regan L. Stimuli-Responsive Smart Gels Realized via Modular Protein Design. Journal of the American Chemical Society. 2010;132:14024–14026. doi: 10.1021/ja106619w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu HD, Soranno DE, Rodell CB, Kim IL, Burdick JA. Secondary Photocrosslinking of Injectable Shear-Thinning Dock-and-Lock Hydrogels. Advanced Healthcare Materials. 2013;2:1028–1036. doi: 10.1002/adhm.201200343. [DOI] [PubMed] [Google Scholar]
- 72.Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 73.McHale MK, Setton LA, Chilkoti A. Synthesis and in vitro evaluation of enzymatically crosslinked elastin-like polypeptide gels for cartilaginous tissue repair. Tissue engineering. 2005;11:1768–1779. doi: 10.1089/ten.2005.11.1768. [DOI] [PubMed] [Google Scholar]
- 74.Trabbic-Carlson K, Setton LA, Chilkoti A. Swelling and mechanical behaviors of chemically cross-linked hydrogels of elastin-like polypeptides. Biomacromolecules. 2003;4:572–580. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- 75.Renner JN, Cherry KM, Su RSC, Liu JC. Characterization of Resilin-Based Materials for Tissue Engineering Applications. Biomacromolecules. 2012;13:3678–3685. doi: 10.1021/bm301129b. [DOI] [PubMed] [Google Scholar]
- 76.Li L, Teller S, Clifton RJ, Jia X, Kiick KL. Tunable Mechanical Stability and Deformation Response of a Resilin-Based Elastomer. Biomacromolecules. 2011;12:2302–2310. doi: 10.1021/bm200373p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moore SJ, Hayden Gephart MG, Bergen JM, Su YS, Rayburn H, Scott MP, et al. Engineered knottin peptide enables noninvasive optical imaging of intracranial medulloblastoma. Proceedings of the National Academy of Sciences. 2013;110:14598–14603. doi: 10.1073/pnas.1311333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kambe Y, Yamamoto K, Kojima K, Tamada Y, Tomita N. Effects of RGDS sequence genetically interfused in the silk fibroin light chain protein on chondrocyte adhesion and cartilage synthesis. Biomaterials. 2010;31:7503–7511. doi: 10.1016/j.biomaterials.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 79.Wang X, Riesberg JJ, Shen W. Reversible regulation of bioactive ligands presented on immobilized gold nanoparticles. Soft Matter. 2012;8:2812–2815. [Google Scholar]
- 80.Yue XS, Murakami Y, Tamai T, Nagaoka M, Cho CS, Ito Y, et al. A fusion protein N-cadherin-Fc as an artificial extracellular matrix surface for maintenance of stem cell features. Biomaterials. 2010;31:5287–5296. doi: 10.1016/j.biomaterials.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 81.Lv S, Bu T, Kayser J, Bausch A, Li H. Towards constructing extracellular matrix-mimetic hydrogels: An elastic hydrogel constructed from tandem modular proteins containing tenascin FnIII domains. Acta Biomaterialia. 2013;9:6481–6491. doi: 10.1016/j.actbio.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 82.Su RSC, Renner JN, Liu JC. Synthesis and Characterization of Recombinant Abductin-Based Proteins. Biomacromolecules. 2013;14:4301–4308. doi: 10.1021/bm401162g. [DOI] [PubMed] [Google Scholar]
- 83.Charati MB, Ifkovits JL, Burdick JA, Linhardt JG, Kiick KL. Hydrophilic elastomeric biomaterials based on resilin-like polypeptides. Soft Matter. 2009;5:3412–3416. doi: 10.1039/b910980c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dickerson MB, Sandhage KH, Naik RR. Protein- and peptide-directed syntheses of inorganic materials. Chemical reviews. 2008;108:4935–4978. doi: 10.1021/cr8002328. [DOI] [PubMed] [Google Scholar]
- 85.Khatayevich D, Gungormus M, Yazici H, So C, Cetinel S, Ma H, et al. Biofunctionalization of materials for implants using engineered peptides. Acta Biomaterialia. 2010;6:4634–4641. doi: 10.1016/j.actbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 86.Wang GC, Cao RY, Chen R, Mo LJ, Han JF, Wang XY, et al. Rational design of thermostable vaccines by engineered peptide-induced virus self-biomineralization under physiological conditions. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7619–7624. doi: 10.1073/pnas.1300233110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: form follows function. Nature Reviews Drug Discovery. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 88.Blondelle SE, Houghten RA. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry. 1992;31:12688–12694. doi: 10.1021/bi00165a020. [DOI] [PubMed] [Google Scholar]
- 89.Torrent M, Valle J, Nogués MV, Boix E, Andreu D. The Generation of Antimicrobial Peptide Activity: A Trade-off between Charge and Aggregation? Angewandte Chemie International Edition. 2011;50:10686–10689. doi: 10.1002/anie.201103589. [DOI] [PubMed] [Google Scholar]
- 90.Gomes SC, Leonor IB, Mano JF, Reis RL, Kaplan DL. Antimicrobial functionalized genetically engineered spider silk. Biomaterials. 2011;32:4255–4266. doi: 10.1016/j.biomaterials.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Multi-component extracellular matrices based on peptide self-assembly. Chem Soc Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rudra JS, Tripathi PK, Hildeman DA, Jung JP, Collier JH. Immune responses to coiled coil supramolecular biomaterials. Biomaterials. 2010;31:8475–8483. doi: 10.1016/j.biomaterials.2010.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rudra JS, Tian YF, Jung JP, Collier JH. A self-assembling peptide acting as an immune adjuvant. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:622–627. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rudra JS, Sun T, Bird KC, Daniels MD, Gasiorowski JZ, Chong AS, et al. Modulating Adaptive Immune Responses to Peptide Self-Assemblies. Acs Nano. 2012;6:1557–1564. doi: 10.1021/nn204530r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Topp S, Prasad V, Cianci GC, Weeks ER, Gallivan JP. A Genetic Toolbox for Creating Reversible Ca2+-Sensitive Materials. Journal of the American Chemical Society. 2006;128:13994–13995. doi: 10.1021/ja064546i. [DOI] [PubMed] [Google Scholar]
- 96.Esser-Kahn AP, Iavarone AT, Francis MB. Metallothionein-Cross-Linked Hydrogels for the Selective Removal of Heavy Metals from Water. Journal of the American Chemical Society. 2008;130:15820-+. doi: 10.1021/ja807095r. [DOI] [PubMed] [Google Scholar]
- 97.Woolley GA. Photocontrolling Peptide α Helices. Accounts of Chemical Research. 2005;38:486–493. doi: 10.1021/ar040091v. [DOI] [PubMed] [Google Scholar]
- 98.Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP. Light-Activated Hydrogel Formation via the Triggered Folding and Self-Assembly of a Designed Peptide. Journal of the American Chemical Society. 2005;127:17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen C-S, Xu X-D, Li S-Y, Zhuo R-X, Zhang X-Z. Photo-switched self-assembly of a gemini [small alpha]-helical peptide into supramolecular architectures. Nanoscale. 2013;5:6270–6274. doi: 10.1039/c3nr01967e. [DOI] [PubMed] [Google Scholar]
- 100.Parisi-Amon A, Mulyasasmita W, Chung C, Heilshorn SC. Protein-Engineered Injectable Hydrogel to Improve Retention of Transplanted Adipose-Derived Stem Cells. Advanced Healthcare Materials. 2012 doi: 10.1002/adhm.201200293. [DOI] [PMC free article] [PubMed] [Google Scholar]