Abstract
Class B G protein-coupled receptors (GPCRs) are important regulators of endocrine physiology, and peptide-based therapeutics targeting some of these receptors have proven effective at treating disorders such as hypercalcemia, osteoporosis, and type 2 diabetes mellitus (T2DM). As next generation efforts attempt to develop novel non-peptide, orally available molecules for these GPCRs, new animal models expressing human receptor orthologs may be required because small molecule ligands make fewer receptor contacts, and thus, the impact of amino acid differences across species may be substantially greater. The objective of this report was to generate and characterize a new mouse model of the human glucagon-like peptide-1 receptor (hGLP-1R), a class B GPCR for which established peptide therapeutics exist for the treatment of T2DM. hGLP-1R knock-in mice express the receptor from the murine Glp-1r locus. Glucose tolerance tests and gastric emptying studies show hGLP-1R mice and their wild-type littermates display similar physiological responses for glucose metabolism, insulin secretion, and gastric transit, and treatment with the GLP-1R agonist, exendin-4, elicits similar responses in both groups. Further, ex vivo assays show insulin secretion from humanized islets is glucose-dependent and enhanced by GLP-1R agonists. To enable additional utility, the targeting construct of the knock-in line was engineered to contain both flanking LoxP sites and a C-terminal FLAG epitope. Anti-FLAG affinity purification shows strong expression of hGLP-1R in islets, lung, and stomach. We crossed the hGLP-1R line with Rosa26Cre mice and generated global Glp-1r−/− animals. Immunohistochemistry of pancreas from humanized and knock-out mice identified a human GLP-1R-specific antibody that detects the GLP-1R in human pancreas as well as in the pancreas of hGLP-1r knock-in mice. This new hGLP-1R model will allow tissue-specific deletion of the GLP-1R, purification of potential GLP-1R partner proteins, and testing of novel therapeutic agents targeting the hGLP-1R.
Introduction
The gluco-regulatory actions of the glucagon-like peptide-1 (GLP-1) and its related therapeutic mimetics include enhancement of glucose-dependent insulin secretion and suppression of glucagon release from pancreatic islets, as well as a decrease in postprandial glucose excursion by delay of gastric emptying and centrally-mediated effects on satiety and energy metabolism [1], [2]. These physiological effects are mediated by the GLP-1 receptor (GLP-1R) and have been determined as a result of iterative basic, translational, and clinical experimentation. Important early findings in incretin biology include proof-of-concept studies showing GLP-1 infusion improves hyperglycemia in patients with type 2 diabetes [3] and cloning of the GLP-1R cDNA [4]. In addition, discovery of the dipeptidyl peptidase-4 (DPP4)-resistant GLP-1R agonist exendin-4 (EX-4) enabled much of our current understanding of GLP-1R activation on glucose homeostasis via studies in animal models [5], [6]. Further, clinical investigations of GLP-1 mimetics demonstrated pleiotropic anti-diabetic pharmacology in humans [7], [8].
While the insulinotropic actions of GLP-1 are well characterized, functional roles of the GLP-1R in extra-pancreatic tissues are less understood. GLP-1 treatment affects hemodynamics in both rodents and humans [9], [10]. However, the GLP-1R-expressing cell types that mediate these effects are not known, although recent studies showed atrial GLP-1R-mediated regulation of blood pressure in mice [11]. In addition, while lung Glp-1r mRNA expression is high in some species [12] and GLP-1R signaling is implicated in surfactant secretion [13]–[15], the role of GLP-1R in pulmonary function is not well established. Further, GLP-1 effects on adiposity and lipid metabolism are not completely understood. Although it is well known that GLP-1R agonists inhibit food intake and reduce body weight, Glp-1r −/− mice are protected from high fat diet-induced obesity and insulin resistance and have decreased hepatosteatosis [16]–[18]. Finally, the role of GLP-1 in the thyroid gland remains to be determined, although GLP-1R expression analyses indicate this is largely a rodent phenomenon [19], [20].
To further characterize GLP-1R, it is important to understand which organs and cell types express the receptor. Glp-1r mRNA is present in many tissues with pancreas, lung, stomach, and small intestine showing the highest levels [21]–[23]. Unfortunately, determining GLP-1R protein expression patterns has been technically difficult due to lack of quality GLP-1R antisera. This has been highlighted in reports demonstrating several commercially available GLP-1R antibodies show non-specific staining in cells not expressing Glp-1r mRNA [16], [24]. Developing better reagents and quality control strategies is needed to determine GLP-1R-positive cell types. Once the expression pattern of GLP-1R is firmly established, targeting strategies to assess GLP-1R function in individual tissues will evaluate the extra-pancreatic physiology of the GLP-1R.
Here, we describe a novel mouse line generated via homologous recombination that expresses human GLP-1R (hGLP-1R) from the murine Glp-1r locus. This approach enables immunohistochemistry (IHC) analyses using an anti-GLP-1R antibody that specifically recognizes hGLP-1R. A unique aspect of this model is the inclusion of an eight amino acid (aa) FLAG epitope fused in-frame at the C-terminus of hGLP-1R. This feature enables anti-FLAG IHC, western blot (WB), and immunoprecipitation (IP)/affinity purification experiments. In addition, the hGLP-1R is flanked by LoxP sites that allow conditional deletion of the hGLP-1R using P1 bacteriophage Cre recombinase [25]. We demonstrate initial utility of this element by crossing animals with Rosa26Cre transgenic mice [26] to generate whole-body Glp-1r−/− animals. Use of anti-human GLP-1R and anti-FLAG immunodetection applications with tissues from hGLP-1R and Glp-1r−/− mice enables well-controlled studies of GLP-1R expression. Future studies using this model will facilitate ex vivo biochemical characterization of GLP-1R regulation via affinity purification procedures and in vivo investigation of GLP-1R function by temporal and conditional gene deletion.
Materials and Methods
Ethics Statement
Animals were studied and maintained in accordance with the Institutional Animal Care and Use Committee (IACUC) of Eli Lilly and Company, the Guide for the Use and Care of Laboratory Animals by the National Institutes of Health, and the German Animal Welfare Act. All animal studies described herein were approved by the IACUC of Eli Lilly and Company.
Animal Husbandry
Mice were singly housed in microisolator cages on wood chip bedding with food (2014 Teklad Global Diet, Harlan, Indianapolis) and deionized water available ad libitum. Lights were on a 12∶12 hour light: dark cycle (except when noted otherwise), and temperature and relative humidity were maintained between 21 and 23°C and 45 and 65%, respectively. Ill or moribund animals were sacrificed using CO2 asphyxiation and post-mortem cardiac blood draw and/or cervical dislocation; the same methods of euthanasia were applied to animals at the end of any in vivo studies.
Generation of hGLP-1R and Glp-1r −/− Mice
Both mouse lines were generated at TaconicArtemis GmbH (Cologne, Germany). The targeting vector to create the models is based on a 10.5-kb genomic fragment from the murine Glp-1r (mGlp-1r) gene encompassing exons 1 to 2 and flanking sequences. This fragment, obtained from the C57BL/6J RP23 BAC library, was modified by replacing the majority of the coding region of mGlp-1r with an engineered hGLP-1R cDNA. The C57BL/6NTac ES cell line was grown on an antibiotic resistant, mitotically inactivated feeder layer comprised of mouse embryonic fibroblasts. 1×107 cells and 30 μg of linearized DNA targeting vector were electroporated (Bio-Rad Gene Pulser) at 240 V and 500 μF. Double positive selection with G418 (200 μg/mL) and 1.5 μg/mL Puromycin started on day 2. Correctly recombined ES cell clones were identified by Southern blot analysis using external and internal probes and were frozen in liquid nitrogen. Superovulated BALB/c females were mated with BALB/c males, and blastocysts were isolated from the uterus at days post coitus 3.5. A microinjection pipette was used to inject 10–15 targeted C57BL/6NTac ES cells into each blastocyst. Eight injected blastocysts were implanted into pseudopregnant NMRI females.
hGLP-1R knock-in and Glp-1r−/− embryos were cryopreserved and shipped to the U.S. for re-derivation at Taconic, USA (Germantown, NY). Genomic DNA was extracted from tail clips using the NucleoSpin Tissue kit (Macherey-Nagel), and PCR was used to genotype animals with PCR amplicons analyzed using a Caliper LabChip GX device. The genotyping primer sequences are listed in Table S1. The genotyping strategies are listed in Figures S1 and S2.
In vivo and in vitro Studies
All animals were randomized into their respective treatment groups based on body weight and were males between 8–12 weeks age. We used n = 7–9 per group to allow for strength of statistical outcomes and each in vivo study was replicated three times. Glucose tolerance tests were performed as previously described [27]. Detailed protocols for islet studies as well as the cAMP accumulation assays are previously described [27], [28]. For gastric emptying, animals housed in reverse light cycle were fasted overnight and then allowed to eat a pre-weighed portion of food for one hour after the start of the dark cycle the next morning. After food consumption was measured, EX-4 (10 nmol/kg body weight (BW)) was injected subcutaneously (SC). The mice were euthanized one hour after EX-4 administration. Stomachs were removed and weighed full and empty for measurement of percent gastric emptying. The mixed meal tolerance test (MMTT) was performed as previously described with some minor modifications [29]. Briefly, all mice were fasted for 4 hours in the morning of the test. At 0 minutes, the animals were orally gavaged with 10 m1/kg BW of Ensure Plus (Abbott). Blood glucose and serum insulin were measured at 0, 10, 30, 60, and 120 minutes post-gavage, and averaged for each group of animals. For IHC, tissues and cells were fixed and processed for paraffin embedding using routine procedures as described previously [30]. Paraffin-embedded human pancreas samples were purchased from Folio Biosciences (Columbus, OH).
IP-WB and mRNA Expression Analyses
Animals were euthanized using CO2 and tissues were harvested and frozen in liquid nitrogen. Frozen tissue (5 mg) was lysed in RIPA buffer using the FastPrep Lysing Matrix system (MpBio). The anti-FLAG M2 antibody (F3165, Sigma) was coupled to MyOne Tosylactivated Dynabeads (65501, Invitrogen by Life Technologies) according to manufacturer protocols. Lysates were incubated with the anti-FLAG M2 antibody-coupled beads overnight at 4°C. Beads were pelleted, washed, PNGase F-treated (G5166, Sigma) (if applicable), and proteins eluted in SDS sample buffer/2.5% mercaptoethanol. WB’s were performed using 10% PAGE followed by dry transfer to nitrocellulose membrane and probed with anti-FLAG L5-HRP antibody (NBP1-06712H, Novus; 1∶10,000 in 0.5% non-fat dry milk) and enhanced chemiluminescence (ThermoFisher). GLP-1R mRNA expression analyses was performed by RT- and qPCR as previously described [27]. The primer and probe sequences are listed in Table S1.
Statistical Analysis
All results are presented as mean±SEM with all data and statistical analyses performed using Prism4 software (GraphPad).
Results
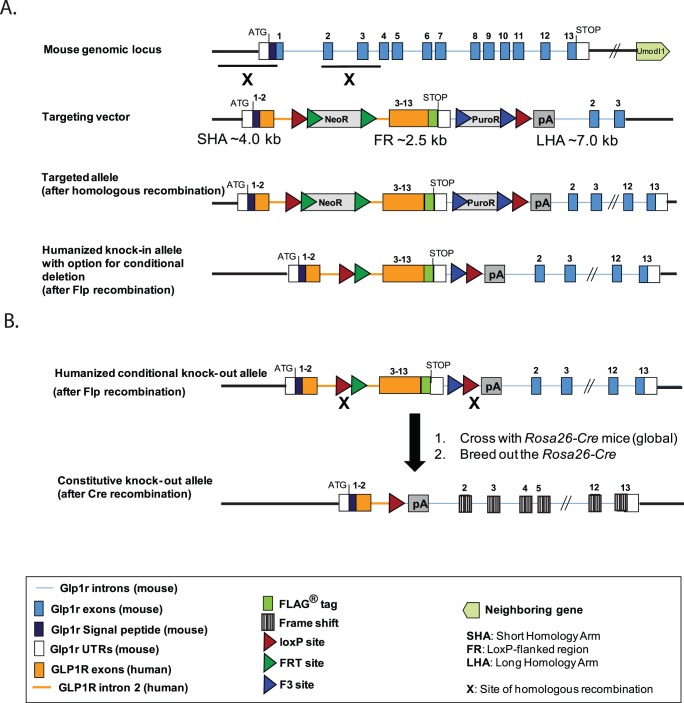
The targeting vector was designed to include several attributes that lend utility and experimental flexibility. The hGLP-1R mouse, generated via homologous recombination, expresses a modified hGLP-1R cDNA containing a C-terminal FLAG epitope ( Figure 1A ). The hGLP-1R is expressed from the murine Glp-1r promoter and endogenous upstream regulatory elements. In an effort to maintain normal processing and intracellular trafficking, the mGlp-1r 5′ UTR, start codon, and signal peptide sequences remain in place and are immediately followed by exon 2 of the hGLP-1R; upon transport to the plasma membrane and subsequent proteolytic cleavage of the murine signal peptide, the mature protein is 100% human GLP-1R with a C-terminal FLAG fusion ( Figure 2B ). The FLAG tag, engineered prior to the stop codon, enables anti-FLAGFLAG antibody methodologies. In order to delete hGLP-1R, flanking LoxP sites were inserted upstream of human exons 3–13 and downstream of the mouse 3′ UTR ( Figure 1A ). Additionally, the polyA signal sequence from the human growth hormone gene was inserted downstream of the distal LoxP site to prevent transcriptional read-through ( Figure 1A ).
Figure 1. Strategies to generate hGLP-1R knock-in and Glp-1r−/− mice.
(A) The targeting vector was injected into ES cells derived from a C57BL/6 line and implanted into BALB/c females, allowing generation of pure C57BL/6 offspring. The targeting construct was designed to insert into the downstream region of exon 1 of the mouse Glp-1r genomic locus. Chimeric male offspring were bred to C57BL/6-Tg (CAG-Flpe)2 Arte females that ubiquitously express Flp recombinase to give rise to the final hGLP-1R mouse line with option for conditional deletion. The humanized allele contains human GLP-1R cDNA with a preserved human intron 2, a region containing potential regulatory elements conserved across species. (B) Glp-1r−/− mice were generated by breeding hGLP-1R animals to Rosa26-Cre-transgenic mice. An additional breeding step was performed to remove the Cre gene, resulting in the Glp-1r−/− line.
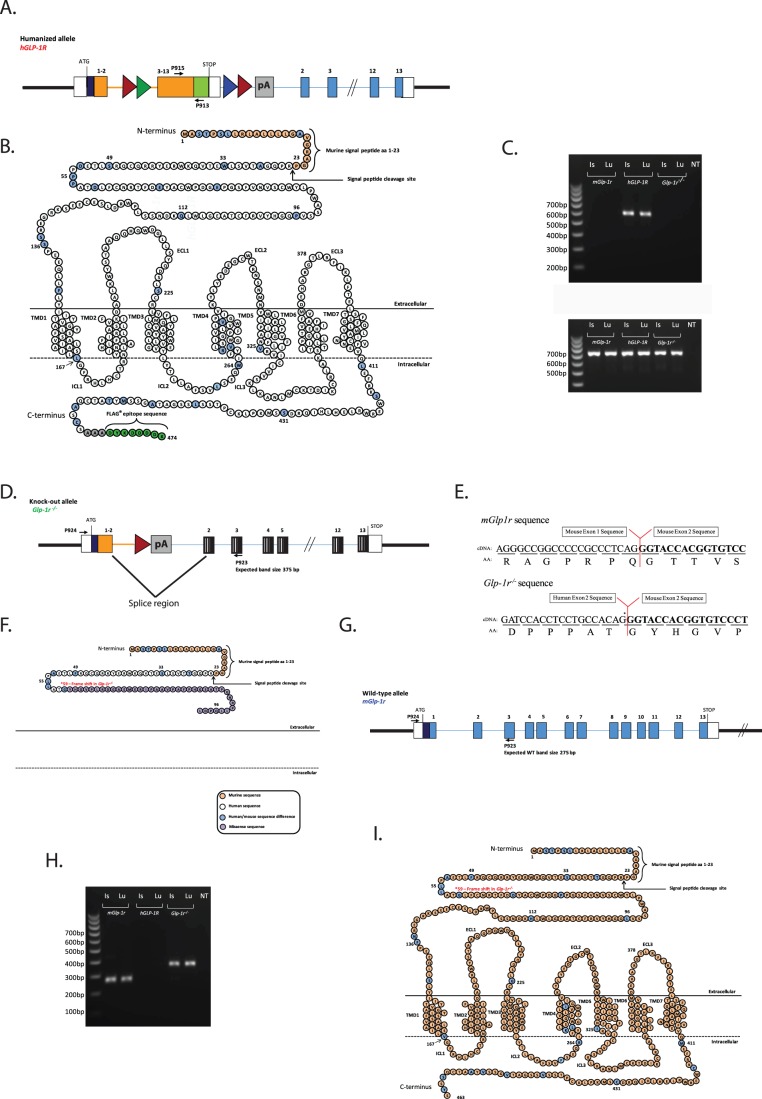
Figure 2. Reverse transcription for PCR validation of hGLP-1R, mGlp-1r and Glp1r−/− lines.
(A) A schematic of the gene that is expressed in the hGLP-1R mice. (B) The protein that is produced from this gene is a fusion of the mouse signal peptide (beige residues) and the human GLP-1R protein (white residues). The blue residues are those that differ between mouse and human GLP-1R. The signaling peptide is cleaved, leaving behind a human GLP-1R protein containing the C-terminal FLAG epitope (green residues). (C) cDNA was generated from total RNA isolated from islet and lung of hGLP-1R, mGlp-1r, and Glp1r−/− mice for RT-PCR. The 5′ P915 primer annealed in human exon 8, while the 3′ P913 primer annealed to the FLAG region, a unique site within the hGLP-1R gene. This PCR product is a 588 bp fragment only observed in the hGLP-1R mice. (D) A schematic of the gene that is expressed in the Glp-1r−/− mice. (E) Once the splice event occurs between human exon 2 and mouse exon 2, a frame-shift mutation nullifies downstream protein expression. (F) The final protein product in Glp-1r−/− mice is a 98-aa truncation mutant. The first 36 aa’s of the mature protein encode a fraction of the GLP-1R extracellular domain, and the remaining 40 aa’s constitute missense sequence that shows no similarity to known proteins. The RT-PCR and DNA sequence analyses of the Glp-1r−/− gene product demonstrate the Glp-1r−/− mouse does not code for a functional GLP-1R. (G) A schematic of the wild-type (mGlp-1r) gene. (H) Using the same primer pair, PCR products from Glp-1r−/− (372 bp) and mGlp-1r (275 bp) mice differ in size by 97 bp. (I) The wild-type GLP-1R protein is 463 aa’s including the signaling peptide.
Once the hGLP-1R mouse line was created, animals were bred with C57BL/6-Gt(ROSA)26Sortm16(Cre) mice for germ-line deletion of hGLP-1R ( Figure 1B ). The Rosa26 locus enables global transgene expression, and Rosa26 Cre mice ubiquitously express the Cre enzyme [31]. Deletion of hGLP-1R results in loss of function by removing most of the coding sequence ( Figure 1B ) and generating a frame-shift mutation in the downstream mouse Glp-1r gene, obviating translation of the remaining mGlp-1r ( Figure 1B ). To remove the Rosa26 Cre locus, Rosa26 Cre:Glp-1r−/− offspring were crossed with wild-type C57BL/6 mice to obtain Glp-1r+/− offspring that were used as breeders to generate all cohorts for these studies. Schematics indicating the GLP-1R aa sequences in each genotype ( Figures 2B, 2F, 2I ) are illustrated similar to the depiction by Doyle and Egan [32].
Total RNA from islets and lung tissue of mGlp-1r, hGLP-1R, and Glp-1r−/− mice was isolated for RT-PCR and sequencing analyses ( Figure 2 ). Expression of the hGLP-1R allele was detected using the primer set P915/P913 that amplifies a region between exon 8 and the FLAG epitope ( Figure 2A ). The resulting product is a single 588 bp band that is only amplified in the hGLP-1R line tissues ( Figure 2C ). The primer set P924/P923 was used to detect mRNA expressed from the mGlp-1r and Glp-1r−/− alleles ( Figure 2C ). By design, this set amplified the region between the murine 5′ UTR and mouse exon 3 and detected the presence of Glp-1r mRNA in both mGlp-1r and Glp1r−/− animals ( Figures 2D and 2G–H ). This primer set amplified a 275 bp band in mGlp-1r cDNA and a 372 bp fragment in cDNA generated from Glp1r−/− mice; the larger amplicon occurred as a result of mRNA processing, shown in Figure 2D. Housekeeping gene 36B4 was detected equally in all genotypes ( Figure 2C ). DNA sequencing of the 372 bp fragment from Glp1r−/− mice confirmed the mRNA transcript expressed in these animals does not code for a functional protein; this transcript does not contain intronic sequence and arises from the splice donor site upstream of the remaining LoxP site and the splice acceptor site located upstream of the intact mouse exon 2 ( Figure 2D ). The size difference between wild-type (mGlp-1r) and Glp-1r−/− sequence is 97 nucleotides, which corresponds to the length of exon 2; the splicing event that generates the fusion of human exon 2 and mouse exon 2 results in a frame-shift early in the Glp-1r open reading frame that gives rise to a predicted 98-aa truncated protein ( Figures 2E–F ).
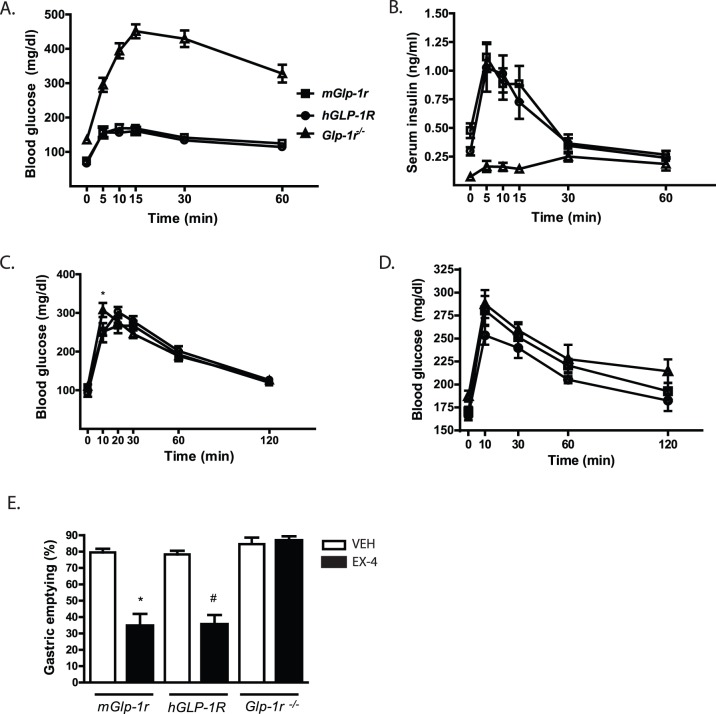
Body weights (mGlp-1r 24.3±0.7; hGLP-1R 22.2±0.4; and Glp-1r−/−23.8±0.8 g) and fasting blood glucose concentrations (mGlp-1r 91.8±9.3; hGLP-1R 106.3±9.9; and Glp-1r−/−110.3±8.6 mg/dl) were similar for each genotype. Because GLP-1R activation helps regulate glucose homeostasis by enhancing postprandial insulin secretion and delaying gastric emptying [33], [34], we performed studies to evaluate these processes. First, an IPGTT was done on all three genotypes to examine the glucose excursion profile 30 minutes after SC EX-4 administration (10 nmol/kg) ( Figure 3A ). As expected, the mGlp-1r and hGLP-1R mice responded with enhanced glucose lowering following EX-4 treatment compared to Glp-1r −/− mice that showed no glucose lowering response to EX-4 during the IPGTT ( Figure 3A ). Glp-1r−/− mice had no insulinotropic response to EX-4 compared to the robust insulin secretion peaks seen in the hGLP-1R and mGlp-1r animals during the IPGTT ( Figure 3B ). Next, OGTTs conducted in mGlp-1r, hGLP-1R, and Glp-1r −/− animals demonstrated no differences in glucose clearance among the groups ( Figure 3C ). However, since lipids and proteins are also potent stimulators of GLP-1 secretion [35], we also conducted a MMTT to assess the impact of GLP-1R ablation on postprandial glucose metabolism ( Figure 3D ). Blood glucose was measured in males given 10 ml/kg BW of Ensure Plus after a 4-hour fast, and glucose excursion during the MMTT did not differ among the three genotypes ( Figure 3D ).
Figure 3. Glucose and mixed meal tolerance, and gastric emptying.
(A) IPGTT. EX-4 (10 nmol/kg SC) was administered 30 minutes before glucose injection (2 g/kg glucose, IP injection). Both mGlp-1r and hGLP-1R mice experienced glucose lowering in response to EX-4, while the Glp-1r−/− mice were refractory to the glucose-lowering effect of EX-4. (B) Insulin secretion during the IPGTT was measured in mGlp-1r, hGLP-1R, and Glp-1r−/− mice. (C) OGTT with 2 g/kg glucose gavage in mGlp-1r, hGLP-1R, and Glp-1r−/− mice after an overnight fast. (D) Glucose excursion and during the MMTT in mGlp-1r, hGLP-1R, and Glp-1r−/− mice after a 4 hour fast (n = 7/group). (E) Gastric emptying in response to EX-4 (10 nmol/kg SC) in mGlp-1r, hGLP-1R, and Glp-1r−/− mice. All animals were fasted overnight and then treated with vehicle (VEH, white bars) or EX-4 (black bars) prior to food intake in order to measure the rate of gastric emptying after a meal. *p<0.001 VEH vs. EX-4 in mGlp-1r mice; #p<0.001 VEH vs. EX-4 in hGLP-1R mice (n = 5–7 per group, one-way ANOVA with Bonferroni post-tests).
Finally, we administered subcutaneous (SC) EX-4 to the three genotypes to assess GLP-1R-mediated gastric emptying. Both mGlp-1r and hGLP-1R mice treated with EX-4 displayed approximately 50% decrease in gastric emptying compared to vehicle-treated animals (PBS) ( Figure 3E ). However, the Glp-1r −/− animals showed no decrease in gastric emptying in response to EX-4. Together, the data show that hGlp1r mice displayed comparable glucose homeostasis to wild-type mGlp-1r mice. Similar to previous reports evaluating Glp-1r −/− mice, the model presented here does not respond to EX-4, consistent with the absence of the GLP-1R [5], [6], [36].
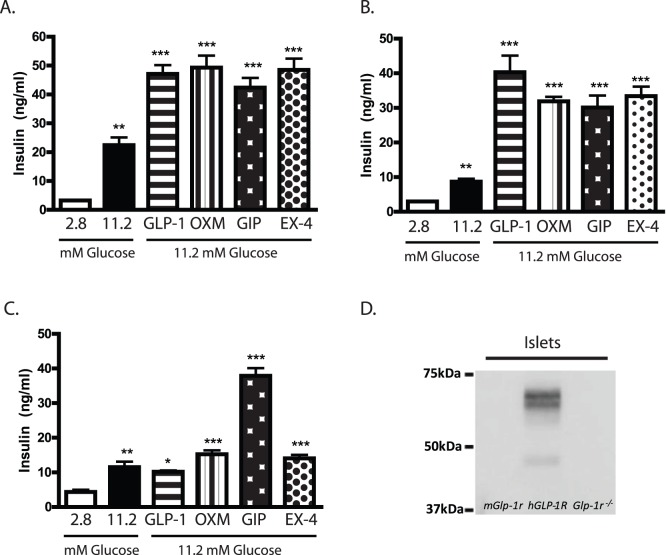
GLP-1R activation in β-cells of pancreatic islets enhances glucose-stimulated insulin secretion. To directly assess GLP-1R-dependent insulin secretion, islets were isolated from mGlp-1r, hGLP-1R, and Glp-1r −/− mice, and static cultures of islets were treated with low or high glucose alone and in combination with GLP-1 (7–36)-NH2, oxyntomodulin (OXM), glucose-dependent insulinotropic peptide (GIP), or EX-4 ( Figures 4A–C ). Insulin concentrations in the medium were measured and compared across groups ( Figures 4A–C ). Both mGlp-1r and hGLP-1R islets showed increased insulin levels in the presence of high glucose ( Figures 4A–B ) and displayed significant increases in insulin secretion in response to high glucose in the presence of GLP-1, OXM, GIP, and EX-4 ( Figures 4A–B ). The Glp-1r −/− islets also displayed glucose-stimulated insulin secretion, but only GIP enhanced insulin release in these cultures, consistent with the specific loss of GLP-1R signaling ( Figure 4C ). Further, the insulinotropic response of Glp-1r−/− islets to GIP was comparable to the effect of GIP in mGlp-1r and hGLP-1R islets, demonstrating that deletion of the GLP-1R does not impact the GIP-GIP receptor pathway ( Figure 4C ). IP-WB using an anti-FLAG antibody confirmed expression of hGLP-1R protein only in islets from hGLP-1R mice; GLP-1R protein migrated to various molecular weights due to glycosylation ( Figure 4D ).
Figure 4. Insulin secretion and GLP-1R protein expression in pancreatic islets.
(A) mGlp-1r and (B) hGLP-1R islets secreted insulin in response to high glucose (11.2 mM) compared to low glucose (2.8 mM) treatment, potentiated by GLP-1, OXM, GIP, and EX-4. (C) Glp-1r−/− islets also secreted insulin in response to high glucose, but no potentiation was observed with GLP-1, OXM, or EX-4 treatment. GIP-induced insulin secretion remained intact in Glp-1r−/− islets. (D) IP-WB of islets from mGlp-1r, hGLP-1R, and Glp-1r−/− mice showed FLAG expression in only the hGLP-1R islets. Hand-selected, size-matched islets were used for all insulin secretion assays, and comparisons are from high (11.2 mM) glucose: **p<0.01; ***p<0.001 (one-way ANOVA with Tukey’s multiple comparison test).
IHC
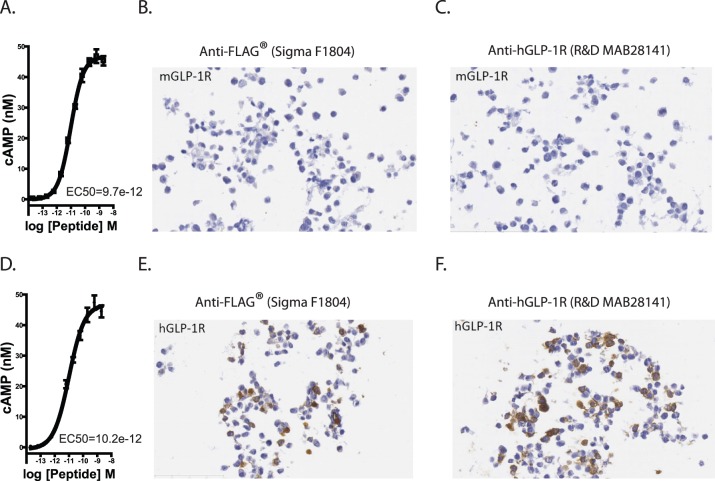
HEK cells were transfected with FLAG-tagged hGLP-1R or untagged, murine Glp-1r constructs to evaluate commercially available GLP-1R and FLAG antibodies. cAMP assays confirmed both expression and activity of mGLP-1R and FLAG-tagged hGLP-1R proteins ( Figures 5A and 5D ). IHC for FLAG showed that mGlp-1r-expressing cells did not stain for FLAG ( Figure 5B ). Furthermore, when these cells were stained with antisera specific for human GLP-1R (R&D MAB 28141), we did not detect any signal, suggesting this antibody does not recognize mGLP-1R ( Figure 5C ). Conversely, hGLP-1R-transfected cells stained strongly for the FLAG epitope ( Figure 5E ), and hGLP-1R was detected by the hGLP-1R-specific antibody from R&D ( Figure 5F ). We evaluated additional GLP-1R antisera (Abcam 39072) which showed only non-specific staining in both mGlp-1r and hGLP-1R tissues (data not shown), consistent with previous work demonstrating non-specific staining of this antibody [16].
Figure 5. Validation of GLP-1R and FLAG antibodies.
(A) mGlp-1r and (D) hGLP-1R were transiently expressed in HEK cells. cAMP accumulation in response to GLP-1 showed similar cAMP accumulation. HEK cells expressing mGLP-1R showed no (B) FLAG or (C) FLAG-tagged hGLP-1R expressing. HEK cells show stained for (E) FLAG and (F) hGLP-1R protein after transient transfection.
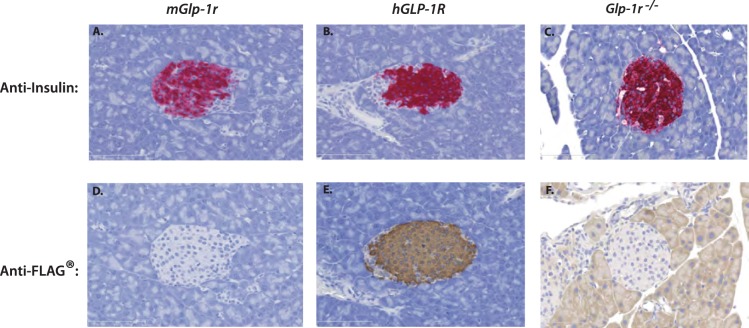
After validation of the anti-FLAG and anti-hGLP-1R antibodies, IHC was performed on pancreata harvested from mGlp-1r, hGLP-1R, and Glp-1r−/− mice. Sections from all animals showed strong insulin staining within the core of islets ( Figures 6A–C ). Staining for FLAG in pancreatic sections of the three genotypes showed strong signal within β-cells of the hGLP-1R mice and none in mGlp-1r or Glp-1r−/− animals ( Figures 6D–F ). Human GLP-1R was present only in the hGLP-1R islets, and therefore, the anti-hGLP-1R antibody did not show staining in mGlp1-r or Glp-1r−/− islets ( Figures 7A–C ). Upon higher magnification, hGLP-1R was observed at the plasma membrane of β-cells in hGLP-1R islets, confirming proper sub-cellular localization ( Figures 7E–G ). We also performed IHC on human pancreas with the hGLP-1R-specific antibody and showed GLP-1R expression within the islets and its localization at the plasma membrane ( Figures 7D and H ).
Figure 6. IHC of pancreata from mGlp-1r, hGLP-1R, and Glp-1r−/− mice.
Pancreata were stained for insulin and showed positive staining in the β-cells of (A) mGlp-1r, (B) hGLP-1R, and (C) Glp-1r−/− islets. FLAG staining was also performed and (E) hGLP-1R islets stained positive for FLAG with none detected in (D) mGlp-1r and (F) Glp-1r−/− islets.
Figure 7. IHC for human GLP-1R expression.
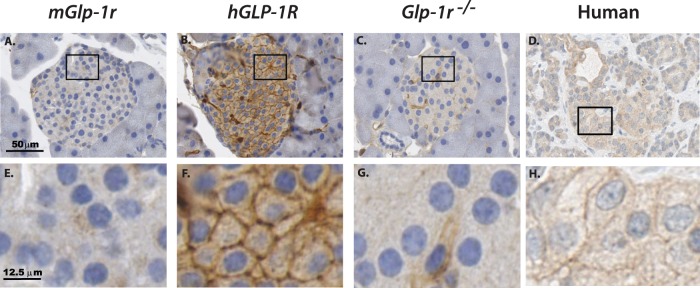
Islets from (A) mGlp-1r, (B) hGLP-1R, (C) Glp-1r−/− mice and (D) human pancreas were stained using an antibody specific for human GLP-1R, and only the (B) hGLP-1R islets and (D) human pancreas showed positive signal. Increased magnification showed that while (E) mGlp-1r and (G) Glp-1r−/− islets display no hGLP-1R expression, the (F) hGLP-1R islets and (H) human islets show plasma membrane staining for the human GLP-1R.
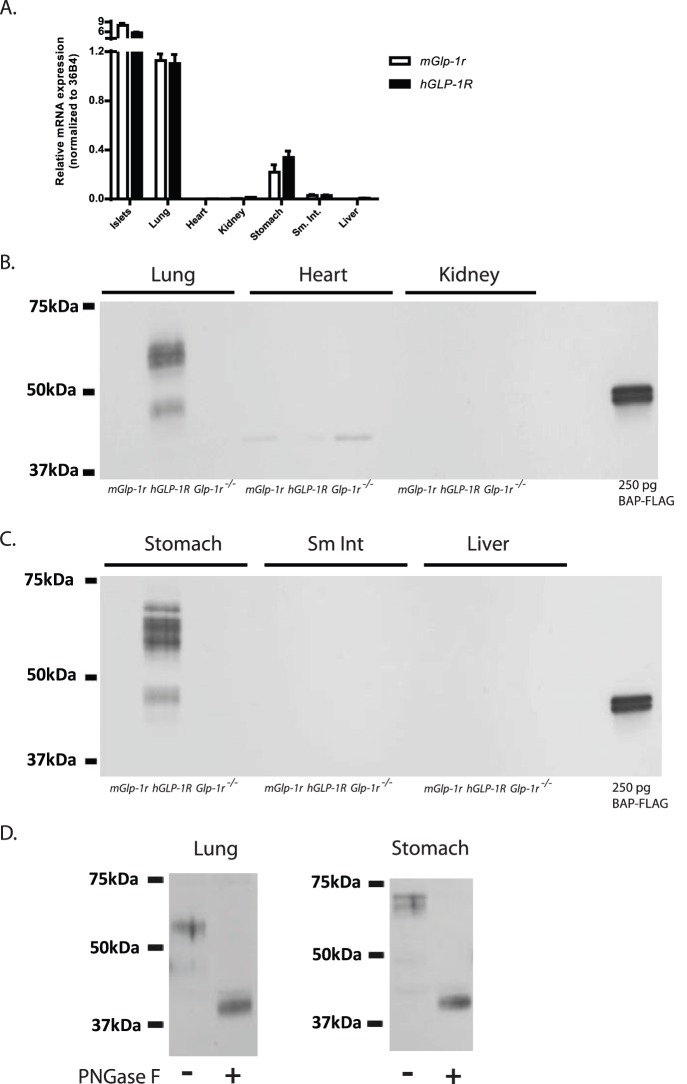
In order to ascertain the relative levels of GLP-1R mRNA and protein expression in vivo, the following tissues were harvested from mGlp-1r, hGLP-1R, and Glp-1r−/− mice: heart, islets, lung, kidney, stomach, small intestine, and liver ( Figure 8A ). qPCR showed abundant GLP-1R mRNA in islets, lung, and stomach, with similar expression levels observed in both hGLP-1R and mGlp-1r tissues ( Figure 8A ). qPCR analyses of both hypothalamus and brain stem of mGlp-1r and hGLP-1R mice revealed moderate GLP-1R mRNA expression, with lower expression detected in cerebral cortex and pituitary (data not shown). We also detected low GLP-1R mRNA expression in heart tissue ( Figure 8A ).
Figure 8. GLP-1R mRNA and protein expression in vivo.
(A) qPCR analyses showed abundant expression in islets, lung, and stomach in both mGlp-1r and hGLP-1R mice. (B)–(C) Western blots using anti-FLAG affinity purification and immunoblotting showed abundant expression of the FLAG-tagged human GLP-1R in the lung and stomach of the hGLP-1R mice. Moreover, there was no FLAG-tagged GLP-1R detected in whole heart, kidney, small intestine (sm. int.), or liver of hGLP-1R mice. Using the FLAG signal as a surrogate for GLP-1R protein, no expression was observed for the receptor in Glp-1r−/− tissues, indicating that LoxP-mediated deletion of the hGLP-1R gene was successful. Purified bacterial alkaline phosphatase (BAP)-FLAG was included in gels as a control for the FLAG antibody. (D) Treatment of samples with PNGase F caused the multiple bands to collapse together from GLP-1R deglycosylation, resulting in a band of smaller molecular weight than the glycosylated form of the receptor.
We first performed traditional WBs on tissue lysates from mGlp-1r, hGLP-1R, and Glp-1r−/− with an anti-FLAG antibody, but a specific signal at the predicted molecular weight for GLP-1R was not detected (data not shown). Therefore, an IP-WB method was implemented using anti-FLAG antibody-conjugated magnetic beads to first pull down the FLAG-tagged protein, followed by cross-blot using a different anti-FLAG antibody ( Figures 4D , 8B–D ). Similar to the hGLP-1R islets ( Figure 4D ), lung and stomach tissue ( Figure 8D ) showed bands of varying molecular weight that likely represent glycosylated forms of GLP-1R [37]–[39].
Using two monoclonal anti-FLAG antibodies to detect the presence of the GLP-1R, we showed that the most abundant expression of GLP-1R occurs in islets, lung, and stomach ( Figures 4D and 8B–C ). The Glp-1r−/− tissues did not show FLAG expression, confirming the absence of functional GLP-1R protein. FLAG-tagged GLP-1R was not detected in whole heart, kidney, small intestine, or liver of hGLP-1R mice, indicating that GLP-1R protein expression is absent or below the limit of detection of our anti-FLAG method ( Figures 8B–C ). The three genotypes show a non-specific band in heart ( Figure 8B ). In addition to islets, the glycosylated forms of GLP-1R were also present in both lung and stomach of the hGLP-1R mice; incubation of PNGase F with these samples collapsed the banding pattern, giving rise to a single band of lower molecular weight ( Figure 8D ).
Discussion
In this study, we show that replacement of the murine GLP-1R with human GLP-1R results in a similar metabolic phenotype. Ex vivo assays of islets isolated from both genotypes demonstrate comparable insulinotropic responses to glucose, as well as enhancement of GLP-1-, GIP-, OXM-, and EX-4-stimulated insulin secretion. Further, the targeting construct used for the knock-in model enabled creation of Glp-1r−/− mice derived via Cre-mediated deletion of the hGLP-1R. Generation of whole-body Glp-1r−/− mice by Rosa26Cre-mediated excision confirms utility of the LoxP sites incorporated within the knock-in locus, a feature that will allow conditional gene deletion strategies in the future. Characterization of this Glp-1r−/− model demonstrates loss of GLP-1R-dependent insulin secretion and gastric emptying; these animals display no response in glucose lowering after EX-4 treatment during IPGTTs, consistent with the established role of the GLP-1R [1], [33].
The emerging controversy surrounding the safety implications of GLP-1 analogues and DPP4 inhibitors [40]–[42] underscores the importance of a thorough characterization of GLP-1 biology. Establishing another Glp-1r −/− model using a different gene deletion strategy is important in fully characterizing GLP-1R function, especially because the targeting approach used for the first model allowed expression of a 186 aa form of GLP-1R [16], [43]. Data from experiments evaluating our new Glp-1r −/− model vary slightly with the published results characterizing the initial Glp-1r null mouse [43]. The first global Glp-1r −/− mouse, reported by Scrocchi et al, showed that whole-body Glp-1r−/− mice generated on the CD1 background had higher basal fasting glucose levels and impaired oral glucose tolerance compared to wild-type littermates [43]. A subsequent study describing the double incretin knock-out (DIRKO) mice showed that male Glp-1r−/− mice are only mildly glucose intolerant while females exhibited normal glucose tolerance after being backcrossed for five generations onto the C57BL/6 strain [5]. In 2013, Wilson-Peréz et al reported another Glp-1r−/− mouse that was also generated by Cre-LoxP-mediated gene deletion on the C57BL/6 background, and showed that on a high-fat diet, loss of the Glp-1r impairs glucose clearance and insulin secretion in response to a mixed meal challenge; however, the authors did not report findings on intraperitoneal or oral glucose tolerance tests [29]. Our studies using the newly generated male Glp-1r−/− mice show similar oral glucose and mixed meal tolerance compared to mGlp-1r mice fed a chow diet, consistent with the phenotype observed in female Glp-1r−/− mice reported by Hansotia et al [5]. Further, the lack of EX-4-induced insulin secretion in the Glp-1r−/− mice during the IPGTT, as well as the absence of GLP-1R protein and hGLP-1R mRNA, confirm complete Glp-1r ablation in the Glp-1r−/− mouse model reported here. Subtle differences in glucose metabolism between Glp-1r−/− mice in our study and those reported previously [5], [29] may be attributable to other factors such as differences in diets or housing conditions.
Recent work aimed at studying GLP-1R function in various tissues revealed concerns about data generated using the existing commercial antisera raised against rodent and human GLP-1R due to the inability of antibodies to specifically detect GLP-1R protein [16], [24]. Panjwani et al tested three antibodies using wild-type and Glp-1r−/− lung tissue extracts, and each failed to detect GLP-1R [16]. In this regard, inclusion of a FLAG epitope at the C-terminus of the hGLP-1R gene in the knock-in construct allows use of well-characterized anti-FLAG antibodies to examine tissue expression of GLP-1R protein. Further, the work here shows an R&D Systems antibody specifically detects human GLP-1R protein via IHC in HEK cells expressing hGLP-1R as well as in islets of the hGLP-1R knock-in mice and human pancreas. Consistent with these results, FLAG staining shows positive signal in islets from hGLP-1R but not in tissues from mGlp-1r or Glp-1r−/− mice.
The use of whole-body Glp-1r−/− mice and GLP-1R agonists such as EX-4 have helped establish many of the physiological roles of the GLP-1R, but determining the whole-body expression pattern of GLP-1R protein is needed to understand specific actions of GLP-1 on various organ systems. Glp-1r mRNA expression has been reported in the pancreas, brain, stomach, lung, small intestine, and colon of rats and mice [21]–[23]. RT-PCR/Southern blot analyses indicates lung has particularly high Glp-1r mRNA levels, as well as the body of the stomach, the small intestine, and the pancreas [23], while whole heart, kidney, and liver have extremely low or nearly undetectable levels of Glp-1r mRNA [22], [23]. The mRNA data presented here are consistent with previous reports on islets, lung, and stomach as being highly enriched for GLP-1R, and our results are further substantiated by western analyses showing strong GLP-1R protein signal in islets, lung, and stomach. We also found low Glp-1r mRNA expression in heart, which supports recent findings on the role of GLP-1R in atrial function [11]. Due to the lack of anti-GLP-1R antisera for immunoblotting, the western results are only possible because of the anti-FLAG technique employed here. Strategies incorporating use of FLAG-tagged proteins for in vivo tissue expression analyses have been used previously [44]–[46]. However, most of these efforts used transgenic systems that promote high expression of tagged proteins. Our humanized mouse model allows investigations of the tissue-specific expression patterns of the GLP-1R under the normal control of the murine Glp-1r locus. In this light, it is important to note that an affinity purification step was required to enrich for the GLP-1R protein before the western blot. Detecting GLP-1R in islets, lung, and stomach in hGLP-1R but not in Glp-1r−/− tissues emphasizes the need to routinely employ rigorous controls when attempting to draw conclusions based on western blot data from poorly characterized antibodies. For example, while some reports showing GLP-1R protein expression in pancreatic β-cells from both mouse and rat have used Glp-1r−/− mouse tissues as negative controls [47], other reports (using different antibodies) claiming GLP-1R in lung, cardiac, and vascular tissues have not been similarly controlled [48]. GLP-1R protein has also been detected using WB in pig kidney tubules, but again, in a study that lacked thorough validation of the GLP-1R antibody [49]. Our humanized mouse model will allow a more comprehensive investigation of expression patterns of the GLP-1R, and thus, may shed more light into its functions in extra-pancreatic tissues.
Although much is now known about the role of GLP-1 in controlling glucose metabolism, improving our understanding of the molecular mechanisms that regulate GLP-1R function in β-cells and other tissues may enable development of improved GLP-1R-based therapies. An emerging area in GPCR biology is identifying partner or accessory proteins and understanding how GPCR interacting proteins help control signaling. Studies using in vitro systems incorporating GPCRs tagged with FLAG or HA have identified interacting protein partners [50]–[52]; tagging the GLP-1R in vivo with FLAG may allow similar approaches. Thus, the FLAG epitope should enable biochemical study of the GLP-1R through IP and mass spectrometry analyses. In summary, we present the first humanized GLP-1R knock-in mouse, a model that allows the exploration of incretin biology of the hGLP-1R in mice. The Glp-1r−/− mouse generated here offers a complementary tool with which to further characterize GLP-1 biology. Together, the hGLP-1R and Glp-1r−/− mice will enable targeted biochemical study and broad tissue expression analysis of GLP-1R protein.
Supporting Information
Genotyping strategy and primer sets for wild-type (mGlp-1r) and humanized knock-in (hGLP-1R) lines. The mGlp-1r PCR product is a single band of 138 bp; the heterozygous PCR product shows the wild-type band (138 bp) and the hGLP-1R band of 482 bp. The hGLP-1R PCR product is a single 482 bp band.
(EPS)
Genotyping strategy and primer sets for the Glp-1r−/− line. The Glp-1r−/− line shows a single 221 bp band. The wild-type band of 138 bp with the 221 bp Glp-1r−/− band denotes Glp-1+/− mice.
(EPS)
Primer and probe sequences used in this study (5′–3′).
(EPS)
Acknowledgments
The authors thank Ralph Gareus, Gloria Esposito and the staff at TaconicArtemis GmbH in Cologne, Germany, and Steve Rashbaum of Taconic in Hudson City, New York, USA for their assistance in generating the mice used in this study. The authors also thank Tao Wei (Bioinformatics) and Daniel A. Briere, C. Parker Siddall, Mark R. Wade, Brian A. Droz, Thomas B. Farb, and Steven M. Bauer (Endocrine Discovery) of Lilly Research Laboratories for their assistance in carrying out the studies described herein.
Funding Statement
This work was supported by Eli Lilly and Company and the Lilly Research Awards Program (LRAP). The LRAP grant enabled immunohistochemistry analyses of the mouse and human pancreas tissues. The funders had no other role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Holst JJ (2007) The physiology of glucagon-like peptide 1. Physiol Rev 87: 1409–1439. [DOI] [PubMed] [Google Scholar]
- 2. Estall JL, Drucker DJ (2006) Glucagon and glucagon-like peptide receptors as drug targets. Curr Pharm Des 12: 1731–1750. [DOI] [PubMed] [Google Scholar]
- 3. Nauck MA, Kleine N, Orskov C, Holst JJ, Willms B, et al. (1993) Normalization of fasting hyperglycaemia by exogenous glucagon-like peptide 1 (7–36 amide) in type 2 (non-insulin-dependent) diabetic patients. Diabetologia 36: 741–744. [DOI] [PubMed] [Google Scholar]
- 4. Thorens B (1992) Expression cloning of the pancreatic beta cell receptor for the gluco-incretin hormone glucagon-like peptide 1. Proc Natl Acad Sci U S A 89: 8641–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansotia T, Baggio LL, Delmeire D, Hinke SA, Yamada Y, et al. (2004) Double incretin receptor knockout (DIRKO) mice reveal an essential role for the enteroinsular axis in transducing the glucoregulatory actions of DPP-IV inhibitors. Diabetes 53: 1326–1335. [DOI] [PubMed] [Google Scholar]
- 6. Lamont BJ, Li Y, Kwan E, Brown TJ, Gaisano H, et al. (2012) Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. J Clin Invest 122: 388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chia CW, Egan JM (2008) Incretin-based therapies in type 2 diabetes mellitus. J Clin Endocrinol Metab 93: 3703–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monami M, Marchionni N, Mannucci E (2009) Glucagon-like peptide-1 receptor agonists in type 2 diabetes: a meta-analysis of randomized clinical trials. Eur J Endocrinol 160: 909–917. [DOI] [PubMed] [Google Scholar]
- 9. Halbirk M, Norrelund H, Moller N, Holst JJ, Schmitz O, et al. (2010) Cardiovascular and metabolic effects of 48-h glucagon-like peptide-1 infusion in compensated chronic patients with heart failure. Am J Physiol Heart Circ Physiol 298: H1096–1102. [DOI] [PubMed] [Google Scholar]
- 10. Mendis B, Simpson E, MacDonald I, Mansell P (2012) Investigation of the haemodynamic effects of exenatide in healthy male subjects. Br J Clin Pharmacol 74: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M, Platt MJ, Shibasaki T, Quaggin SE, Backx PH, et al.. (2013) GLP-1 receptor activation and Epac2 link atrial natriuretic peptide secretion to control of blood pressure. Nat Med. [DOI] [PubMed]
- 12. Korner M, Stockli M, Waser B, Reubi JC (2007) GLP-1 receptor expression in human tumors and human normal tissues: potential for in vivo targeting. J Nucl Med 48: 736–743. [DOI] [PubMed] [Google Scholar]
- 13. Vara E, Arias-Diaz J, Garcia C, Balibrea JL, Blazquez E (2001) Glucagon-like peptide-1(7–36) amide stimulates surfactant secretion in human type II pneumocytes. Am J Respir Crit Care Med 163: 840–846. [DOI] [PubMed] [Google Scholar]
- 14. Benito E, Blazquez E, Bosch MA (1998) Glucagon-like peptide-1-(7–36)amide increases pulmonary surfactant secretion through a cyclic adenosine 3′,5′-monophosphate-dependent protein kinase mechanism in rat type II pneumocytes. Endocrinology 139: 2363–2368. [DOI] [PubMed] [Google Scholar]
- 15. Richter G, Feddersen O, Wagner U, Barth P, Goke R, et al. (1993) GLP-1 stimulates secretion of macromolecules from airways and relaxes pulmonary artery. Am J Physiol 265: L374–381. [DOI] [PubMed] [Google Scholar]
- 16. Panjwani N, Mulvihill EE, Longuet C, Yusta B, Campbell JE, et al. (2013) GLP-1 Receptor Activation Indirectly Reduces Hepatic Lipid Accumulation But Does Not Attenuate Development of Atherosclerosis in Diabetic Male ApoE−/− Mice. Endocrinology 154: 127–139. [DOI] [PubMed] [Google Scholar]
- 17. Turton MD, O’Shea D, Gunn I, Beak SA, Edwards CM, et al. (1996) A role for glucagon-like peptide-1 in the central regulation of feeding. Nature 379: 69–72. [DOI] [PubMed] [Google Scholar]
- 18. Tushuizen ME, Bunck MC, Pouwels PJ, van Waesberghe JH, Diamant M, et al. (2006) Incretin mimetics as a novel therapeutic option for hepatic steatosis. Liver Int 26: 1015–1017. [DOI] [PubMed] [Google Scholar]
- 19. Gier B, Butler PC, Lai CK, Kirakossian D, DeNicola MM, et al. (2012) Glucagon like peptide-1 receptor expression in the human thyroid gland. J Clin Endocrinol Metab 97: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bjerre Knudsen L, Madsen LW, Andersen S, Almholt K, de Boer AS, et al. (2010) Glucagon-like Peptide-1 receptor agonists activate rodent thyroid C-cells causing calcitonin release and C-cell proliferation. Endocrinology 151: 1473–1486. [DOI] [PubMed] [Google Scholar]
- 21. Bullock BP, Heller RS, Habener JF (1996) Tissue distribution of messenger ribonucleic acid encoding the rat glucagon-like peptide-1 receptor. Endocrinology 137: 2968–2978. [DOI] [PubMed] [Google Scholar]
- 22. Campos RV, Lee YC, Drucker DJ (1994) Divergent tissue-specific and developmental expression of receptors for glucagon and glucagon-like peptide-1 in the mouse. Endocrinology 134: 2156–2164. [DOI] [PubMed] [Google Scholar]
- 23. Dunphy JL, Taylor RG, Fuller PJ (1998) Tissue distribution of rat glucagon receptor and GLP-1 receptor gene expression. Mol Cell Endocrinol 141: 179–186. [DOI] [PubMed] [Google Scholar]
- 24. Pyke C, Knudsen LB (2013) The Glucagon-Like Peptide-1 Receptor–or Not? Endocrinology 154: 4–8. [DOI] [PubMed] [Google Scholar]
- 25. Schwenk F, Baron U, Rajewsky K (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23: 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otto C, Fuchs I, Kauselmann G, Kern H, Zevnik B, et al. (2009) GPR30 does not mediate estrogenic responses in reproductive organs in mice. Biol Reprod 80: 34–41. [DOI] [PubMed] [Google Scholar]
- 27. Sloop KW, Cao JX, Siesky AM, Zhang HY, Bodenmiller DM, et al. (2004) Hepatic and glucagon-like peptide-1-mediated reversal of diabetes by glucagon receptor antisense oligonucleotide inhibitors. J Clin Invest 113: 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sloop KW, Willard FS, Brenner MB, Ficorilli J, Valasek K, et al. (2010) Novel small molecule glucagon-like peptide-1 receptor agonist stimulates insulin secretion in rodents and from human islets. Diabetes 59: 3099–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson-Perez HE, Chambers AP, Ryan KK, Li B, Sandoval DA, et al. (2013) Vertical sleeve gastrectomy is effective in two genetic mouse models of glucagon-like Peptide 1 receptor deficiency. Diabetes 62: 2380–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kojima K, Bowersock GJ, Kojima C, Klug CA, Grizzle WE, et al. (2012) Validation of a robust proteomic analysis carried out on formalin-fixed paraffin-embedded tissues of the pancreas obtained from mouse and human. Proteomics 12: 3393–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soriano P (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71. [DOI] [PubMed] [Google Scholar]
- 32. Doyle ME, Egan JM (2007) Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol Ther 113: 546–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drucker DJ (2006) The biology of incretin hormones. Cell Metab 3: 153–165. [DOI] [PubMed] [Google Scholar]
- 34. Imeryuz N, Yegen BC, Bozkurt A, Coskun T, Villanueva-Penacarrillo ML, et al. (1997) Glucagon-like peptide-1 inhibits gastric emptying via vagal afferent-mediated central mechanisms. Am J Physiol 273: G920–927. [DOI] [PubMed] [Google Scholar]
- 35. Baggio LL, Drucker DJ (2007) Biology of incretins: GLP-1 and GIP. Gastroenterology 132: 2131–2157. [DOI] [PubMed] [Google Scholar]
- 36. Hansotia T, Drucker DJ (2005) GIP and GLP-1 as incretin hormones: lessons from single and double incretin receptor knockout mice. Regul Pept 128: 125–134. [DOI] [PubMed] [Google Scholar]
- 37. Chen Q, Miller LJ, Dong M (2010) Role of N-linked glycosylation in biosynthesis, trafficking, and function of the human glucagon-like peptide 1 receptor. Am J Physiol Endocrinol Metab 299: E62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goke R, Just R, Lankat-Buttgereit B, Goke B (1994) Glycosylation of the GLP-1 receptor is a prerequisite for regular receptor function. Peptides 15: 675–681. [DOI] [PubMed] [Google Scholar]
- 39. Whitaker GM, Lynn FC, McIntosh CH, Accili EA (2012) Regulation of GIP and GLP1 receptor cell surface expression by N-glycosylation and receptor heteromerization. PLoS One 7: e32675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler AE, Campbell-Thompson M, Gurlo T, Dawson DW, Atkinson M, et al.. (2013) Marked Expansion of Exocrine and Endocrine Pancreas with Incretin Therapy in Humans with increased Exocrine Pancreas Dysplasia and the potential for Glucagon-producing Neuroendocrine Tumors. Diabetes. [DOI] [PMC free article] [PubMed]
- 41. Heine RJ, Fu H, Kendall DM, Moller DE (2013) Comment on: Butler et al. Marked Expansion of Exocrine and Endocrine Pancreas With Incretin Therapy in Humans With Increased Exocrine Pancreas Dysplasia and the Potential for Glucagon-Producing Neuroendocrine Tumors. Diabetes 2013; 62: 2595–2604. Diabetes 62: e16–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Butler AE, Campbell-Thompson M, Gurlo T, Dawson DW, Atkinson M, et al. (2013) Reponse to Comments on: Butler et al. Marked Expansion of Exocrine and Endocrine Pancreas With Incretin Therapy in Humans With Increased Exocrine Pancreas Dysplasia and the Potential for Glucagon-Producing Neuroendocrine Tumors. Diabetes 2013; 62: 2595–2604. Diabetes 62: e19–e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scrocchi LA, Brown TJ, MaClusky N, Brubaker PL, Auerbach AB, et al. (1996) Glucose intolerance but normal satiety in mice with a null mutation in the glucagon-like peptide 1 receptor gene. Nat Med 2: 1254–1258. [DOI] [PubMed] [Google Scholar]
- 44. Lobbestael E, Reumers V, Ibrahimi A, Paesen K, Thiry I, et al. (2010) Immunohistochemical detection of transgene expression in the brain using small epitope tags. BMC Biotechnol 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Romani-Perez M, Outeirino-Iglesias V, Gil-Lozano M, Gonzalez-Matias LC, Mallo F, et al. (2013) Pulmonary GLP-1 Receptor Increases at Birth and Exogenous GLP-1 Receptor Agonists Augmented Surfactant-Protein Levels in Litters From Normal and Nitrofen-Treated Pregnant Rats. Endocrinology 154: 1144–1155. [DOI] [PubMed] [Google Scholar]
- 46. Suzuki H, Dann CT, Rajkovic A (2013) Generation of a germ cell-specific mouse transgenic CHERRY reporter, Sohlh1-mCherryFlag. Genesis 51: 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tornehave D, Kristensen P, Romer J, Knudsen LB, Heller RS (2008) Expression of the GLP-1 receptor in mouse, rat, and human pancreas. J Histochem Cytochem 56: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ban K, Noyan-Ashraf MH, Hoefer J, Bolz SS, Drucker DJ, et al. (2008) Cardioprotective and vasodilatory actions of glucagon-like peptide 1 receptor are mediated through both glucagon-like peptide 1 receptor-dependent and -independent pathways. Circulation 117: 2340–2350. [DOI] [PubMed] [Google Scholar]
- 49. Schlatter P, Beglinger C, Drewe J, Gutmann H (2007) Glucagon-like peptide 1 receptor expression in primary porcine proximal tubular cells. Regul Pept 141: 120–128. [DOI] [PubMed] [Google Scholar]
- 50. Maurel D, Comps-Agrar L, Brock C, Rives ML, Bourrier E, et al. (2008) Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat Methods 5: 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Persani L, Calebiro D, Bonomi M (2007) Technology Insight: modern methods to monitor protein-protein interactions reveal functional TSH receptor oligomerization. Nat Clin Pract Endocrinol Metab 3: 180–190. [DOI] [PubMed] [Google Scholar]
- 52. Zvonok N, Yaddanapudi S, Williams J, Dai S, Dong K, et al. (2007) Comprehensive proteomic mass spectrometric characterization of human cannabinoid CB2 receptor. J Proteome Res 6: 2068–2079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotyping strategy and primer sets for wild-type (mGlp-1r) and humanized knock-in (hGLP-1R) lines. The mGlp-1r PCR product is a single band of 138 bp; the heterozygous PCR product shows the wild-type band (138 bp) and the hGLP-1R band of 482 bp. The hGLP-1R PCR product is a single 482 bp band.
(EPS)
Genotyping strategy and primer sets for the Glp-1r−/− line. The Glp-1r−/− line shows a single 221 bp band. The wild-type band of 138 bp with the 221 bp Glp-1r−/− band denotes Glp-1+/− mice.
(EPS)
Primer and probe sequences used in this study (5′–3′).
(EPS)