Abstract
Integrative analyses of multiple genomic datasets for selected samples can provide better insight into the overall data and can enhance our knowledge of cancer. The objective of this study was to elucidate the association between copy number variation (CNV) and gene expression in colorectal cancer (CRC) samples and their corresponding non-cancerous tissues. Sixty-four paired CRC samples from the same patients were subjected to CNV profiling using the Illumina HumanOmni1-Quad assay, and validation was performed using multiplex ligation probe amplification method. Genome-wide expression profiling was performed on 15 paired samples from the same group of patients using the Affymetrix Human Gene 1.0 ST array. Significant genes obtained from both array results were then overlapped. To identify molecular pathways, the data were mapped to the KEGG database. Whole genome CNV analysis that compared primary tumor and non-cancerous epithelium revealed gains in 1638 genes and losses in 36 genes. Significant gains were mostly found in chromosome 20 at position 20q12 with a frequency of 45.31% in tumor samples. Examples of genes that were associated at this cytoband were PTPRT, EMILIN3 and CHD6. The highest number of losses was detected at chromosome 8, position 8p23.2 with 17.19% occurrence in all tumor samples. Among the genes found at this cytoband were CSMD1 and DLC1. Genome-wide expression profiling showed 709 genes to be up-regulated and 699 genes to be down-regulated in CRC compared to non-cancerous samples. Integration of these two datasets identified 56 overlapping genes, which were located in chromosomes 8, 20 and 22. MLPA confirmed that the CRC samples had the highest gains in chromosome 20 compared to the reference samples. Interpretation of the CNV data in the context of the transcriptome via integrative analyses may provide more in-depth knowledge of the genomic landscape of CRC.
Introduction
Colorectal cancer is a major health concern, with more than a million individuals diagnosed every year worldwide [1]. This cancer is among the top three of all cancers that lead to death worldwide [2]. In Malaysia, it ranks as the second most common cancer in both sexes [3].
One form of genetic instability that is observed in at least 85% of sporadic CRC cases is chromosomal instability (CIN) [4]. Aneuploidy is a consequence of CIN that leads to the gain or loss of whole or parts of chromosomal regions [5], and it may cause structural complexity that leads to genomic instability. One common form of structural variants due to CIN is known as copy number variations (CNVs), which is defined as a gain or loss of copies of DNA segments that are larger than 1 kb in length when compared to a reference genome [6]. CNVs can affect gene expression and have been associated with disease susceptibility. It has been suggested that transcriptional changes correspond to CNVs and alterations in gene dosage can be correlated with changes in expression level [7].
Thousands of CNV sites have been documented using microarray technology. Previous studies on colorectal cancers have revealed gains at chromosome 8q, 13 and 20q and losses at chromosome 8p, 17p and 18q [8], [9], [10], [11], [12]. These aberrations lead to the deletion or amplification of tumor suppressor genes, oncogenes, or non-coding RNAs such as miRNAs, which result in aberrant expression of genes that affect cancer-related biological processes [13], [14].
A gene that has one duplicated allele (a copy number of 3) has a higher level of expression than the wild-type [15]. Conversely, a gene that has one allele deleted (a copy number of 1) will have a lower level of expression [15]. Integrative analyses in CRC showed that expression levels of certain oncogenes and tumor suppressor genes were related to CNV [16]. For example, amplification or gain of the MYC gene at position 8q24 results in over-expression of this gene in CRC. Furthermore, deletion or loss of the APC gene at position 5q21 leads to its deregulated expression in CRC [17]. Similar patterns of correlation have also been observed in breast and lung cancers. Approximately 12% of changes in gene expression levels were reported to be in concordance with copy number in breast cancer [18], and approximately 78% genes showed a positive correlation between CNV and gene expression level in a lung cancer study [19].
The goal of this study was to obtain an insight into the molecular mechanisms of CRC via the analyses of CNV and genome-wide expression profiling of a primary tumor and its corresponding non-cancerous colonic epithelium. We also wanted to identify the relationship between the CNV profile and the transcriptome in CRC.
Materials and Methods
Patient recruitment
The study was performed with approval from the ethics committee of Universiti Kebangsaan Malaysia (UKM 1.5.3.5/244/UMBI-004-2012), and written informed consent was taken from each of the 64 patients who underwent surgery. None of the patients had received chemotherapy or radiotherapy treatment prior to surgery. Primary tumor and non-cancerous tissues (10 cm away from the tumor) were obtained immediately after surgery and snap-frozen in liquid nitrogen for storage.
Genomic DNA and RNA isolation
Frozen sectioning was performed on all samples using a cutting thickness of 5 to 7 µm. The sections were mounted onto glass slides and stained with hematoxylin and eosin (H&E). The stained slides were then evaluated by a histopathologist to confirm the presence (>80% cancer cells) or absence of tumor cells and their corresponding non-cancerous cells. DNA was isolated using the Qiagen DNeasy Blood and Tissue Kit according to the manufacturer's protocol. Total RNA was extracted from 15 paired samples using the Qiagen QIAmp Mini Plus Kit. The quality of the isolated DNA and RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and only samples with a purity of 1.8 to 2.1 (A260/A280) were selected. The integrity of the isolated DNA samples was evaluated on 1.0% agarose gels, and the integrity of the isolated RNA was determined using a Bioanalyzer (Agilent Technologies, CA, USA). Samples with a RNA Integrity Number (RIN) of 6.0 and above were selected for gene expression profiling.
CNV and gene expression profiling
All 64 paired samples were assayed using the Illumina HumanOmni1-Quad Bead Chip, which contains 1,140,419 single nucleotide polymorphism (SNP) loci, based on the Illumina Infinium II assay protocol (Illumina, San Diego, CA, USA). Gene expression profiling was performed on the 15 paired samples using the Affymetrix GeneChip Human Gene 1.0 ST array, which contains 36,079 transcripts with 28,869 well-annotated genes (Affymetrix Inc., Santa Clara, CA, USA). The RNA was prepared using the Applause WT-Amp ST System protocol before the hybridization process (NuGen Technologies Inc., San Carlos, CA, USA). The arrays were stained and scanned based on the GeneChip Whole Transcript (WT) Sense Target Labeling Assay protocol that was outlined by Affymetrix.
Submission of microarray data to the ArrayExpress database
The raw and normalized microarray data were loaded into the ArrayExpress database: http://www.ebi.ac.uk/arrayexpress. The ArrayExpress accession is E-MEXP-3980.
Statistical analysis of CNV profiling
The binary files (.idat) that were produced by the Illumina scanning software (Bead Scan Array Reader) were analyzed using the Illumina Genome Studio Genotyping Module version 3.2.33 (Illumina, San Diego, CA, USA) to obtain normalized genotype data. The genotype call rate threshold was set at ≥90%, and the final report of the normalized genotype data was transferred to a third-party program, Partek Genomic Suite version 6.6 (Partek Inc., St. Louis, MO, USA), to determine the CNV profiles.
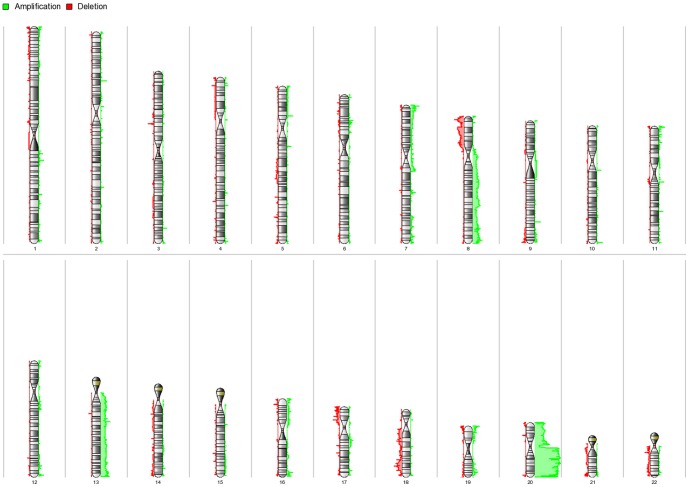
Paired CNV analysis was carried out by comparing the intensity of each hybridization signal from a tumor sample to that of its matched non-cancerous epithelium. The genomic segmentation algorithm was used to detect CNV gains and losses. The following stringent parameters were set to reduce any false-positive alteration: each segment must contain a minimum of 10 consecutive filtered probe sets, a p-value threshold of 0.001 when compared to the neighboring adjacent regions and a signal-to-noise threshold of 0.5. The cut-off value for the gain was set at above 2.3, while loss was set at below 1.7. CNV was called for the gains or losses that occurred in at least 10% (7 samples) of the total samples. A full listing of all CNV gains and losses are included in Table S1. The chromosomal locations of the copy number gains and losses of the 22 autosomes are shown by karyograms (Figure 1).
Figure 1. Karyogram view of detected gains and losses regions across autosomes.
Gains are shown in green and losses are shown in red. The length of the horizontal bar corresponds to number of samples involved at the respective cytoband. Most of the gains were found at the long arm of chromosome 20 and losses were mainly observed at the short arm of chromosome 8.
Statistical analysis of gene expression profiling
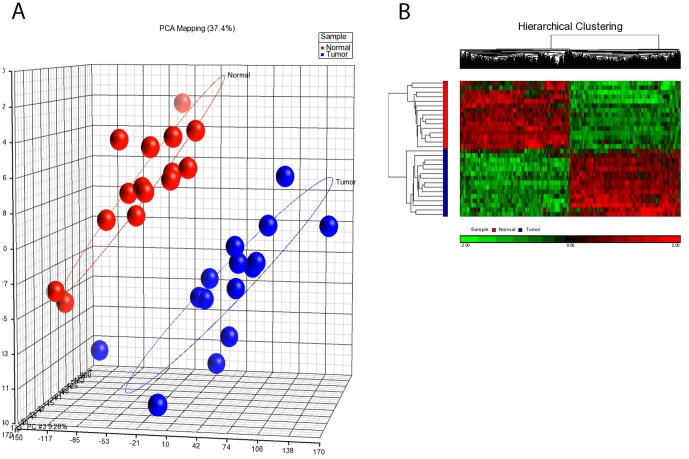
The Affymetrix CEL files were imported to Partek Genomic Suite 6.6 (Partek Inc., St. Louis, MO, USA) to perform gene expression profiling analysis. Raw CEL files were processed for background correction and quantile normalization (median scaling) using the robust multi-array averaging (RMA) method. A principal component analysis (PCA) plot was generated as the quality control step (Figure 2A), and the batch effect was removed as a source of variation. A three-way ANOVA was performed across all samples. Statistically significantly expressed genes were identified using the mixed model analysis of variance with a false discovery rate (Benjamini–Hochberg test) adjusted p value of ≤0.05 and fold-change values of −2 to 2. Hierarchical clustering was generated to visualize patterns of expression in the data (Figure 2B). Gene ontology enrichment analysis was performed using DAVID (Database for Annotation, Visualization and Integration Discovery) Bioinformatic tools.
Figure 2. Plots of principal components analysis (PCA) and hierarchical clustering of gene expression datasets.
(A) PCA scatter plot of CRC data. Each point represents sample. Points are colored by group status with blue representing non-cancerous epithelium and red representing tumor tissue. (B) Hierarchical clustering of mRNA profiles. Samples are indicated along the horizontal axis and grouped by the color bar between the dendogram and the heat map. Blue represents non-cancerous epithelium and red represents tumor tissue. Overall, there was a clear separation between non-cancerous epithelium and tumor tissue group when examined by both PCA (Figure 2A) and hierarchical clustering (Figure 2B).
Integration of copy number variation and genome-wide expression analyses
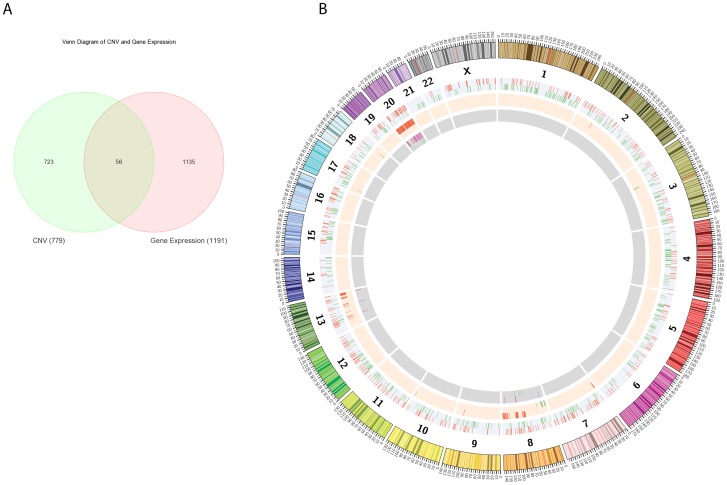
Data from CNV and genome-wide expression analyses were analyzed individually. To identify the significant genes that exhibited CNV and gene expression changes, we overlapped the two datasets as presented in the Venn diagram (Figure 3A). The chromosomal locations of the overlapping genes between the two datasets are shown in a circular map (Figure 3B).
Figure 3. Overlapped genes of integrated CRC datasets.
(A) Venn-diagram representing the common genes in CNV and gene expression datasets revealed 56 overlapping genes. (B) Circular map showing overview of CNVs and gene expression data. Chromosomes are shown in the color coded of the outer most ring. The second ring shows the distribution of gene expression profile. (red indicates up-regulated genes and green indicates down-regulated genes). The inner ring represents CN changes (red denotes gain in CN and green denotes loss in CN). The innermost ring shows the distribution of the two overlapping datasets.
Sub-analysis of the data from the copy number variation and genome-wide expression microarray for the 15 paired cases
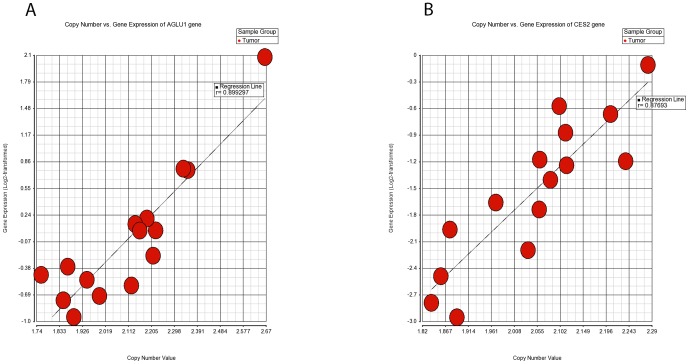
We analyzed the data that were obtained from copy number and genome-wide expression profiling of the 15 paired samples using Partek Genomic Suite 6.6 (Partek Inc., St. Louis, MO, USA). The genomic segmentation algorithm was applied with the parameters that were mentioned in the copy number analysis section. The resulting spreadsheet contained individual markers that displayed the aberrant levels of DNA in each tumor relative to its paired normal. Expression data were normalized to the baseline and the ratios were log2-transformed prior to analysis. Both datasets were correlated using Pearson's linear correlation method and we generated a scatter plot for viewing the results (Figure 4A and 4B).
Figure 4. Correlation of gene expression and CNV datasets in 15 paired subsets of CRC patients.
Scatter plots of gene expression (y-axis) correlating to copy number (x-axis) with differential expression & CN change in CRC for ARGLU1 (Figure 4A) and UGGT2 (Figure 4B) genes. Each dot represents one sample.
Multiplex Ligation Probe Amplification (MLPA)
To validate the CNV profiling results, we selected chromosome 20 for the MLPA assay using the SALSA MLPA P157 20q probe mix according to the manufacturer's protocol (MRC-Holland, Amsterdam, and The Netherlands). The probe mix contains 34 probes for 26 different genes that are located on 20q. Fragment analysis of the PCR products was performed using the GeneScan-500 LIZ Size Standard on the Applied Biosystems 3130 DNA Analyzer (Applied Biosystems, Foster City, CA). The fluorescence data were collected during fragment separation and imported to the Coffalyser.Net software (MRC-Holland, Amsterdam, and The Netherlands) for analysis.
Results
Demographic Data
Of the 64 patients, 39 (60.94%) were females and 25 (39.06%) were males (Table 1). The majority were Malays (48.44%), followed by Chinese (46.88%) and Indians (4.69%). Based on the Dukes' classification, 33 patients (51.56%) were of Dukes' B, 27 patients (42.19%) were of Dukes' C and 4 patients (6.25%) were of Dukes' A. The mean age was 56±13.35 years old.
Table 1. Distribution of clinicopathological features among 64 paired CRC sample.
| Characteristics | Number (n) | Percentage (%) |
| Gender | ||
| Male | 25 | 39.06 |
| Female | 39 | 60.94 |
| Age | ||
| <50 years old | 11 | 17.19 |
| >50 years old | 53 | 82.81 |
| Race | ||
| Malay | 31 | 48.44 |
| Chinese | 30 | 46.88 |
| India | 3 | 4.69 |
| Stage | ||
| Duke's A | 4 | 6.25 |
| Duke's B | 33 | 51.56 |
| Duke's C | 27 | 42.19 |
| Differentiation | ||
| Well Differentiated | 31 | 48.44 |
| Moderately Differentiated | 28 | 43.75 |
| Poorly Differentiated | 5 | 7.81 |
| Location | ||
| Right | 15 | 23.43 |
| Left | 49 | 76.56 |
Copy number gains were frequently found in the q arm of chromosome 20
Of the 8722 genomic segments in all samples, we narrowed down our analysis to 2212 CNV regions that corresponded to the autosomes that showed amplifications (Table S1). The CNVs were scattered across chromosome 1 to 22 with the highest amplification found in chromosome 20, which had 388 segments that represented 17.5% of all gains. The second highest number of gains was documented at chromosome 7, with 369 genomic segments, followed by chromosome 8, with 338 genomic segments. The longest length for the CNV gain segments was between 8q22.1 and 8q22.2 corresponding to 4,070,488 bp. A total of 1,638 genes were amplified in all samples and 765 of these were unique or non-redundant genes. Cytoband 20q12 had the highest frequency of gains involving at least 45.31% of the tumor samples. Eight genes were identified at this locus and three of them, namely PTPRT, CHD6 and EMILIN3, have been previously described in colorectal cancer. Figure 1 shows the distribution of these genes at chromosome 20, with gains shown in green. Details on the top 10 significant genes according to the RefSeq database are provided in Table 2.
Table 2. Top 10 significant copy number gain regions.
| Cytoband | Genes | Frequency (%) | Start | Stop | Length (bps) | Gain value |
| 20q12 | PTPRT | 45.31 | 40848490 | 40858418 | 9929 | 2.50 |
| 20q12 | CHD6, EMILIN3, LPIN3, PLCG1, TOP1, ZHX3, MAFB | 43.75 | 39344635 | 41684451 | 2339816 | 2.50 |
| 20q13.12 | EYA2, MIR3616, LOC100131496, ZMYND8 | 43.75 | 45757009 | 45948813 | 191805 | 2.49 |
| 20q13.13 | LOC284749, PREX1, CSE1L, KCNB1, SNAI1, SATA2, STAU1 | 43.75 | 46984676 | 48922757 | 1938081 | 2.49 |
| 20q11.23 | BLCAP, CTNNBL1, RBL1, SRC, MANBAL | 43.75 | 35626170 | 36460629 | 834460 | 2.48 |
| 20q11.21 | DEFB115, DEFB116, DEFB118, DEFB119, DEFB121, DEFB122, FRG1B | 42.19 | 29466797 | 30046837 | 580040 | 2.50 |
| 20q11.23 – 12 | ACTR5, ADIG, DHX35, TGM2, VSTM2L | 42.19 | 36484397 | 39212610 | 2728214 | 2.49 |
| 20q13.11–13.12 | IFT2, L3MBTL1, MYBL2, SGK2, SRSF6 | 42.19 | 41917787 | 42308263 | 390477 | 2.51 |
| 20q11.21 | BCL2L1, COX412, DEFB124, HM13, ID1 | 42.19 | 30049554 | 30310687 | 261134 | 2.50 |
| 20q11.22 | AHCY, DYNLRB1, MYH7B, ITCH, NCOA6 | 40.63 | 32766406 | 33610899 | 844493 | 2.49 |
Copy number losses were mainly found in the p arm of chromosome 8
A deletion or loss of copy number value less than 1.7 was observed throughout the chromosome 1 to 22 autosomes, with the majority detected at chromosome 8, which had 225 CNV affected segments. The specific region for the losses was observed at position 8p23.3 with 17.9% occurrence in all tumor samples. The second-highest frequency of losses was detected in chromosome 17, with 213 genomic segments, followed by chromosome 19, with 184 genomic segments. The longest loss of a genomic segment was at 8p23.1 to 8p22, which consisted of 3,093,282 bp. We identified only 14 unique or non-redundant genes (Table 3). Analysis of the individual genes in chromosome 8 revealed that only 5 of the genes were previously reported to be related to CRC; these genes were CSMD1, DLC1, TUSC3, SGCZ and LONRF1. The distribution of the losses, shown in red, can be observed in the karyotype diagram as shown in Figure 1.
Table 3. The top most significant CN loss regions.
| Cytoband | Genes | Frequency (%) | Start | Stop | Length (bps) | Loss value |
| 8p23.2 | CSMD1 | 17.18 | 4014635 | 4023153 | 8519 | 1.60 |
| 3p14.2 | FHIT | 11.0 | 60252758 | 60260269 | 7512 | 1.45 |
| 18q22.1 | LOC284294, LOC400654 | 11.0 | 61918208 | 61919976 | 1769 | 1.63 |
| 17p12 | SHISA6 | 11.0 | 11352448 | 11354727 | 2280 | 1.37 |
| 8p23.1–8p22 | DLC1, C8orf48, KIAA1456, LOC340357, LONRF1, TUSC3, MIR383, SGCZ | 11.0 | 12601480 | 15694761 | 3093282 | 1.65 |
| 8p12 | TEX15 | 11.0 | 30675344 | 30698971 | 23628 | 1.59 |
Analysis of gene expression profiling
Analysis revealed significant differences in gene expression between the primary tumor and non-cancerous epithelial samples. Classification by PCA showed a clear separation of two distinctive groups according to the tissue type (Figure 2A). A total of 1408 genes were differentially expressed by at least two-fold with statistical significance (p<0.05). However, a single gene is represented by multiple probe sets called ‘siblings probes’ in the Affymetrix GeneChip. Therefore, any redundant probe sets were filtered, which left 1191 unique genes for further analysis. Of these, 584 genes were found to be up-regulated while another 607 genes were found to be down-regulated. Most of the up-regulated genes were found in chromosomes 7 (n = 48, 8.22%), 2 (n = 44, 7.53%) and 12 (n = 42, 7.19%). Down-regulated genes were mainly located at chromosomes 1 (n = 73, 12%), 2 (n = 44, 7.25%), 3 and 4 (n = 43, 7.08%). Among the up-regulated genes were MYC, CD44, ABC22, TIMP1, and BIRC5, while the down-regulated genes included FAS, KLF4, UGTIA1 and CA2. A full list of the differentially expressed genes and their corresponding fold-changes in expression and p values are provided in Table S2. Hierarchical clustering was generated and visualized via a heat map, where two distinctive expression patterns can be observed (Figure 2B). The functional characterization of the significant genes were performed using GO analysis, and they were found to be distributed throughout the top five classes, which included cell cycle, cell division, chromosome segregation, nucleoside binding and ATP binding (p<0.05).
Effect of CNV on the expression of colorectal cancer-related genes
Integration of the two profiling datasets showed 56 overlapping genes (Figure 3A and 3B), and a full list of these overlapping genes is presented in Table S3. A total of 1,135 genes with only gene expression changes and 723 genes with CNV changes but no changes in transcript levels were observed. Integration of CNV and gene expression analyses showed a positive association in 48 genes (85.7%) and a negative association in the remaining 8 genes (14.3%) (Table 4).
Table 4. Categories of overlapping genes.
| CNV | Gene Expression | No of overlapping genes |
| Gain | Up-regulated | 47 |
| Gain | Down-regulated | 8 |
| Loss | Up-regulated | 0 |
| Loss | Down-regulated | 1 |
| Total | 56 | |
To further understand further the biological function of these genes, the 56 genes were subjected to functional annotation and classification analysis using DAVID v6.7. We found the genes to be related to biological processes and cellular components.
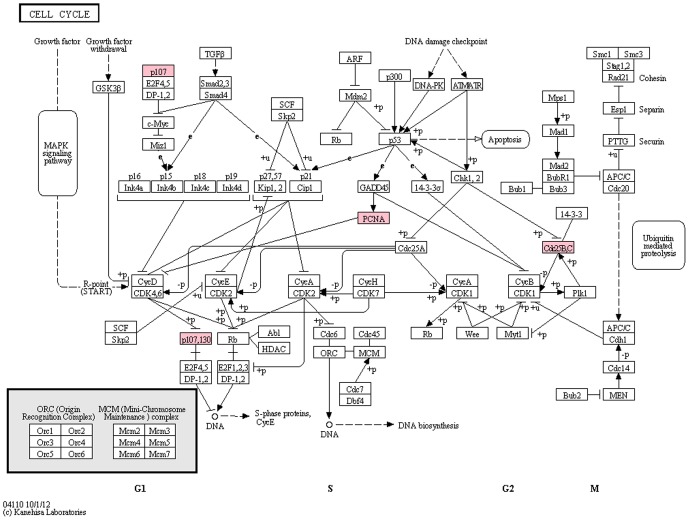
These genes were further mapped to the KEGG pathway database to determine the interactions of the candidate oncogenes or tumor suppressor genes that were identified by CNV and the expression array. The analysis revealed that the cell cycle was the most significantly enriched pathway and CDC25B, PCNA and p107/RBL1 were the key, involved genes (Figure 5).
Figure 5. Cell cycle map from KEGG pathway.
Cell cycle was found to be the most significant enriched pathway (p<0.05). Genes involved (CDC25B, PCNA and p107/RBL1) shown in red color box indicates the genes to have gain in CN and increased level of expression.
Correlation of CNV with gene expression of colorectal cancer-related genes on a subset of 15 paired samples
To determine the relationship between DNA copy number and gene expression, we applied the Pearson's linear correlation model (Figure 4A & 4B). The correlation values were found between −0.85 to 0.89. Using a cut-off of p<0.05, 2159 genes showed significant correlations between copy number and gene expression. A total of 914 transcripts from 616 genes showed a good correlation (r>0.6) between copy number and gene expression. The top five-correlated genes were ARGLU1, UGGT2, CES2, FUT10 and PAOX. A full list of the correlated genes with their Pearson's correlation and p values, can be viewed in Table S4.
Validation of CNV profiling using MLPA
We performed the MLPA assay and targeted 26 genes in 150 samples (50 normal tissues and 100 tumors) to validate the CNV profiling data. We chose probes that covered the q arm of chromosome 20 and the results were considered acceptable if the control peak fell within the range of 0.8 to 1.2. A deletion was scored if the mean dosage of the test to the internal control peaks was less than 0.7, and duplication was scored if the mean dosage was 1.3 or greater. A normal reference sample showed no copy number alterations in any of the genes, and MLPA analysis showed that copy number gain was detected in all twenty-six tested genes. Table 5 summarizes the MLPA analysis with the mean values of each gene.
Table 5. Validation of copy number profiling by MLPA.
| Gene | Frequency (%) | Mean±SD |
| BCL2L1 | 27 | 1.61±0.51 |
| TPX2 | 16 | 1.43±0.45 |
| HCK | 31 | 1.62±0.52 |
| DNMT3B | 32 | 1.53±0.47 |
| E2F1 | 16 | 1.50±0.50 |
| NCAO6 | 41 | 1.58±0.54 |
| SRC | 21 | 1.64±0.54 |
| NNAT | 25 | 1.44±0.44 |
| TOP1 | 36 | 1.50±0.49 |
| MYBL2 | 24 | 1.54±0.51 |
| HNF4A | 25 | 1.50±0.51 |
| UEB2C | 18 | 1.55±0.52 |
| MMP9 | 24 | 1.49±0.50 |
| CSE1L | 16 | 1.41±0.42 |
| PTPN1 | 42 | 1.50±0.43 |
| NFATC2 | 30 | 1.47±0.42 |
| ZNF217 | 39 | 1.47±0.56 |
| BCAS1 | 36 | 1.46±0.46 |
| AURKA | 38 | 1.47±0.46 |
| STX16 | 29 | 1.47±0.44 |
| GNAS | 22 | 1.35±0.40 |
| ADRM1 | 20 | 1.60±0.55 |
| KCNQ2 | 17 | 1.55±0.50 |
| EEF1A2 | 19 | 1.60±0.57 |
| TNFRSF6B | 6 | 1.67±0.60 |
| OPRL1 | 24 | 1.57±0.51 |
Discussion
We performed an integrated analysis using multiple datasets in colorectal tissues to identify the differentially expressed genes with alteration in genomic segments. We analyzed the CNVs and gene expression profiling in 64 paired and 15 paired CRC tissues respectively. The use of adjacent non-cancerous tissues from the same individual reduced the variations that were caused by inter-individual heterogeneity.
The present study identified a number of focal genomic gains and losses in CRC, which showed some concordance with the results from previous studies [20], [21], [22]. Individual analysis of the CNV dataset revealed significant gains in the chromosome 20q that were also highly consistent with the previous studies [23], [24]. Similar gains had also been observed in breast cancer and primary gastric cancer [25]. Cytoband 20q12 showed the highest frequency of gains that spanned from 39239931 to 41684451 bp with the involvement of eight genes, including PTPRT, CHD6, EMILIN3, LPIN3, PLCG1, TOP1, ZHX3 and MAFB. Of the eight genes, TOP1, PLCG1 and PTPRT were related to CRC.
Topoisomerase 1 (TOP1) is an oncogene that catalyzes the unwinding of DNA and creates single-strand molecules, which are required in numerous biological processes such as DNA replication, transcription and DNA repair [26]. A study of TOP1 using array CGH found gains in its copy number in CRC samples [9]. An increased copy number of TOP1 was also detected in Stage III CRC patients with an average of four gene copies for every cell using a fluorescence in situ hybridization (FISH) method [27], [28].
The second gene that was related to CRC identified in this study was phospholipase C gamma 1 (PLCG1), which is a signaling molecule and is a neighboring gene of TOP1. PLCG1 is activated in response to growth factor stimulation and is involved in the regulation of a variety of cellular functions such as cell migration, invasion and metastasis [29], [30], [31].
Protein tyrosine phosphatase receptor-T (PTPRT) is situated within the amplified region of 20q12 between 39344635 to 41684451 bp. It is a tumor suppressor gene and has been shown to play integral roles in cell adhesion and intracellular signaling [32]. Gain of copy number involving the PTPRT gene has been identified in a previous study on ovarian clear cell carcinoma by using array CGH [33]. The gain in copy number of this gene may be as a result of the ‘passenger gene’ effect because we could not detect any changes in its gene expression.
Other significantly amplified cytobands in the q arm of chromosome 20 encoded well-established oncogenes that were associated with CRC, and these included 20q11.21 (BCL2L1), 20q13.2–20q13.31 (AURKA), 20q11.23 (SRC & CTNNBL1), 20q11.21–20q11.22 (DNMT3B) and 20q11.22 (DYNLRB1). These findings suggest the important involvement of multiple candidate genes within the long arm of chromosome 20 in cancer development and progression. The MLPA appears to be a reliable and efficient method to evaluate DNA copy number changes because majority of these tested genes revealed concordance with the microarray results.
Copy number losses were mainly found at the p arm of chromosome 8. The highest loss was found at cytoband 8p23.2 from 4008230 to 4027339 bp. Among the genes that reside at this region is the CUB and Sushi Multiple Domains 1 gene (CSMD1), which is a tumor suppressor gene that codes for multiple domain complement regulatory and adhesion protein. Focal copy number loss of the CSMD1 gene has been observed in CRC [34], breast cancer [35] and gastric cancer [36], and decreased level of CSMD1 expression was reported to be significantly associated with high-tumor grade and reduced overall survival in a breast cancer study [37]. Another region that was noted to exhibit copy number loss that was identified in this study was cytoband 8p23.1–p22, that included a region that covered 12601480 to 15694761 bp. A gene that is located within this region is the Deleted in Liver Cancer 1 (DLC1), which was reported to be a tumor suppressor gene and has been shown to undergo copy number loss in several cancers, such as hepatocellular carcinoma and breast cancer [38], [39].
We did a sub-analysis of 15 paired samples of CRC to evaluate the relationship between copy number changes and gene expression. Genes involved in CRC, such as MYC (v-myc avian myelocytomatosis viral oncogene homolog) [40] CCNB1 (cyclin B1) [41] and PLK1 (polo-like kinase 1) [42], were up-regulated and concordantly amplified in copy number in our study. Seven genes were found to be down-regulated with loss in copy number but only two genes, MUC17 (mucin 7) [43] and CES (carboxylesterase 2) [44] have been shown to be related to CRC.
Integrated analysis using a Venn diagram showed that 85.7% of genes had a positive association. When mapped to the KEGG pathway, the cell cycle was identified as the most significantly enriched pathway (p<0.05). The cell cycle is a critical regulator of cell proliferation, and growth and cell division after DNA damage. The cell cycle pathway is mainly driven by the cyclin-dependent kinase (CDKs) family and their regulatory subunits, the cyclins. The cell cycle has four phases and the two major checkpoints at the G1-S and G2-M transitions to maintain the correct order of events [45]. The loss of cell cycle checkpoint control promotes genetic instability, which leads to uncontrolled cell proliferation and could promote cancer development [46].
Cell division cycle 25B (CDC25B) is a member of the cell division cycle (CDC) phosphatase family that functions as activators of CDKs and cyclin complexes to regulate progression of the cell cycle [47]. CDC25B is responsible for the initial dephosphorylation and activation of the CDKs, thus initiating the sequence of events that leads to entry into mitosis [48], [49]. Over-expression of CDC25B has been observed in 43% of CRC patients and is correlated with poor prognosis [50]. Increased CDC25B level is sufficient to impair the DNA damage checkpoints, which in turn, increases spontaneous mutagenesis and interferes with the entry into mitosis [51], [52], [53].
Proliferating cell nuclear antigen (PCNA) was reported to be essential for DNA replication, DNA repair and cell cycle regulation [54]. Retinoblastoma-like 1 (p107/RBL1) is a member of the retinoblastoma gene family (RB), and the genes in this family have been identified as tumor suppressors. RBL1 and other RB proteins cooperate to regulate cell cycle progression through G1 phase of the cell cycle [55]. However, the mechanism of PCNA and RBL1 involvement in the cell cycle pathway of CRC is still unclear and requires further exploration.
We also found genes with negative associations between copy number and gene expression levels. The genes include BCAS1, EDN3, FABP4, MATN2, SDCBP2, SPTLC3, TRPA1 and WFDC2. This paradox of a negative relationship between copy number status and gene expression has also been observed in a previous study on CRC [56] and might be attributed to the multiple mechanisms that are responsible for normal and abnormal control of gene expression, including those related to mutation, promoter methylation and miRNA expression [57]. To understand this phenomenon, an approach using deep sequencing technology will most likely probably provide an answer to these unexpected findings.
In conclusion, by integrating the datasets from two different profiling studies, we successfully identified 56 overlapping genes with changes in copy number and gene expression. The cell cycle was identified as the key signaling pathway from this integrated analysis. However, future studies are necessary to determine the impact of these genes on the outcome of the disease.
Supporting Information
Full information on copy number variation profile of 64 colorectal cancer patients.
(XLSX)
Full information on gene expression profile of 15 colorectal cancer patients.
(XLSX)
Full list of 56 overlapped genes following integration analysis of both datasets.
(XLSX)
Full list of sub-analysis between CNV and gene expression of 15 paired subset colorectal cancer patients.
(XLSX)
Acknowledgments
The authors wish to thank Mrs. Ezanee Azlina Mohamad Haniff and Mr. Muhiddin Ishak for their technical assistance throughout the project.
Funding Statement
The project was funded by the the Ministry of Science, Technology & Innovation (07-05-MGI-GMB016) and Higher Institution Centre of Excellence grant from the Ministry of Higher Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, et al. (2010) Colorectal cancer. Lancet 375: 1030–1047. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA: a cancer journal for clinicians 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 3.Zainal Ariffin O, Nor Saleha IT (2011) National Cancer Registry Report 2007. Kuala Lumpur: Ministry of Health. [Google Scholar]
- 4. Issa JP (2008) Colon cancer: it's CIN or CIMP. Clinical cancer research : an official journal of the American Association for Cancer Research 14: 5939–5940. [DOI] [PubMed] [Google Scholar]
- 5. Gordon DJ, Resio B, Pellman D (2012) Causes and consequences of aneuploidy in cancer. Nature reviews Genetics 13: 189–203. [DOI] [PubMed] [Google Scholar]
- 6. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. (2006) Global variation in copy number in the human genome. Nature 444: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaignat E, Yahya-Graison EA, Henrichsen CN, Chrast J, Schutz F, et al. (2011) Copy number variation modifies expression time courses. Genome research 21: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakao M, Kawauchi S, Furuya T, Uchiyama T, Adachi J, et al. (2009) Identification of DNA copy number aberrations associated with metastases of colorectal cancer using array CGH profiles. Cancer genetics and cytogenetics 188: 70–76. [DOI] [PubMed] [Google Scholar]
- 9. Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, et al. (2007) Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. Journal of molecular medicine 85: 293–304. [DOI] [PubMed] [Google Scholar]
- 10. Jones AM, Douglas EJ, Halford SE, Fiegler H, Gorman PA, et al. (2005) Array-CGH analysis of microsatellite-stable, near-diploid bowel cancers and comparison with other types of colorectal carcinoma. Oncogene 24: 118–129. [DOI] [PubMed] [Google Scholar]
- 11. Alcock HE, Stephenson TJ, Royds JA, Hammond DW (2003) Analysis of colorectal tumor progression by microdissection and comparative genomic hybridization. Genes, chromosomes & cancer 37: 369–380. [DOI] [PubMed] [Google Scholar]
- 12. Lipska L, Visokai V, Levy M, Svobodova S, Kormunda S, et al. (2007) Tumor markers in patients with relapse of colorectal carcinoma. Anticancer research 27: 1901–1905. [PubMed] [Google Scholar]
- 13. Wu X, Zhang D, Li G (2012) Insights into the regulation of human CNV-miRNAs from the view of their target genes. BMC genomics 13: 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poptsova M, Banerjee S, Gokcumen O, Rubin MA, Demichelis F (2013) Impact of constitutional copy number variants on biological pathway evolution. BMC evolutionary biology 13: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woodwark C, Bateman A (2011) The characterisation of three types of genes that overlie copy number variable regions. PloS one 6: e14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Camps J, Nguyen QT, Padilla-Nash HM, Knutsen T, McNeil NE, et al. (2009) Integrative genomics reveals mechanisms of copy number alterations responsible for transcriptional deregulation in colorectal cancer. Genes, chromosomes & cancer 48: 1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, et al. (2006) Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, chromosomes & cancer 45: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 19. Lu TP, Lai LC, Tsai MH, Chen PC, Hsu CP, et al. (2011) Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PloS one 6: e24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jasmine F, Rahaman R, Dodsworth C, Roy S, Paul R, et al. (2012) A genome-wide study of cytogenetic changes in colorectal cancer using SNP microarrays: opportunities for future personalized treatment. PloS one 7: e31968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie T, G DA, Lamb JR, Martin E, Wang K, et al. (2012) A comprehensive characterization of genome-wide copy number aberrations in colorectal cancer reveals novel oncogenes and patterns of alterations. PloS one 7: e42001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, et al. (2009) Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut 58: 79–89. [DOI] [PubMed] [Google Scholar]
- 24. Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, et al. (2006) Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer research 66: 2129–2137. [DOI] [PubMed] [Google Scholar]
- 25. Kimura Y, Noguchi T, Kawahara K, Kashima K, Daa T, et al. (2004) Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 17: 1328–1337. [DOI] [PubMed] [Google Scholar]
- 26. Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews Molecular cell biology 3: 430–440. [DOI] [PubMed] [Google Scholar]
- 27. Romer MU, Nygard SB, Christensen IJ, Nielsen SL, Nielsen KV, et al. (2013) Topoisomerase 1(TOP1) gene copy number in stage III colorectal cancer patients and its relation to prognosis. Molecular oncology 7: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith DH, Christensen IJ, Jensen NF, Markussen B, Romer MU, et al. (2013) Mechanisms of Topoisomerase I (TOP1) Gene Copy Number Increase in a Stage III Colorectal Cancer Patient Cohort. PloS one 8: e60613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomas SM, Coppelli FM, Wells A, Gooding WE, Song J, et al. (2003) Epidermal growth factor receptor-stimulated activation of phospholipase Cgamma-1 promotes invasion of head and neck squamous cell carcinoma. Cancer research 63: 5629–5635. [PubMed] [Google Scholar]
- 30. Jones NP, Peak J, Brader S, Eccles SA, Katan M (2005) PLCgamma1 is essential for early events in integrin signalling required for cell motility. Journal of cell science 118: 2695–2706. [DOI] [PubMed] [Google Scholar]
- 31. Wells A, Grandis JR (2003) Phospholipase C-gamma1 in tumor progression. Clinical & experimental metastasis 20: 285–290. [DOI] [PubMed] [Google Scholar]
- 32. Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411: 355–365. [DOI] [PubMed] [Google Scholar]
- 33. Tan DS, Iravani M, McCluggage WG, Lambros MB, Milanezi F, et al. (2011) Genomic analysis reveals the molecular heterogeneity of ovarian clear cell carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research 17: 1521–1534. [DOI] [PubMed] [Google Scholar]
- 34. Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, et al. (2009) Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America 106: 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma C, Quesnelle KM, Sparano A, Rao S, Park MS, et al. (2009) Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer biology & therapy 8: 907–916. [DOI] [PubMed] [Google Scholar]
- 36. Deng N, Goh LK, Wang H, Das K, Tao J, et al. (2012) A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 61: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamal M, Shaaban AM, Zhang L, Walker C, Gray S, et al. (2010) Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast cancer research and treatment 121: 555–563. [DOI] [PubMed] [Google Scholar]
- 38. Jia D, Wei L, Guo W, Zha R, Bao M, et al. (2011) Genome-wide copy number analyses identified novel cancer genes in hepatocellular carcinoma. Hepatology 54: 1227–1236. [DOI] [PubMed] [Google Scholar]
- 39. Hawthorn L, Luce J, Stein L, Rothschild J (2010) Integration of transcript expression, copy number and LOH analysis of infiltrating ductal carcinoma of the breast. BMC cancer 10: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lagerstedt KK, Staaf J, Jonsson G, Hansson E, Lonnroth C, et al. (2007) Tumor genome wide DNA alterations assessed by array CGH in patients with poor and excellent survival following operation for colorectal cancer. Cancer informatics 3: 341–355. [PMC free article] [PubMed] [Google Scholar]
- 41. Huang V, Place RF, Portnoy V, Wang J, Qi Z, et al. (2012) Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic acids research 40: 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodel F, Keppner S, Capalbo G, Bashary R, Kaufmann M, et al. (2010) Polo-like kinase 1 as predictive marker and therapeutic target for radiotherapy in rectal cancer. The American journal of pathology 177: 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Senapati S, Ho SB, Sharma P, Das S, Chakraborty S, et al. (2010) Expression of intestinal MUC17 membrane-bound mucin in inflammatory and neoplastic diseases of the colon. Journal of clinical pathology 63: 702–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang X, Wu H, Wu Z, Wang G, Wang Z, et al. (2008) Carboxylesterase 2 is downregulated in colorectal cancer following progression of the disease. Cancer investigation 26: 178–181. [DOI] [PubMed] [Google Scholar]
- 45. Walworth NC (2000) Cell-cycle checkpoint kinases: checking in on the cell cycle. Current opinion in cell biology 12: 697–704. [DOI] [PubMed] [Google Scholar]
- 46. Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nature reviews Cancer 9: 153–166. [DOI] [PubMed] [Google Scholar]
- 47. Kristjansdottir K, Rudolph J (2004) Cdc25 phosphatases and cancer. Chemistry & biology 11: 1043–1051. [DOI] [PubMed] [Google Scholar]
- 48. Goldstone S, Pavey S, Forrest A, Sinnamon J, Gabrielli B (2001) Cdc25-dependent activation of cyclin A/cdk2 is blocked in G2 phase arrested cells independently of ATM/ATR. Oncogene 20: 921–932. [DOI] [PubMed] [Google Scholar]
- 49. Loffler H, Rebacz B, Ho AD, Lukas J, Bartek J, et al. (2006) Chk1-dependent regulation of Cdc25B functions to coordinate mitotic events. Cell cycle 5: 2543–2547. [DOI] [PubMed] [Google Scholar]
- 50. Takemasa I, Yamamoto H, Sekimoto M, Ohue M, Noura S, et al. (2000) Overexpression of CDC25B phosphatase as a novel marker of poor prognosis of human colorectal carcinoma. Cancer research 60: 3043–3050. [PubMed] [Google Scholar]
- 51. Lindqvist A, Kallstrom H, Karlsson Rosenthal C (2004) Characterisation of Cdc25B localisation and nuclear export during the cell cycle and in response to stress. Journal of cell science 117: 4979–4990. [DOI] [PubMed] [Google Scholar]
- 52. Aressy B, Bugler B, Valette A, Biard D, Ducommun B (2008) Moderate variations in CDC25B protein levels modulate the response to DNA damaging agents. Cell cycle 7: 2234–2240. [DOI] [PubMed] [Google Scholar]
- 53. Bugler B, Quaranta M, Aressy B, Brezak MC, Prevost G, et al. (2006) Genotoxic-activated G2-M checkpoint exit is dependent on CDC25B phosphatase expression. Molecular cancer therapeutics 5: 1446–1451. [DOI] [PubMed] [Google Scholar]
- 54. Strzalka W, Ziemienowicz A (2011) Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Annals of botany 107: 1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henley SA, Dick FA (2012) The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell division 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Platzer P, Upender MB, Wilson K, Willis J, Lutterbaugh J, et al. (2002) Silence of chromosomal amplifications in colon cancer. Cancer research 62: 1134–1138. [PubMed] [Google Scholar]
- 57. Kotliarov Y, Kotliarova S, Charong N, Li A, Walling J, et al. (2009) Correlation analysis between single-nucleotide polymorphism and expression arrays in gliomas identifies potentially relevant target genes. Cancer research 69: 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full information on copy number variation profile of 64 colorectal cancer patients.
(XLSX)
Full information on gene expression profile of 15 colorectal cancer patients.
(XLSX)
Full list of 56 overlapped genes following integration analysis of both datasets.
(XLSX)
Full list of sub-analysis between CNV and gene expression of 15 paired subset colorectal cancer patients.
(XLSX)