Abstract
The biocontrol strain Pseudomonas sp. Cab57 was isolated from the rhizosphere of shepherd’s purse growing in a field in Hokkaido by screening the antibiotic producers. The whole genome sequence of this strain was obtained by paired-end and whole-genome shotgun sequencing, and the gaps between the contigs were closed using gap-spanning PCR products. The P. sp. Cab57 genome is organized into a single circular chromosome with 6,827,892 bp, 63.3% G+C content, and 6,186 predicted protein-coding sequences. Based on 16S rRNA gene analysis and whole genome analysis, strain Cab57 was identified as P. protegens. As reported in P. protegens CHA0 and Pf-5, four gene clusters (phl, prn, plt, and hcn) encoding the typical antibiotic metabolites and the reported genes associated with Gac/Rsm signal transduction pathway of these strains are fully conserved in the Cab57 genome. Actually strain Cab57 exhibited typical Gac/Rsm activities and antibiotic production, and these activities were enhanced by knocking out the retS gene (for a sensor kinase acting as an antagonist of GacS). Two large segments (79 and 115 kb) lacking in the Cab57 genome, as compared with the Pf-5 genome, accounted for the majority of the difference (247 kb) between these genomes. One of these segments was the complete rhizoxin analog biosynthesis gene cluster (ca. 79 kb) and another one was the 115-kb mobile genomic island. A whole genome comparison of those relative strains revealed that each strain has unique gene clusters involved in metabolism such as nitrite/nitrate assimilation, which was identified in the Cab57 genome. These findings suggest that P. protegens is a ubiquitous bacterium that controls its biocontrol traits while building up strain-specific genomic repertoires for the biosynthesis of secondary metabolites and niche adaptation.
Introduction
Many plant commensal strains have been classified into the Pseudomonas fluorescens group, which currently includes more than fifty named species [1]. Among the strains in the P. fluorescens group, Pseudomonas protegens strains Pf-5 and CHA0 (previously called P. fluorescens Pf-5 and CHA0, respectively) have been used as model strains in studies on the biosynthesis of several extracellular enzymes such as AprA protease, and secondary metabolites such as 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin (Prn), and pyoluteorin (Plt), with antibiotic activity in the rhizosphere [2]. These exoproducts contribute to plant protection by these strains and other root-colonizing Pseudomonas species with biocontrol activity. P. protegens CHA0 was isolated from the roots of tobacco in Swiss soil which was naturally suppressive to black root rot in tobacco caused by Thielaviopsis basicola [3]. The fully sequenced strain Pf-5, which is very closely related to strain CHA0, was isolated from the rhizosphere soil surrounding cotton seedlings in Texas, USA [4], [5]. The complete genome data of strain Pf-5 are available at the Pseudomonas.com website (http://www.pseudomonas.com) and provide detailed annotation data of this strain. The genome of strain Pf-5, which is 7.07 Mbp, is the largest Pseudomonas genome among Pseudomonas spp. (whose genome sizes range from 4.17 Mbp) sequenced to date (http://www.pseudomonas.com). The complete genome data of strain CHA0 have recently been deposited and its size is 6.87 Mbp. Such large genomes should confer an environmental fitness advantage on these strains.
The expression of biocontrol factors depends on the Gac/Rsm signal transduction pathway ([6] and the references cited therein). The Gac/Rsm cascade is initiated by the GacS/GacA two-component system. The GacS sensor kinase is known to be phosphorylated and activates the cognate GacA response regulator by phosphotransfer during slow growth and in the presence of unidentified signal molecules. In P. protegens CHA0, activated GacA promotes the transcription of non-coding small RNAs (sRNAs) termed RsmX, RsmY, and RsmZ. These sRNAs have high affinity for the RNA-binding protein RsmA and also for the RsmA paralogue RsmE in strain CHA0 [7]–[10]. The RsmA and RsmE proteins repress the translation of genes involved in secondary metabolism during trophophase. When RsmX/Y/Z sRNAs are induced in idiophase, they relieve the translational repression of target genes by sequestering the RsmA and RsmE proteins, thereby allowing the synthesis of secondary metabolites [9], [11], [12]. Thus, mutants defective in the Gac/Rsm signal transduction pathway have a reduced ability to produce such biocontrol factors and to suppress plant diseases [11], [13]. Two additional sensors termed RetS and LadS were shown to provide input into the Gac/Rsm pathway in P. fluorescens and Pseudomonas aeruginosa [14], [15]. RetS inhibits and LadS activates the activity of the Gac/Rsm pathway and, in this way, both sensors affect the expression of target genes and biocontrol factors [15].
In aspects of the effective and ecological application of biocontrol agents, screening for plant beneficial pseudomonads has led to advances in biocontrol research. Moreover, the genomic sequence of strain Pf-5 has provided new insights into the molecular basis underlying the pseudomonad-mediated suppression of plant disease. For example, novel natural products including the cyclic lipopeptide orfamide A [16], rhizoxin analogs [17] and the insect toxin FitD [18] have been discovered through genomics-guided approaches. Comparative genome analysis among strains in the P. fluorescens group has also contributed to the understanding of other different strains [19]. P. fluorescens strain A506 is now commercially available in the USA and is sold as BlightBan A506 [20], which is primarily used to suppress bacterial disease fire blight on pears and apples. The complete genome data of strain A506 revealed distinct features that classified this strain into subgroup 3 of the P. fluorescens group, whereas strain Pf-5 was classified into subgroup 1 [19].
Although P. protegens has been isolated from North America, Europe, and Africa [21], [22], little has been reported on the characterization of P. protegens in Asia. In the present study, we aimed to screen biocontrol pseudomonad strains relative to strains Pf-5 and CHA0 from fields in Japan, with expectations that their genome data should contribute to the study of core and accessory genes of the three strains geographically dispersed. We showed that one of the isolates producing DAPG exhibited similar biocontrol potential against Pythium damping-off and root rot in the cucumber to that of strain CHA0. We named this isolate Cab57, which was fully sequenced and identified as P. protegens. Cab57 exhibited typical Gac/Rsm activities such as the expression of Rsm sRNA RsmZ and antibiotic production. We also showed that the genomic comparison and characterization of each strain provides new insights into the identity and diversity of this species.
Results and Discussion
Isolation of DAPG-producing Pseudomonads from Rhizosphere Soil in Japan
Approximately 2,800 fluorescent pseudomonads were obtained from plant roots. Based on PCR analysis of this collection to screen the phlD + strain, 48 isolates were selected as candidates of DAPG-producing isolates and their production activities were confirmed by thin layer chromatography (TLC) (data not shown). Of the 48 strains, five were PCR-positive for all of the three other antibiotic biosynthetic genes (for pyrrolnitrin, pyoluteorin, and hydrogen cyanide) that are typically found in P. protegens. One of the five strains named Cab57, which was isolated from the rhizosphere of shepherd’s purse [Capsella bursa-pastoris (L.) Medik.] growing in a field in Hokkaido, was selected for further analysis.
DAPG production levels by strain Cab57 were determined by HPLC. The concentration of DAPG was 248±69 nmol per optical density at 600 nm (OD600) in the strain grown in nutrient broth supplemented with 1% glucose to OD600 = 4.0 (one OD600 unit corresponds to approximately 1×109 cells/mL). The concentration of DAPG in strain CHA0 under the same growth conditions was 208±62 nmol per OD600, which suggested that strain Cab57 is a prominent producer of DAPG, similar to strain CHA0.
Biocontrol Efficacy of Strain Cab57
To investigate the biocontrol activity of strain Cab57 in a natural habitat, we adopted a cucumber-Pythium ultimum pathosystem, which enabled us to evaluate plant protection efficacy by measuring the root and shoot weights. Strain Cab57 was as effective a biocontrol agent as strain CHA0, with 95% confidence (Table 1). Furthermore, this strain had a similar root colonization capacity to that of CHA0, both in the presence and absence of Pythium (Table 1).
Table 1. The suppression of Pythium damping-off and root rot in the cucumber by Pseudomonas protegens CHA0 and Cab57.
| Bacterial strain added* | Pythium added* | Surviving plants per flask (%)** | Shoot fresh weight per flask (g)** | Root fresh weight per flask (g)** | Colonization by P. protegens (log10 CFU/g of root) *** |
| None | – | 100 a | 0.75 a | 0.48 a | ND |
| CHA0 | – | 100 a | 0.73 a | 0.50 a | 7.43±0.22 |
| Cab57 | – | 100 a | 0.73 a | 0.48 a | 7.40±0.18 |
| None | + | 6 c | 0.18 d | 0.10 c | ND |
| CHA0 | + | 50 b | 0.35 c | 0.24 b | 8.43±0.07 |
| Cab57 | + | 73 b | 0.43 b | 0.30 b | 8.29±0.10 |
*P. protegens strains were added at 107 CFU per g of soil contained within 100-mL flasks (30 g of soil per flask), after planting three 92-h-old, sterile-grown cucumber seedlings per flask. P. ultimum was added as a millet-seed inoculum at 2.5 g/kg of soil before planting. Plants were harvested after 7 days.
**Data represent the averages of 10 replicates (flasks containing three cucumber plants) per treatment without P. ultimum and 16 replicates per treatment with P. ultimum. Means within the same column followed by different letters (a–d) are significantly different (P<0.05) according to Fisher’s protected LSD test.
***The rhizosphere-stable plasmid pME6031 containing a tetracycline-resistant determinant [49] was introduced as a selective marker into the bacterial strains to determine their root colonization capacity in soil. ND, not detected.
The Genome Structure of Cab57 Showed Properties Typical of P. protegens
We conducted whole-genome sequencing to obtain information on strain Cab57. The Cab57 genome is organized into a single circular chromosome with 6,827,892 bp (Fig. S1) and 63.3% G+C content. The genome is predicted to encode 6,186 proteins. The general genomic features of strain Cab57 as well as those of strains CHA0 and Pf-5 are listed in Table 2.
Table 2. General genomic features of P. protegens strains Cab57, CHA0, and Pf-5.
To identify species, the 16S rRNA gene (position 125095.126636) of strain Cab57 was applied to a BLAST search and was found to be 100% identical to that of P. protegens CHA0 and Pf-5. Strain Cab57 was also characterized using API 20 NE, which revealed the same profile as that of strain CHA0 with the code 0156557. Strain Cab57 was deposited in the MAFF Genebank, National Institute of Agrobiological Sciences as MAFF212077.
Recently, the JSpecies program has been commonly used to compare two genomes for species definition [23]. The whole-genome sequences of P. fluorescens group strains are publicly available. Strains of the P. fluorescens group used in the genomic comparison are as follows; P. protegens Pf-5 [5], P. protegens CHA0 (http://www.pseudomonas.com), P. chlororaphis subsp. aureofaciens 30–84 [19], P. fluorescens Pf0-1 [24], P. fluorescens SBW25 [24], and P. fluorescens A506 [19]; covering all the three sub-clades of P. fluorescens group [19]. The average nucleotide identity calculated with BLAST algorithm (ANIb) values, which provides a numerical and stable species boundary [23], confirmed the adscription of strain Cab57 to P. protegens (Table 3); values higher than 96% were found for strains of the same species. Together with ANIb, tetranucleotide frequency correlation coefficients (TETRA) reinforced the objective boundary for species circumscription when TETRA values >0.99 [23]. As shown in Table 3, both strains of P. protegens showed values of >0.999, whereas the other strains showed the values of <0.988. A BLASTp search of the three strains revealed that strain Pf-5 was most abundant in its unique coding sequences (CDSs) and strain Cab57 shared more CDSs with strain CHA0 than with strain Pf-5 (Fig. S2). Of the 234 genes specific to strain CHA0, 64 genes belong to the regions associated with mobile genetic elements as mentioned later in Table 4 and 100 genes encode hypothetical proteins of unknown functions.
Table 3. Whole genome comparison of strain Cab57 with those of the Pseudomonas fluorescens group.
| Strains within the Pseudomonas fluorescens group | ||||||
| CHA0 | Pf-5 | 30–84 | Pf0-1 | SBW25 | A506 | |
| Average Nucleotide Identity to Cab57 (%) | 98.47 | 98.22 | 84.29 | 81.06 | 80.55 | 80.32 |
| TETRA value to Cab57 | 0.99992 | 0.99984 | 0.98795 | 0.9256 | 0.94548 | 0.94512 |
Table 4. Mobile genetic elements in the P. protegens Cab57 genome.
| Type | Gene range | 5′ end | 3′ end | Size (bp) | %GC | Presence of integrase | Similarity to Pf-5 Prophage |
| Prophage I | 1248 to 1284 | 1398706 | 1430358 | 31653 | 62.7 | No | Hybrid of Prophage 01 and 03 |
| Prophage II | 2134 to 2148 | 2346251 | 2359004 | 12754 | 56.6 | Yes | Partly homologous to Prophage 04 and 06 |
| Prophage III | 3451 to 3525 | 3840966 | 3895710 | 54745 | 57.6 | Yes | Homologous to Prophage 06 |
| Prophage IV | 3558 to 3607 | 3923029 | 3966962 | 43934 | 58.1 | Yes | Homologous to Prophage 03 |
| Transposon 1 | 1981 to 1998 | 2176361 | 2198710 | 22350 | 50.3 | No | None |
| Transposon 2 | 3883 to 3893 | 4318849 | 4328514 | 9666 | 55.5 | Yes | None |
Four gene clusters (hcn, plt, prn, and phl) encoding the typical antibiotic metabolites in P. protegens were conserved in the genomic sequence of strain Cab57 (Gene ID 2622–2624, 2829–2845, 3743–3746, and 5898–5905, respectively, in Table S1), which supported the results obtained by screening the five strains that were PCR-positive for all four antibiotic biosynthetic genes described above. In strain CHA0, the production of the major extracellular protease AprA, which functions as a biocontrol factor, was also shown to be controlled by the Gac/Rsm signal transduction pathway [25], [26]. Each of ten genomes of the P. fluorescens group, including P. protegens, has a conserved cluster for this protease [19]. The aprA gene cluster is also conserved in strain Cab57 (Gene ID 3232–3236 in Table S1). The reported genes associated with the Gac/Rsm signal transduction pathway were fully conserved in the Cab57 genome with >99% homology to those of Pf-5 (Table S2).
Other typical gene clusters encoding biocontrol factors were also explored (Table S3). Gene clusters for pyoverdine, whose product has been commonly reported in strain Pf-5 and CHA0, were conserved in the Cab57 genome in four different loci (Gene ID 2934–2935, 4179–4198, 4266–4276, and 4286–4288) as reported in Pf-5 [27]. Gene clusters for Enantio-pyochelin, which has been isolated from strain CHA0 and its gene cluster is fully conserved in Pf-5 [28], and for orfamide A, which has been identified in strain Pf-5 from mining of Pseudomonas genomes [16], were also conserved in the Cab57 genome (Gene ID 3642–3651 and 2162–2164, respectively). Phenazine is an antibiotic produced by some Pseudomonas strains (excluding P. protegens Pf-5 and CHA0). The homology search of the gene cluster over the entire genome suggested that the known pathways for the synthesis of phenazine may not be present in the Cab57 strain.
Rsm sRNA Expression in P. protegens Cab57
To determine whether P. protegens Cab57 conserved the function of the Gac/Rsm cascade, we analyzed the expression of the rsmZ sRNA gene. As DNA sequences of the upstream region of rsmZ are highly conserved between Cab57 and CHA0 strains (four nucleotides differ in a 200-nucleotide sequence upstream of +1 site, with absolute conservation in the upstream activating sequence (UAS), and in −35, −10, and +1 sites), the transcriptional fusion rsmZ-lacZ containing the promoter region of CHA0 was assumed to be applicable to Cab57 by introducing the construct into the strain. As shown in Figure 1, the expression of this fusion developed in a growth-dependent manner, which is consistent with that previously reported for CHA0 [7]. We also speculated as to whether knocking out the retS gene (for a sensor kinase acting as an antagonist of GacS) in strain Cab57 would affect the Gac/Rsm cascade. The retS mutant showed an enhanced level of expression (Fig. 1), as reported in CHA0 [15], confirming the conserved activity of the Gac/Rsm cascade.
Figure 1. Expression of the rsmZ CHA0 and aprA CHA0 genes in P. protegens Cab57.
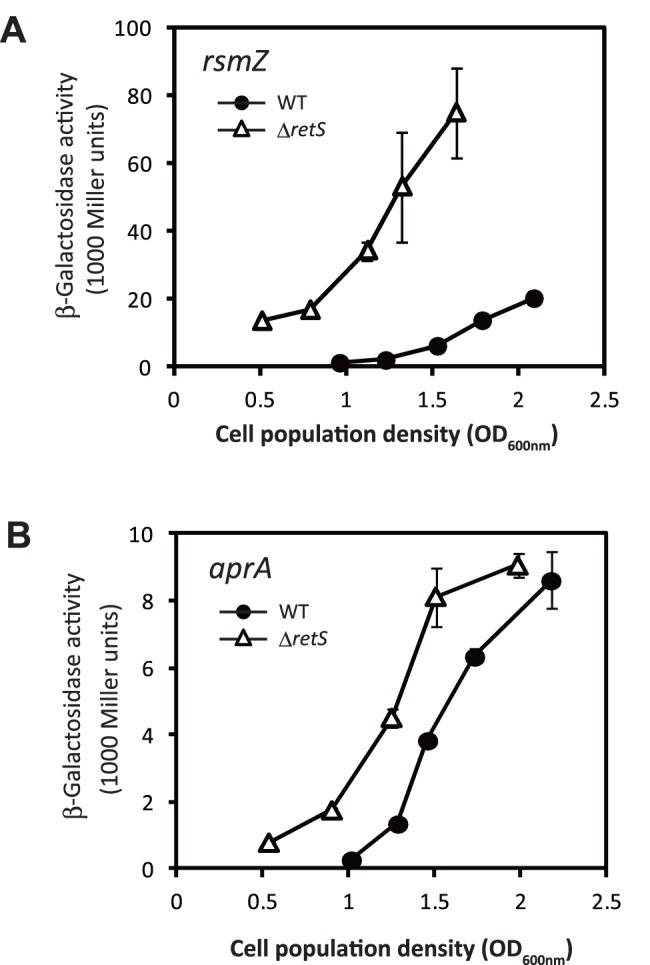
The expression of a rsmZ CHA0 -lacZ fusion carried by pME6091 (A) and aprA CHA0 ’-‘lacZ fusion carried by pME6060 (B) was determined in P. protegens Cab57 wild type (WT, closed circles) and the retS mutant (ΔretS, open triangles). The expression of these hybrid proteins was measured according to LacZ activity. Incubation was performed in Erlenmeyer flasks as described in the Materials and Methods. Measurements were conducted in triplicate. Symbols indicate averages and the error bars indicate standard deviations. OD600 nm, optical density at 600 nm.
A retS Mutant Exhibited Increased Expression of aprA and Antibiotic Activity
The retS mutant had been previously reported to cause increased antibiotic production in P. protegens CHA0 [15]. In CHA0, the Gac/Rsm system has been shown to positively control the translational expression of aprA, the gene for the major exoprotease [29]. As DNA sequences of the upstream region of aprA are known to be highly conserved between Cab57 and CHA0 strains (no nucleotide differs in a 200-nucleotide sequence upstream of the start codon), the translational fusion aprA’-‘lacZ containing the promoter region of CHA0 was assumed to be applicable to Cab57 by introducing the construct into the strain. Therefore, we followed the expression of aprA’-‘lacZ fusion in P. protegens Cab57 and the retS mutant (Fig. 1). The expression of this fusion again developed in a growth-dependent manner in Cab57 and its expression level was enhanced in the retS mutant.
We also investigated whether RetS would affect the antibiotic activity of P. protegens Cab57. In the retS mutant, the production of antibiotics with activity against Bacillus subtilis was higher than that of the wild type (Fig. 2A), as reported in CHA0. We also tested the antibiotic activity of this strain and the retS mutant toward the phytopathogenic oomycete Pythium ultimum on a PDA plate. The growth of P. ultimum was inhibited by the wild type Cab57 strain, and was markedly inhibited by the retS mutant (Fig. 2B). Growth of the phytopathogenic fungus Fusarium oxysporum was also inhibited in a similar manner (Fig. 2C). The pigmentation observed in the retS mutant on PDA plates may be attributed to the overproduction of secondary metabolites.
Figure 2. Effects of the retS mutation on antibiotic activity.
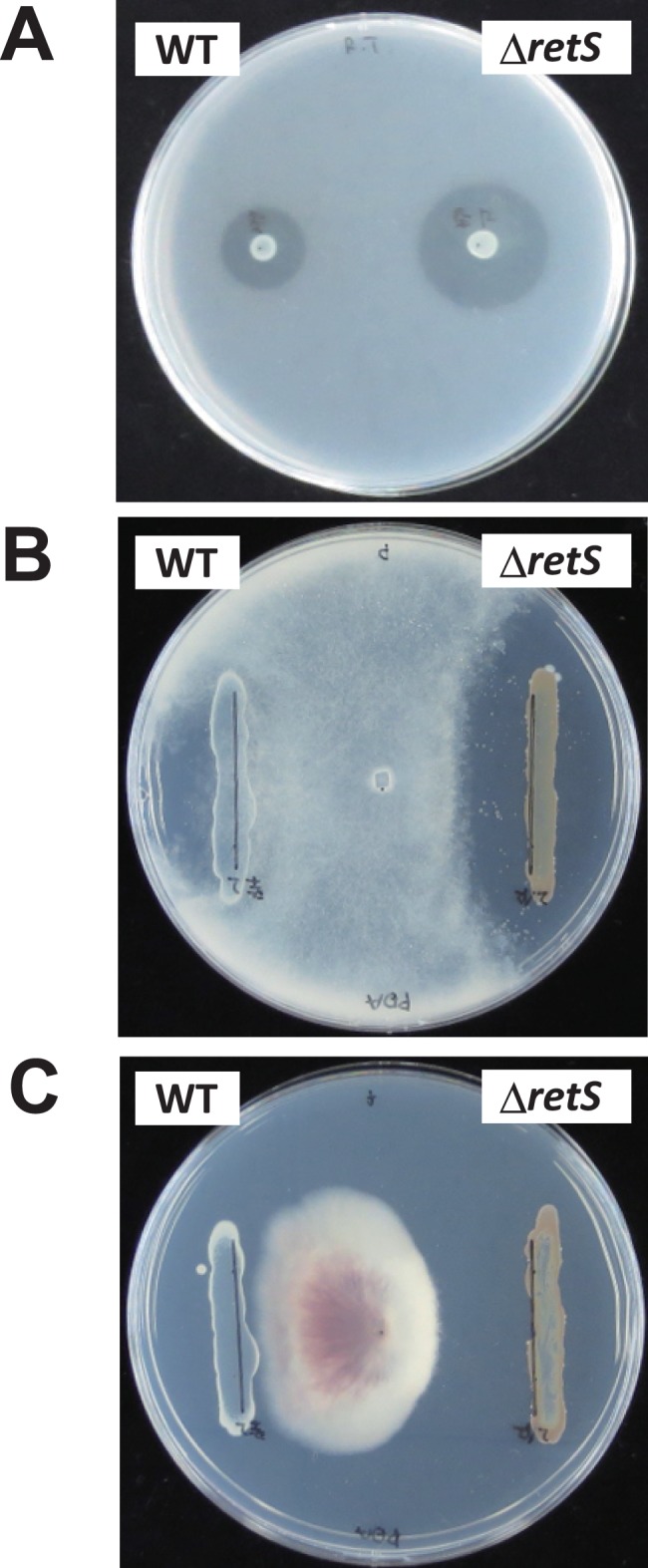
Antibiotic activities of P. protegens strains toward B. subtilis (A), P. ultimum (B) and F. oxysporum (C) were evaluated by the size of the growth inhibition zone. Antibiotic activities of P. protegens Cab57 wild type (WT) and the retS mutant (ΔretS) were compared.
Whole Genome Comparison
Absence of the rhizoxin analogue biosynthesis gene cluster in the Cab57 genome
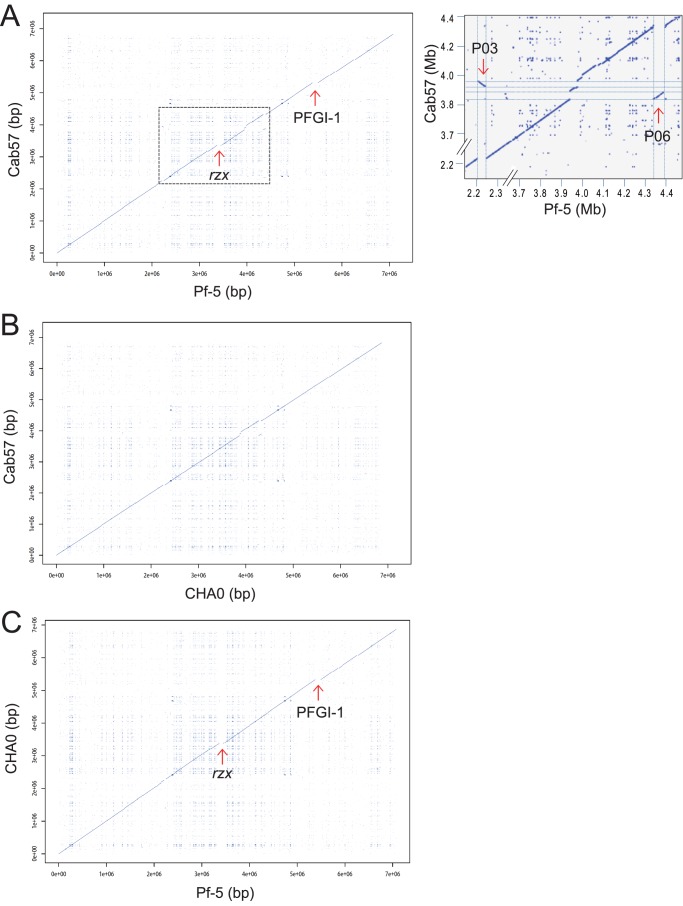
We performed dotplot analysis to obtain an overview of genome comparisons among the strains of P. protegens (Fig. 3). Compared to the Pf-5 genome, whose size is 7,074,893 bp, two large segments were absent in the Cab57 genome (Fig. 3A). These two segments spanning about 195 kb accounted for the majority of the difference (247 kb) between the Cab57 and Pf-5 genomes. These two segments were also absent from the CHA0 genome, the size of which was 6,867,980 (Fig. 3C).
Figure 3. Syntenic dot plot of P. protegens strains.
Dot plot comparison of the Cab57 genome versus the Pf-5 genome (A), the Cab57 genome versus the CHA0 genome (B) and the CHA0 genome versus the Pf-5 genome (C). The blue line represents the regions of similarities between the two genomes while discontinuities in this syntenic line represent regions of genomic variations at a given locus between the two strains. Positions of phage-related elements and genomic island reported in the Pf-5 genome [34] are marked by arrows (P, Prophage; PFGI, Genomic island). The position of the biosynthetic gene cluster for rhizoxin analogs in the Pf-5 genome [17] is also indicated by an arrow (rzx). Regions corresponding to Pf-5 Prophage 03 (P03) and Prophage 06 (P06) in the Cab57 genome are surrounded by a dashed line in panel A and the expanded view is shown in the right.
One of these segments was the complete rzx gene cluster (ca. 79 kb, Fig. S3), which has been reported to encode genes for the biosynthesis of analogues of the antimitotic macrolide rhizoxin in P. protegens Pf-5 [17]. Rhizoxin was firstly isolated from Rhizopus chinensis, a fungus causing a disease in rice seedlings, as its virulence factor [30]. Rhizoxin was then found to be produced by Burkholderia rhizoxinica, the bacterial endosymbiont of the fungus, but not by Rhizopus [31]. The rhizoxin biosynthesis gene cluster (rhi gene cluster) was reported in B. rhizoxinica [32], and its full genome sequencing has revealed that this cluster is encoded on the chromosome but not in the megaplasmid carried by this strain [33]. However, as the rhi gene cluster is flanked by transposase genes, its potential as a mobile region of the genome has been suggested. Furthermore, a trace showed this cluster was originally on one of the megaplasmids of B. rhizoxinica [33]. The high conservation of this cluster between two distant species, P. protegens Pf-5 and B. rhizoxinica, suggests the evolutionary traces of horizontal gene transfer. In the Cab57 genome, the adjacent genes of this cluster (Gene ID 3017 to 3027) were fully conserved with >95% amino acid sequence homologies. Although whether strain Pf-5 acquired this segment, or strain Cab57 discarded this segment after its acquisition remains unclear, such differences in antibiotic production potential indicate diversity among strains.
At the upstream region of this deletion, the fit cluster encoding a functional insect toxin reported in P. protegens CHA0 [18] was conserved between Cab57 and Pf-5 (Fig. S3, Table S3). This cluster is also conserved in the distant species, Photorhabdus luminescens. However, the transmissibility of this cluster has not yet been demonstrated and its evolutionary origin remains unknown.
Absence of mobile genomic islands PFGI-1 and -2 of Pf-5 in the Cab57 genome
A 115-kb mobile genomic island 01 of Pf-5 (PFGI-1, [5]) was revealed as another large segment absent from the Cab57 genome (Fig. 3 and Fig. S4). This island was found to be site-specifically integrated into one of the two tRNA genes flanking both sides of PFGI-1 as attL and attR [5], and its corresponding locus in Cab57 represents the tRNALys gene, which is predicted to be a target for the insertion of this island (Fig. S4). PFGI-1 has elements of both temperate phages and conjugative plasmids showing similarity to the mobile genomic island pKLC102 from P. aeruginosa C [5], [34]. The pil cluster that spans over 10 kb is contained in this island and should encode mating pili rather than type IV pili, and PFGI-1 also carries putative genes involved in conjugal DNA transfer [34]. This pil cluster is shared by the pathogenicity islands of P. aeruginosa and genomic islands of P. syringae. Other notable genes encoded by PFGI-1 such as a cluster of cyo genes, which encode components of the cytochrome o ubiquinol oxidase complex, have also been predicted to contribute to the survival of P. protegens Pf-5; however, their actual function remains unclear [34]. Another genomic island of Pf-5 PFGI-2, which spans 16.8 kb, was also absent from the Cab57 genome. Both of these islands are also absent from the CHA0 genome.
Phage-related elements and transposon
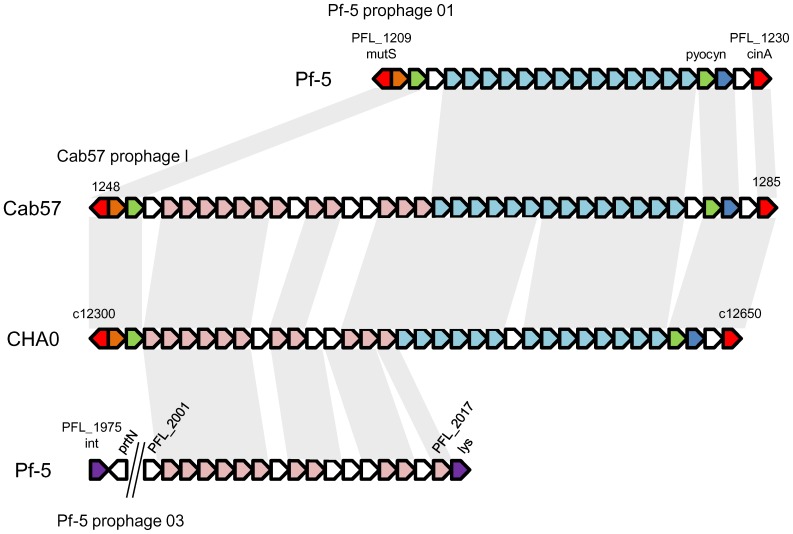
Six prophage regions have been reported as Prophage 01 to 06 in the Pf-5 genome [34]. We identified four prophage regions as Prophage I to IV in the Cab57 genome (Table 4, Fig. 4, Fig. S5, Fig. S6 and Fig. S7). Each prophage showed homology to one or two of the prophages of Pf-5 and was identified occasionally in different integrated sites. For example, Prophage IV, a homologue of Prophage 03 of Pf-5, was identified in a different location of the Cab57 genome where Prophage 05 was identified in the Pf-5 genome (Fig. S7). Two large prophages (Prophage III and IV) were identified in the genomic region of Gene ID 3451 to 3607 (Fig. 3A, Fig. S6 and Fig. S7). The homologue of Prophage 06 was identified in the CHA0 genome in the same integrated sites of the Pf-5 genome (Fig. S6 and Fig. S8), reflecting the continuity in the syntenic line observed by dot plot analysis in the corresponding region (Fig. 3C). The homologue of Prophage 03 was not detected in the CHA0 genome (Fig. S7).
Figure 4. Genetic organization of the region surrounding Prophage I in P. protegens Cab57 and the corresponding regions in P. protegens CHA0 and Pf-5.
The common genes in the three strains are in the same color and conserved genes are connected with gray shading, whereas genes that were not common in all strains are colored white. The sizes of genes are not to scale.
The prophage integrated in the mutS/cinA region, which has been reported in ten genomes of the P. fluorescens group including Pf-5 [19], was also conserved in the Cab57 genome in the same position at which Prophage 01 was identified in the Pf-5 genome, and was subsequently named Prophage I. Prophage I consists not only of most of the corresponding prophage of Pf-5 Prophage 01 (PFL_1209 to PFL_1230), which closely resembles their counterparts from the long contractile tail of the serotype-converting bacteriophage SfV of Shigella flexneri [34], but also the P2-like tail assembly region of Prophage 03 of Pf-5 (PFL_2002 to PFL_2017 of Prophage 03) (Fig. 4). Such a hybrid structure has also been found in a homologous region from P. fluoresens Pf0-1, which contains both lambda-like and P2-like tail clusters [34]. The genome of CHA0 also consists of the homologue of Prophage I as a hybrid of Prophage 01 and 03 in the same position as that of Cab57 (Fig. 4).
Prophage II shows homology to the 3′ end of Prophage 04 of Pf-5 and its homologue of CHA0, and three ORFs in Prophage II showed homology to those of Prophage 06 (Fig S5), suggesting the chimeric structure of this prophage. The homologue of Prophage 02 was absent from the Cab57 and CHA0 genomes. The homologue of Prophage 05 was absent from the Cab57 genome, whereas it was present in the CHA0 genome (data not shown).
Unlike the genome of Pf-5, that of Cab57 possesses two IS elements in the genomic regions of Gene ID 1981 to 1998 (Transposon 1) and Gene ID 3883 to 3893 (Transposon 2) with putative transposases (Table S4). Transposon 2 also contains a bacteriophage host lysis protein, suggesting it is a mixture of transposon and prophage. Transposon 2 was identified in the same location at which Prophage 06 of Pf-5 was integrated (Fig. S8). Both of these regions were conserved in the CHA0 genome, whereas several genes including the transposase genes were absent from each region (Table S4).
Overall, the integrated sites of prophages were more restricted in the Cab57 genome than in the Pf-5 genome. These strain-specific segments of the genome were found in limited chromosomal locations, referred to as regions of genomic plasticity.
Regions unique to the Cab57 genome
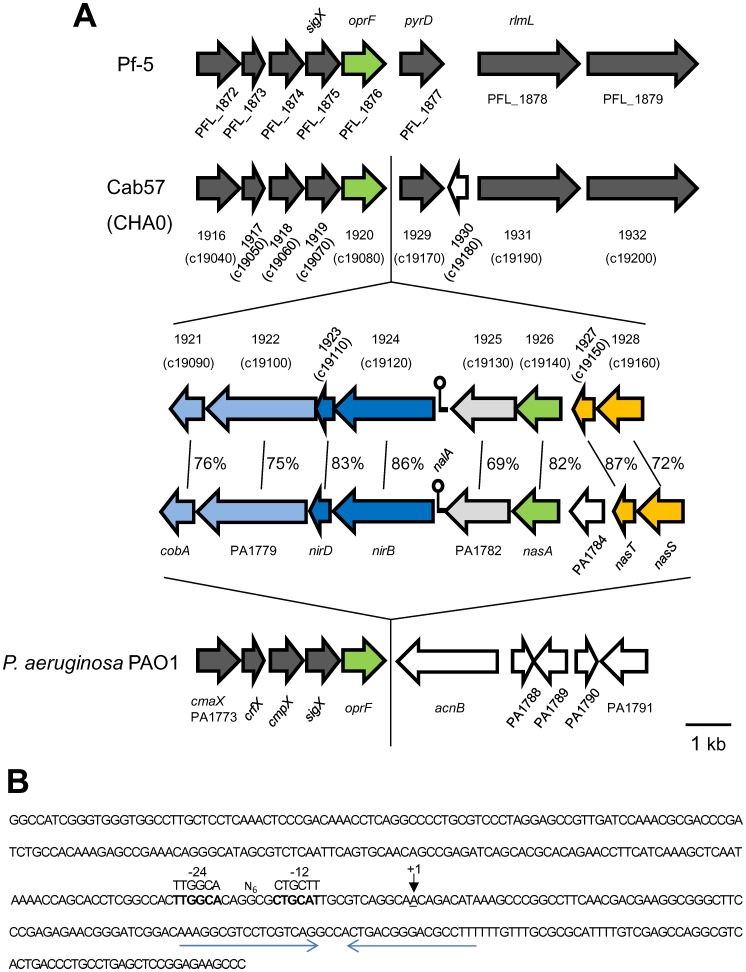
Apart from the different large genomic islands and prophages found in each genome as described above, we listed regions including at least four contiguous ORFs missing in the Pf-5 genome in order to identify regions unique to Cab57 (as Cluster S1 to S14 in Table S5). One prominent difference in the genome of strain Cab57 from that of Pf-5 was the presence of a putative nitrite/nitrate assimilation operon (Cluster S2: Fig. 5 and Table S5). In P. aeruginosa PAO1, the corresponding locus PA1778 to PA1786 was inferred to encode functions required for a nitrite/nitrate assimilation operon, due to homologies with genes reported in other bacteria, and its function has already been demonstrated [35], [36]. For example, the deletion mutant of PA1785, the product of which is required for transcription of the nitrate/nitrite reductase operon, failed to grow on nitrate as the sole nitrogen source [36]. In that study, they found a small RNA termed nitrogen assimilation leader A (NalA) in the intergenic region of PAO1, which is required for the expression of this operon. The NalA homologue was identified in the same locus in the Cab57 genome. The −24 and −12 boxes of the σ 54 -dependent promoter, both of which are indispensable for promoter activity upon nitrogen depletion, were conserved in its promoter sequence. Unlike PAO1 and Salmonella typhimurium, putative palindromic NtrC binding motifs, which are involved in the full expression of the promoter activity necessary [36], [37], were not found. The nalA terminator sequence was conserved as a palindromic sequence (Fig. 5). This operon was also present in the CHA0 genome.
Figure 5. Genetic organization of the nitrite/nitrate assimilation operons in P. aeruginosa PAO1 [36], [53] and the corresponding regions in P. protegens strains Cab57, CHA0, and Pf-5.
(A) ORFs encoding different functional classes of proteins are indicated in each color and the ORFs conserved among the strains are indicated in the same color. The percentage of amino acid identity is indicated. Other conserved genes are colored gray, and strain-specific genes are colored white. (B) Sequence of the intergenic region between Gene ID 1924 and 1925 in P. protegens Cab57. The putative −24 and −12 boxes of the σ 54 -dependent promoter are shown in bold type and the putative transcriptional start site of nalA is underlined. The σ 54 consensus sequence is shown above the sequence. The putative nalA terminator sequence is denoted by horizontal arrows.
P. protegens Pf-5 has been reported to be defective in reducing nitrates, which was attributed to the absence of this operon [19]. Only three of the ten sequenced strains in the P. fluorescens group (strains O6, Q8r1-96, and Q2-87) possess this operon [19]. The presence of this operon in strain Cab57 suggests that this is not a core component in the same species; therefore, it may provide diversity at the species level.
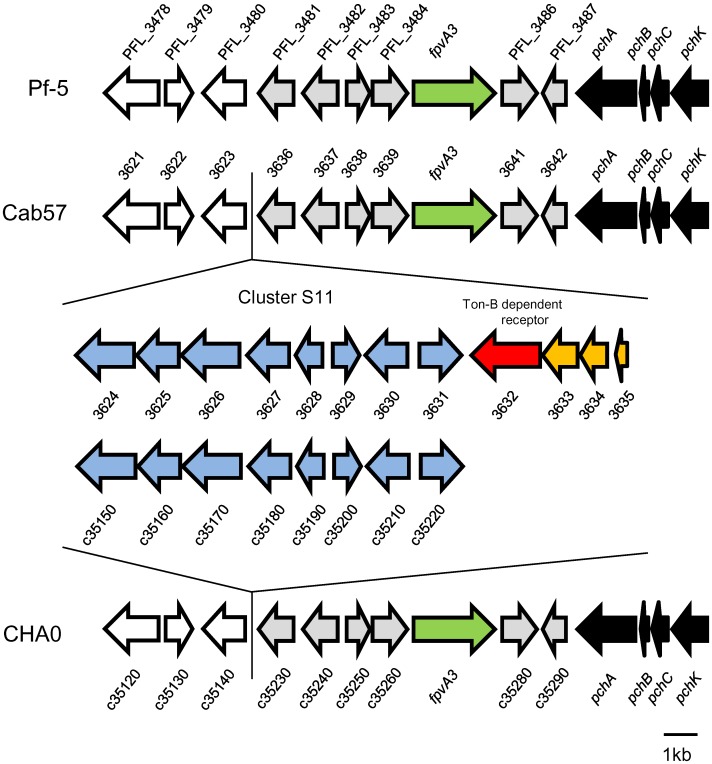
Other gene clusters include the following; Cluster S3 encoding the penicillin binding protein, Cluster S10 encoding formate dehydrogenase, Cluster S13 encoding the McrBC restriction enzyme, and Cluster S14 involved in the O-antigen (Table S5). Cluster S11 and Cluster S12 were identified as secondary metabolite clusters by the antiSMASH program. From an ecological viewpoint, such gene clusters may also contribute to niche adaptation by this strain. Cluster S11 (Gene ID 3624 to 3635) was located near the pch cluster (Gene ID 3642 to 3651, for enantio-pyochelin), and we identified the PFL_3485 homologue (Gene ID 3640) which was annotated as the ferripyoverdine receptor, between these two clusters (Fig. 6). Cluster S11 possessed a specific region (Gene ID 3632 to 3635) that was also absent from strain CHA0. One of the genes (Gene ID 3632) was annotated as the TonB-dependent siderophore receptor, which suggests different potentials for iron acquisition among the strains.
Figure 6. Genetic organization of Cluster S11 in P. protegens Cab57 and the corresponding regions in P. protegens strains Pf-5 and CHA0.
Cluster S11 ORFs conserved in Cab57 and CHA0 genomes are indicated in blue, whereas those specific to the Cab57 genome are indicated in red and yellow. ORFs for the pch cluster are in black. The ORF for the ferripyoverdine receptor (fpvA3) is in green, and that for the TonB siderophore receptor is in red. The ORFs conserved among the strains are indicated in the same color. This figure was based on sequence data from the Pseudomonas.com website (http://www.pseudomonas.com) and from Youard et al [28].
Materials and Methods
Ethics Statement
For the isolation of fluorescent pseudomonads, plant roots were sampled on private lands in diverse locations (Hokkaido, Ibaraki, Kanagawa, Nagano, Aichi and Nagasaki) in Japan after the land owners gave permission to conduct sampling on the sites. None of these species is an endangered or protected species.
Isolation of DAPG-producing Pseudomonads from Rhizosphere Soil and Selection for Whole Genome Analysis
We isolated approximately 2,800 fluorescent pseudomonads based on a method described previously [38]. DAPG-producing isolates were pre-screened by PCR using the DAPG biosynthesis gene phlD-specific primers Phl2a and Phl2b (Table S6). phlD + isolates were incubated on dNBYG medium [39] and DAPG production was evaluated by TLC [40]. DAPG production was also evaluated in strain Cab57 by HPLC [41]. Other antibiotic biosynthetic genes were also explored by PCR analysis using specific sets of primers; PRND1/PRND2 for pyrrolnitrin, PltBf/PltBr for pyoluteorin, and PM2/PM7-26R for hydrogen cyanide (Table S6). Selection by this method revealed five isolates were PCR-positive for all four antibiotic biosynthetic genes and one of the five strains named Cab57, which was isolated from the rhizosphere of shepherd’s purse [Capsella bursa-pastoris (L.) Medik.] growing in a field in Hokkaido, was selected for whole genome analysis.
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used are listed in Table 5. E. coli and P. protegens strains were routinely grown in NYB (2.5% [w/v] nutrient broth, 0.5% [w/v] yeast extract) and Luria-Bertani (LB) medium with shaking, or on nutrient agar plates (4% [w/v] blood agar base, 0.5% [w/v] yeast extract) amended with the following antibiotics when required: ampicillin, 100 μg/mL (only for E. coli); kanamycin, 25 μg/mL; or tetracycline, 25 μg/mL (125 μg/mL for selection of P. fluorescens). The inoculation temperatures were 28°C for P. protegens and 37°C for E. coli. P. protegens was grown at 33°C to improve its capacity to accept heterologous DNA (e.g., in electrotransformation or in matings with E. coli). Bacteria were grown in glycerol-casamino acid medium (GCM) for other assays [42].
Table 5. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
| Strains | ||
| Bacillus subtilis | ||
| M168 | Wild type | C. Keel |
| Escherichia coli | ||
| DH5α, HB101 | Laboratory strains | [50] |
| Pseudomonas protegens | ||
| CHA0 | Wild type | [3] |
| Cab57 | Wild type | This study |
| Cab57retS | ΔretS | This study |
| Plasmids | ||
| pCR-Blunt II-TOPO | Cloning vector, pUC ori; Kmr | Invitrogen |
| pME497 | Mobilizing plasmid, IncP-1, Tra, RepA(Ts); Apr | [51] |
| pME3087 | Suicide vector, ColE1 replicon, Mob; Tcr | [52] |
| pME3087retS | pME3087 containing a BamHI/HindIII 1.3-kb retS region containing a deletion of the retS gene; Tcr | This study |
| pME6031 | pACYC177-pVS1 shuttle vector; Tcr | [49] |
| pME6060 | Translational aprA’-‘lacZ fusion; Tcr | [25] |
| pME6091 | Transcriptional rsmZ-lacZ fusion; Tcr | [7] |
DNA Manipulation
Small-scale plasmid extraction was performed with a QIAprep spin miniprep kit (QIAGEN); large-scale preparations were obtained with a QIAGEN plasmid midi kit (QIAGEN). Chromosomal DNA from P. protegens was prepared with a QIAGEN Genomic DNA buffer set and Genomic-tip (QIAGEN). DNA fragments were purified from agarose gels with a QIAquick gel extraction kit (QIAGEN). Oligonucleotides used are listed in Table S6.
Whole Genome Sequencing and Assembly
Whole genome sequencing was performed using Roche Genome Sequencer FLX Titanium (for 8 kb long paired-end sequencing) and FLX+ (for shotgun sequencing) technology provided by Operon Biotechnologies K.K. (Tokyo, Japan). A shotgun library was prepared and subsequently sequenced, generating 361,203 reads in 211.0 Mbp of sequencing data. An 8 kb long paired-end library was also prepared and sequenced, generating 394,536 reads in 78.5 Mbp of sequencing data. Co-assembly of the results of both shotgun and paired-end sequencing was performed by newbler 2.6, and were fully assembled into 14 large contigs defined as >1 kb. These contigs were then integrated only one scaffold, the size of which was 6.8 Mbp with several gaps. Gaps between the contigs were filled in by sequencing the PCR products using Applied Biosystems 3100×l DNA Analyzer. High-fidelity DNA polymerase KOD Plus™ (Toyobo) was used for PCR amplification. The prediction of putative coding sequences and gene annotation was performed using the Microbial Genome Annotation Pipeline (http://migap.lifesciencedb.jp/). The circular map that illustrates the general genomic feature as well as its comparison with other two strains was plotted by using the software BRIG [43].
Accession Number
The complete genome sequence of P. protegens Cab57 has been deposited in the DDBJ/EMBL/GenBank database under accession no. AP014522.
Species Identification
To identify species, the 16S rRNA gene of strain Cab57 was applied to a BLAST search. The strain was also characterized using API 20 NE strips (BioMerieux Vitek, Inc.) incubated at 28°C for 48 hours. Whole genome comparisons of strain Cab57 with those of the P. fluorescens group were performed using the JSpecies program [23]. This program is commonly used to compare two genomes and uses BLAST software [44].
Comparative Genome Analysis
To compare the whole genome of P. protegens Cab57 with the P. protegens Pf-5 or CHA0 strain, the alignment data between each two strains’ genome sequences were prepared using the LASTZ program (Release 1.02.00), and dot plots were then illustrated from the alignment data by R (version 3.0.1). Genomic regions that were unique to each strain were searched for using the Island Viewer program [45] and verified manually. Secondary metabolite production clusters were examined using the antiSMASH program [46].
Plant Disease Suppression and Root Colonization Assays
Flasks containing vermiculite were planted with three cucumber seedlings each and treated with Pythium ultimum MAFF425494 and/or P. protegens. After seven days of incubation, the biocontrol activity and root colonization of each strain were estimated as described previously [47].
Generation of retS-negative Mutant
Primers are listed in Supplementary Table S6. An in-frame deletion in the chromosomal retS gene of P. protegens Cab57 was created as follows. Fragments of 620-bp located upstream and 640-bp located downstream of the retS gene, respectively, were amplified by PCR with the primer pairs RetSUF/RetSUR and RetSDF/RetSDR, high-fidelity DNA polymerase KOD Plus™ (Toyobo), and genomic DNA of P. protegens Cab57 as a template. Each two corresponding fragments were annealed and amplified as a 1.3-kb fragment using the primer pair RetSUF/RetSDR. This 1.3-kb fragment was cloned into pCR-Blunt II-TOPO™ (Invitrogen). The insert obtained was confirmed by sequencing and digested with BamHI and HindIII. After sequencing, these fragments were subcloned into pME3087 cleaved at the BamHI and HindIII sites to give pME3087retS. This plasmid was mobilized from E. coli DH5α to P. protegens Cab57 by triparental mating with E. coli HB101/pME497. Excision of the vector via a second crossing-over was obtained after the enrichment of tetracycline-sensitive cells, generating the retS mutant.
β-Galactosidase Assays
β-Galactosidase activities were quantified by the Miller method [48]. P. protegens strains were grown at 30°C in 50-mL flasks containing 20 mL of NYB amended with 0.05% Triton X-100 with shaking at 180 rpm. Triton X-100 was required to prevent cell aggregation.
Detection of Antibiotic Activity
The antibiotic activities of P. protegens strains were determined with B. subtilis M168, Pythium ultimum MAFF425494, or Fusarium oxysporum MAFF105034 as the reporter. Cultures of P. protegens were adjusted to OD600 nm = 1.5 for the assay with B. subtilis, and 5-μL samples were spotted onto the GCM plate. After overnight incubation at 30°C, cells were killed by UV irradiation on a transilluminator for 5 min. An overlay of B. subtilis revealed antibiotic production by growth inhibition zones. Cultures of P. protegens were adjusted to OD600 nm = 1.5 for the assay with P. ultimum or F. oxysporum, 20 μL samples were streaked around the edge of the potato dextrose agar (PDA) plate, and an inoculum of P. ultimum or F. oxysporum was transferred to the center of the plate. The plate was incubated at room temperature until P. ultimum or F. oxysporum reached the edge of the plate.
Supporting Information
Circular genome map of P. protegens Cab57. From the inside out, circle 1 represents the mean centered G+C content (bars facing outside-above mean, bars facing inside-below mean); circle 2 shows the GC skew (G–C)/(G+C); 3 (blue) and 4 (red) represent comparison of gene content with P. protegens strain CHA0 and Pf-5, respectively. The outermost ring highlights the regions of mobile genetic elements (P, Prophage shown in green; T, Transposon shown in purple), and gene clusters which are absent from Pf-5 genome (S, shown in black) as described in Table 4 and, respectively. The inner black circle shows the scale line in Mbps.
(EPS)
Venn diagram comparing the coding sequence sets of P. protegens strains Cab57, CHA0, and Pf-5. The number of orthologous coding sequences (CDSs) shared by all strains is in the center. The numbers in the non-overlapping portions of each circle represent the number of CDSs unique to each strain. The total number of CDSs within each genome is listed below the strain name. Each set of genes was compared to the other two sets using BLASTp, and sequence identity cut-off was set at 60% to identify common genes. Note that due to multiple hits, gene numbers did not add up to the strain totals.
(TIF)
Genetic organization of fit cluster (for FitD toxin) and rzx cluster (for rhizoxin analogs) in P. protegens Pf-5 and the corresponding regions in P. protegens Cab57 and CHA0. The conserved genes are colored gray, and strain-specific genes are colored white.
(TIF)
Genetic organization of the region surrounding the genomic island PFGI-1 in P. protegens Pf-5 and the corresponding regions in P. protegens Cab57 and CHA0. The conserved genes are colored gray, and strain-specific genes are colored white.
(TIF)
Genetic organization of the region surrounding Prophage II in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. Homologous genes are connected with gray shading. The genes in prophage are colored white and genes outside prophage are colored gray. The sizes of genes are not to scale.
(TIF)
Genetic organization of the region surrounding Prophage III in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. Homologous genes are connected with gray shading. The genes in prophage are colored white and genes outside prophage are colored gray. The sizes of genes are not to scale.
(TIF)
Genetic organization of the region surrounding Prophage IV in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. Homologous genes are connected with gray shading. The genes in prophage are colored white and genes outside prophage are colored gray. The sizes of genes are not to scale.
(TIF)
Genetic organization of the region surrounding Transposon 2 in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. The conserved genes are colored gray, and strain-specific genes are colored white.
(TIF)
Sequence analysis of gene clusters for the synthesis of antibiotics and exoenzyme in P. protegens Cab57 and similarities to those in P. protegens Pf-5.
(DOCX)
Sequence analysis of Gac/Rsm homologues in P. protegens Cab57 and similarities to those in P. protegens Pf-5.
(DOCX)
Sequence analysis of gene clusters for the synthesis of cyclic lipopeptide, siderophores, and toxin in P. protegens Cab57 and similarities to those in P. protegens Pf-5.
(DOCX)
Sequence analysis of the transposon of the P. protegens Cab57 genome.
(DOCX)
Sequence analysis of gene clusters in P. protegens Cab57 which are absent from Pf-5 genome.
(DOCX)
Oligonucleotides used in this study.
(DOCX)
Acknowledgments
We thank Dieter Haas for providing the bacterial strains and plasmids, Kosumi Yamada for help with the HPLC analysis, Yuichi Katayose and Hisataka Numa for help with the bioinformatic analysis, Hiroyuki Sawada for help with the API test, Kumiko Oka for help with the DNA manipulation, and Chie Setoyama for help with the plant experiments. This work was supported partly by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and by the NIAS technical support system.
Funding Statement
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mulet M, Lalucat J, García-Valdés E (2010) DNA-sequence-based analysis of the Pseudomonas species. Environ Microbiol 12: 1513–1530. [DOI] [PubMed] [Google Scholar]
- 2. Haas D, Keel C (2003) Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41: 117–153. [DOI] [PubMed] [Google Scholar]
- 3. Stutz EW, Défago G, Kern H (1986) Naturally occurring fluorescent Pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76: 181–185. [Google Scholar]
- 4. Howell CR, Stipanovic RD (1979) Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with and antibiotic produced by the bacterium. Disease Control and Pest Management 69: 480–482. [Google Scholar]
- 5. Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, et al. (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23: 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lapouge K, Schubert M, Allain FH-T, Haas D (2008) Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67: 241–253. [DOI] [PubMed] [Google Scholar]
- 7. Heeb S, Blumer C, Haas D (2002) Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol 184: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valverde C, Heeb S, Keel C, Haas D (2003) RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol 50: 1361–1379. [DOI] [PubMed] [Google Scholar]
- 9. Kay E, Dubuis C, Haas D (2005) Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci USA 102: 17136–17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reimmann C, Valverde C, Kay E, Haas D (2005) Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol 187: 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. [DOI] [PubMed] [Google Scholar]
- 12. Kay E, Humair B, Dénervaud V, Riedel K, Spahr S, et al. (2006) Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa . J Bacteriol 188: 6026–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim YC, Leveau J, McSpadden Gardener BB, Pierson EA, Pierson III LS, et al. (2011) The multifacterorial basis for plant health promotion by plant-associated bacteria. Appl Environ Microbiol 77: 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, et al. (2009) Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Humair B, González N, Mossialos D, Reimmann C, Haas D (2009) Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J 3: 955–965. [DOI] [PubMed] [Google Scholar]
- 16. Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, et al. (2007) The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol 14: 53–63. [DOI] [PubMed] [Google Scholar]
- 17. Loper JE, Henkels MD, Shaffer BT, Valeriote FA, Gross H (2008) Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl Environ Microbiol 74: 3085–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Péchy-Tarr M, Bruck DJ, Maurhofer M, Fischer E, Vogne C, et al. (2008) Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. . Environ Microbiol 10: 2368–2386. [DOI] [PubMed] [Google Scholar]
- 19. Loper JE, Hassan KA, Mavrodi DV, Davis EWII, Lim CK, et al. (2012) Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8: e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stockwell VO, Stack JP (2007) Using Pseudomonas spp. for integrated biological control. Phytopathology 97: 244–249. [DOI] [PubMed] [Google Scholar]
- 21. Keel C, Weller DM, Natsch A, Défago G, Cook RJ, et al. (1996) Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol 62: 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer JM, et al. (2011) Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin, Syst Appl Microbiol 34: 180–188. [DOI] [PubMed] [Google Scholar]
- 23. Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, et al. (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens . Genome Biol 10: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blumer C, Heeb S, Pessi G, Haas D (1999) Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 96: 14073–14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siddiqui IA, Haas D, Heeb S (2005) Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Appl Environ Microbiol 71: 5646–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gross H, Loper JE (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26: 1408–1466. [DOI] [PubMed] [Google Scholar]
- 28. Youard ZA, Mislin GL, Majcherczyk PA, Schalk IJ, Reimmann C (2007) J Biol Chem. 282: 35546–35553. [DOI] [PubMed] [Google Scholar]
- 29. Takeuchi K, Kiefer P, Reimmann C, Keel C, Dubuis C, et al. (2009) Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J Biol Chem 284: 34976––34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwasaki S, Kobayashi H, Furukawa J, Namikoshi M, Okuda S, et al. (1984) Studies on macrocyclic lactone antibiotics. VII. Structure of a phytotoxin″rhizoxin″ produced by Rhizopus chinensis . J Antibiot 37: 354–362. [DOI] [PubMed] [Google Scholar]
- 31. Partida-Martinez LP, Hertweck C (2005) Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437: 884–888. [DOI] [PubMed] [Google Scholar]
- 32. Partida-Martinez LP, Hertweck C (2007) A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”. the bacterial endosymbiont of the fungus Rhizopus microspores Chembiochem 8: 41–45. [DOI] [PubMed] [Google Scholar]
- 33. Lackner G, Moebius N, Partida-Martinez L, Hertweck C (2011) Complete genome sequence of Burkholderia rhizoxinica, the endosymbiont of Rhizopus microsporus . J Bacteriol 193: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mavrodi DV, Loper JE, Paulsen IT, Thomashow LS (2009) Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noriega C, Hassett DJ, Rowe JJ (2005) The mobA gene is required for assimilatory and respiratory nitrate reduction but not xanthine dehydrogenase activity in Pseudomonas aeruginosa. . Curr Microbiol 51: 419–424. [DOI] [PubMed] [Google Scholar]
- 36. Romeo A, Sonnleitner E, Sorger-Domenigg T, Nakano M, Eisenhaber B, et al. (2012) Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology 158: 1543–1552. [DOI] [PubMed] [Google Scholar]
- 37. Ferro-Luzzi Ames G, Nikaido K (1985) Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J 4: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Someya N, Morohoshi T, Ikeda T, Tsuchiya K, Ikeda S (2012) Genetic diversity and ecological evaluation of fluorescent pseudomonads isolated from the leaves and roots of potato plants. Microbes Environ 27: 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duffy BK, Défago G (1999) Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 65: 2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, et al. (1992) Suppression of root diseases by Pseudomonas fluorescens CHA0: Importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant- Microbe Interact 5: 4–13. [Google Scholar]
- 41. Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, et al. (2000) Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol 182: 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, et al. (1998) Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88: 678–684. [DOI] [PubMed] [Google Scholar]
- 43. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 45. Langille MG, Hsiao WW, Brinkman FS (2010) Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol 8: 373–382. [DOI] [PubMed] [Google Scholar]
- 46. Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, et al. (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucl. Acids Res 39: W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takeuchi K, Yamada K, Haas D (2012) ppGpp controlled by the Gac/Rsm regulatory pathway sustains biocontrol activity in Pseudomonas fluorescens CHA0. Mol Plant- Microbe Interact 25: 1440–1449. [DOI] [PubMed] [Google Scholar]
- 48.Miller JH (1972) Experiments in Molecular Genetics: New York: Cold Spring Harbor Laboratory Press.
- 49. Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, et al. (2000) Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant-Microbe Interact 13: 232–237. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Russell DW, Irwin N (2001) Molecular Cloning: A Laboratory Manual, 3rd ed. New York: Cold Spring Harbor Laboratory Press.
- 51. Voisard C, Rella M, Haas D (1988) Conjugative transfer of plasmid RP1 to soil isolates of Pseudomonas fluorescens is facilitated by certain large RP1 deletions. FEMS Microbiol Lett 55: 9–13. [Google Scholar]
- 52.Voisard C, Bull CT, Keel C, Laville J, Maurhofer M, et al. (1994) Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: Molecular Ecology of Rhizosphere Microorganisms (O’Gara, F., Dowling, D.N., and Boesten, B., eds): VCH Weinheim, Germany. 67–89.
- 53.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, et al.. (2009) Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acid Res 37 (Database issue): D483–D488. [DOI] [PMC free article] [PubMed]
- 54. Raaijmakers JM, Weller DM, Thomashow LS (1997) Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol 63: 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Souza JT, Raaijmakers JM (2003) Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol Ecol 43: 21–34. [DOI] [PubMed] [Google Scholar]
- 56. Mavrodi OV, McSpadden Gardener BB, Mavrodi DV, Bonsall RF, Weller DM, et al. (2001) Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology 91: 35–43. [DOI] [PubMed] [Google Scholar]
- 57. Svercel M, Duffy B, Défago G (2007) PCR amplification of hydrogen cyanide biosynthetic locus hcnAB in Pseudomonas spp. J Microbiol Methods 70: 209–213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circular genome map of P. protegens Cab57. From the inside out, circle 1 represents the mean centered G+C content (bars facing outside-above mean, bars facing inside-below mean); circle 2 shows the GC skew (G–C)/(G+C); 3 (blue) and 4 (red) represent comparison of gene content with P. protegens strain CHA0 and Pf-5, respectively. The outermost ring highlights the regions of mobile genetic elements (P, Prophage shown in green; T, Transposon shown in purple), and gene clusters which are absent from Pf-5 genome (S, shown in black) as described in Table 4 and, respectively. The inner black circle shows the scale line in Mbps.
(EPS)
Venn diagram comparing the coding sequence sets of P. protegens strains Cab57, CHA0, and Pf-5. The number of orthologous coding sequences (CDSs) shared by all strains is in the center. The numbers in the non-overlapping portions of each circle represent the number of CDSs unique to each strain. The total number of CDSs within each genome is listed below the strain name. Each set of genes was compared to the other two sets using BLASTp, and sequence identity cut-off was set at 60% to identify common genes. Note that due to multiple hits, gene numbers did not add up to the strain totals.
(TIF)
Genetic organization of fit cluster (for FitD toxin) and rzx cluster (for rhizoxin analogs) in P. protegens Pf-5 and the corresponding regions in P. protegens Cab57 and CHA0. The conserved genes are colored gray, and strain-specific genes are colored white.
(TIF)
Genetic organization of the region surrounding the genomic island PFGI-1 in P. protegens Pf-5 and the corresponding regions in P. protegens Cab57 and CHA0. The conserved genes are colored gray, and strain-specific genes are colored white.
(TIF)
Genetic organization of the region surrounding Prophage II in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. Homologous genes are connected with gray shading. The genes in prophage are colored white and genes outside prophage are colored gray. The sizes of genes are not to scale.
(TIF)
Genetic organization of the region surrounding Prophage III in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. Homologous genes are connected with gray shading. The genes in prophage are colored white and genes outside prophage are colored gray. The sizes of genes are not to scale.
(TIF)
Genetic organization of the region surrounding Prophage IV in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. Homologous genes are connected with gray shading. The genes in prophage are colored white and genes outside prophage are colored gray. The sizes of genes are not to scale.
(TIF)
Genetic organization of the region surrounding Transposon 2 in P. protegens Cab57 and the corresponding regions in P. protegens Pf-5 and CHA0. The conserved genes are colored gray, and strain-specific genes are colored white.
(TIF)
Sequence analysis of gene clusters for the synthesis of antibiotics and exoenzyme in P. protegens Cab57 and similarities to those in P. protegens Pf-5.
(DOCX)
Sequence analysis of Gac/Rsm homologues in P. protegens Cab57 and similarities to those in P. protegens Pf-5.
(DOCX)
Sequence analysis of gene clusters for the synthesis of cyclic lipopeptide, siderophores, and toxin in P. protegens Cab57 and similarities to those in P. protegens Pf-5.
(DOCX)
Sequence analysis of the transposon of the P. protegens Cab57 genome.
(DOCX)
Sequence analysis of gene clusters in P. protegens Cab57 which are absent from Pf-5 genome.
(DOCX)
Oligonucleotides used in this study.
(DOCX)