This study investigated whether it is possible to isolate pancreatic endocrine progenitors from differentiating human embryonic stem cell (hESC) cultures by lineage tracing of NGN3. Results indicate that NGN3+ cells represent pancreatic endocrine progenitors in humans. This hESC reporter line is a unique tool that may aid in gaining insight into the developmental mechanisms underlying fate choices in human pancreas and in developing cell-based therapies.
Keywords: Diabetes, Progenitor cells, Embryonic stem cells, Gene targeting, Differentiation
Abstract
Pancreatic endocrine progenitors obtained from human embryonic stem cells (hESCs) represent a promising source to develop cell-based therapies for diabetes. Although endocrine pancreas progenitor cells have been isolated from mouse pancreata on the basis of Ngn3 expression, human endocrine progenitors have not been isolated yet. As substantial differences exist between human and murine pancreas biology, we investigated whether it is possible to isolate pancreatic endocrine progenitors from differentiating hESC cultures by lineage tracing of NGN3. We targeted the 3′ end of NGN3 using zinc finger nuclease-mediated homologous recombination to allow selection of NGN3eGFP+ cells without disrupting the coding sequence of the gene. Isolated NGN3eGFP+ cells express PDX1, NKX6.1, and chromogranin A and differentiate in vivo toward insulin, glucagon, and somatostatin single hormone-expressing cells but not to ductal or exocrine pancreatic cells or other endodermal, mesodermal, or ectodermal lineages. This confirms that NGN3+ cells represent pancreatic endocrine progenitors in humans. In addition, this hESC reporter line constitutes a unique tool that may aid in gaining insight into the developmental mechanisms underlying fate choices in human pancreas and in developing cell-based therapies.
Introduction
Human embryonic stem cells (hESCs) are considered a promising source of β cells to treat diabetes mellitus type 1. Growth factors and/or small molecules have been used to generate insulin-producing β cells from hESCs [1–5]. Evidence exists that hESC-derived β cells, like primary islets, can reverse hyperglycemia. For example, several groups have grafted mixed embryonic stem cell (hESC) progeny consisting chiefly of PDX1- and NKX6.1-expressing cells to reverse hyperglycemia in diabetic mice. However, development of normoglycemia required long-term in vivo maturation of the hESC-derived pancreatic endoderm [2, 3]. Kelly et al. selected pancreatic endoderm progenitors with an anti-CD142 antibody or endocrine cells with an anti-CD200 and -CD318 antibody. CD142-selected cells yielded islet-like clusters, ductal tissue, and exocrine tissue after implantation, whereas the CD200- and CD318-enriched population gave rise mainly to GCG+ cells [6]. All these studies relied on the transplantation of mixed or not completely pure end-stage cell populations, which in some cases led to the generation of teratomas [2, 3]. Therefore, understanding the developmental mechanisms involved in human pancreas specification and developing cell therapies from stem cells are hampered by the heterogeneity of their differentiated progenies. Although many studies have already been performed in mice, it appears that developmental regulation is not always conserved among species. The few studies performed in human tissue using immunohistological analyses [7, 8], gene expression profiling [9, 10], and transplantation experiments of fetal pancreatic tissue of no less than 7 weeks and up to 23 weeks of age [11, 12] support this idea. For example, the secondary transition, a restricted period within mouse islet development when β cells are generated, is not apparent in human development but instead occurs throughout the whole period of gestation. Thus, there is need for a more precise and reliable tool to address specific questions related to human pancreatic development.
In addition, in terms of clinical applications, it is unclear whether endocrine progenitors or mature β cells will be the best cell population for β-cell replacement therapies. Currently, generation of mature β cells from pluripotent stem cells (PSCs) in vitro remains challenging and still requires complex in vivo signals to fully reach maturation [13]. Endocrine pancreas progenitors can be used to dissect the cues needed to generate functional pancreatic islet cells.
For these reasons, we hypothesized that the isolation of a specific pancreatic endocrine progenitor population would provide a useful tool for the characterization of mechanisms that underlie generation of β cells from hESCs in vitro and/or in vivo and could serve eventually as a valuable and safe source for cell therapy. Ngn3-expressing cells are known to give rise to all hormone-secreting endocrine cell types in the mouse pancreas [14]. Upon maturation of islet cells, Ngn3 is no longer expressed [15]. Therefore, NGN3 might be a useful marker for the identification of human pancreatic endocrine progenitors. In this study, we explored this possibility by selecting NGN3-expressing cells from differentiating hESCs in vitro. Zinc finger nuclease (ZFN)-mediated homologous recombination was used to target a selectable cassette at the C terminus of NGN3, allowing the isolation, purification, and characterization of NGN3-expressing cells. Pure NGN3eGFP+ cells gave rise in vivo to single hormone-expressing endocrine cells, confirming their pancreatic endocrine commitment for the first time in humans.
Materials and Methods
Cell Culture
hESC lines H9 and H1 were purchased from WiCell Research Institute (Madison, WI, http://www.wicell.org) and cultured according to the manufacturer’s recommendations on feeder layers. Experiments were performed at KU Leuven with approval from the Medical Ethics Committee (UZ Leuven, Gasthuisberg). For differentiation purposes, cells were harvested through enzymatic dissociation and plated on 2% Matrigel-coated wells (Becton, Dickinson and Company, Franklin Lakes, NJ, http://www.bd.com) in mTESR medium (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com). After the cells reached 60%–70% confluence, differentiation was initiated. Pancreatic differentiation medium consisted of 55% Dulbecco’s modified Eagle’s medium with 1,000 mg/l d-glucose, sodium pyruvate, 2% fetal bovine serum, 0.1% (0.1 mM) β-mercaptoethanol, 1% penicillin-streptomycin liquid (Life Technologies, Rockville, MD, http://www.lifetech.com), 40% MCDB 201 medium with trace elements, l-glutamine, 30 mM HEPES, 1% ITS+1 (100×), and 1% (0.1 mM) l-ascorbic acid (all from Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). Wnt3a (50 ng/ml) and Activin A (100 ng/ml) were used for stage 1. Noggin (100 ng/ml) and anti-Sonic hedgehog (SHH) (2.5 μg/ml) (both from R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com), nicotinamide (NA) (10 mM), and retinoic acid (RA) (10 μM) (both from Sigma-Aldrich) were added from day 4 until day 16 (stage 2).
The NGN3eGFP+ enriched cells were cultured as clusters: 10,000 cells per well were plated in 96-well low-attachment plates with noggin, anti-SHH, RA, and NA. To allow cluster formation, plates were spun at 200g for 5 minutes. After 2 days, medium was replaced with RPMI 1640 medium (Life Technologies) with 0.05 wt% bovine serum albumin (BSA) containing NA (10 mM) (Sigma-Aldrich) and insulin-like growth factor II (IGF-II) (50 ng/ml) (R&D Systems) for an additional 24 days.
NGN3 Gene Targeting and Cell Sorting
A specific NGN3 ZFN set was generated to target the gene at the C-terminal region of the protein (Sigma-Aldrich). The different components for the NGN3 gene-targeting vector were cloned in the pCR2.1 plasmid (Life Technologies; sequence shown in supplemental online Table 4). Two to 3 million hESCs were nucleofected with 20 μg of gene targeting vector and 5 μl of ZFNs mRNA using hESC Nucleofector Solution 2, program A13 (Lonza, Walkersville, MD, http://www.lonza.com) following the manufacturer’s instructions. Cells were plated on inactivated DR4 mouse embryonic fibroblasts (GlobalStem, Rockville, MD, http://globalstem.com) in media containing 10 µM ROCK inhibitor (Sigma-Aldrich). Selection with hygromycin (Sigma-Aldrich; H9, 50 μg/ml; H1, 25 μg/ml) for up to 2 weeks was performed. Surviving colonies were individually picked and expanded. Genotyping polymerase chain reactions (PCRs) were performed according to standard procedures.
Southern Blot
The Southern blots were performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Indianapolis, IN, http://www.roche.com) according to the manufacturer’s instructions. The sets of primers used for the generation of the probes can be found in supplemental online Table 2.
Teratoma Formation and Analysis
hESCs were collected through enzymatic dissociation, resuspended in 120 μl of phosphate-buffered saline, and injected with 120 μl of Matrigel subcutaneously in the back of severe combined immunodeficient RAG2γc-knockout mice. Tumors generally developed within 4–8 weeks. Animals were sacrificed for dissection, and teratomas were fixed in 4% paraformaldehyde (overnight) and subsequently embedded in paraffin. After sectioning, the presence of cells from three germ layers was assessed following hematoxylin and eosin staining.
Array Comparative Genomic Hybridization and Karyotyping
Genomic DNA was isolated from NGN3eGFP-H9 (three different clones, n = 2 of each clone) and wild-type (WT) hESCs (n = 2), all having passage numbers between 40 and 57 using the QiaAmp DNA mini kit (Qiagen, Hilden, Germany, http://www.qiagen.com), and subjected to copy number variation analysis on 180k Cytosure ISCA v2 arrays (Oxford Gene Technology, Oxford, U.K., http://www.ogt.co.uk). One representative clone of each NGN3eGFP-hESC H9 or H1 line and their WT counterparts were further analyzed by standard cytogenetic procedures at passage numbers of 50 for WT and 60 for transgenic cell lines.
Immunocytochemistry of NGN3eGFP+ Cells
Because of the autofluorescence and multilayered nature of the day 16 differentiation hESC cultures, a thorough optimization of the staining procedure for NGN3 and green fluorescent protein (GFP) was needed. The human hepatocarcinoma cell line Huh7.5 (American Type Culture Collection, Manassas, VA, http://www.atcc.org), transfected with an NGN3eGFP-Puromycin expression vector, was used to optimize the NGN3 and GFP staining procedure. The NGN3eGFP-Puromycin cassette, amplified by reverse transcription (RT)-PCR using day 16 differentiated progeny as template, was cloned into a vector containing the CAGGS constitutive promoter and confirmed by sequencing. Huh7.5-transfected cells and day 16 progeny were fixed with 10% neutral buffered formalin, incubated with 10% donkey serum blocking solution, and permeabilized with 0.2% Triton X-100 (Sigma-Aldrich). Optimized staining conditions established by using the NGN3eGFP-Puromycin-expressing Huh7.5 cells as control cells required an amplification step for anti-NGN3 antibody (R&D Systems) using the Tyramide Signal Amplification kit (PerkinElmer Life and Analytical Sciences, Waltham, MA, http://www.perkinelmer.com). This was not needed for the staining with the anti-GFP antibody (Abcam, Cambridge, U.K., http://www.abcam.com).
Fluorescence-activated cell sorting (FACS)-selected GFP+ cells were stained with an anti-NGN3 antibody (R&D Systems) without need of amplification. A list of all antibodies used on the NGN3eGFP+ cells can be found in supplemental online Tables 3 and 4. DAPI was used as the nuclear counterstain in all cases. Images of the day 16 differentiation hESC cultures were obtained using the AxioImager-Z1, and images of the sorted cells were obtained using the Nikon A1R Eclipse Ti.
FACS
Day 16 hESC differentiated progeny was sorted using the FACSAria III cell sorter (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com).
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted using the RNeasy microkit (Qiagen), and cDNA was synthesized from 1 μg of total RNA using Superscript III reverse transcriptase (Life Technologies). Quantitative RT-PCR (qRT-PCR) was performed on a Mastercycler ep Realplex instrument using specific primers (listed in supplemental online Table 2) and Platinum SyBRGreen qPCR SuperMix-UDG (Life Technologies). PPIA was used as a housekeeping control gene, and results are shown in fold change using the 2−ΔΔCt method for relative quantification.
In Vitro C-Peptide Production
C-peptide release was challenged by incubating the further differentiated NGN3eGFP+ clusters in Krebs-Ringer solution with bicarbonate and HEPES (KRBH; 129 mM NaCl, 4.8 mM KCl, 2.5 mM CaCl2, 1.2 mM, KH2PO4, 1.2 mM MgSO4, 5 mM NaHCO3, 10 mM HEPES, 0.1% (wt/vol) BSA (all from Sigma-Aldrich). The clusters were washed three times using KRBH. A 30-minute exposure with KRBH was considered a non-stimulating condition; this medium was kept for normalization purposes and was followed by a 1-hour incubation with stimulation medium containing either 2.5 mM d-glucose, 20 mM d-glucose, 2.5 mM d-glucose with 1 mM 3-isobutyl-1-methylxanthine (IBMX) and 30 mM KCl or 20 mM d-glucose with 50 μM nifedepine (all from Sigma-Aldrich). Samples were analyzed for C-peptide release using the ultra-sensitive C-peptide (Mercodia) and C-peptide ELISA kit (Mercodia).
Transplantation Under the Kidney Capsule
NGN3eGFP+ cells were transplanted under the kidney capsule of RAG2γc-knock-out mice.
Immunohistochemistry and Fluorescence In Situ Hybridization
Kidney grafts were fixed with 4% paraformaldehyde, embedded in paraffin and stained according to standard procedures using 10% of donkey serum blocking solution and 0.2% Triton X-100 (Sigma-Aldrich) permeabilization solution. Staining of the grafts with anti-GFP (Clontech, Mountain View, CA, http://www.clontech.com) required amplification of the signal using the Tyramide Signal Amplification kit (PerkinElmer). This kit was used according to manufacturer’s instructions with fluorescein isothiocyanate fluorophores. A list of all antibodies can be found in supplemental online Tables 3 and 4.
After immunofluorescence staining, grafts were further processed for fluorescence in situ hybridization (FISH). The human CotI DNA probe (Life Technologies) was labeled using the Bioprime Total Genomic labeling system (Life Technologies), diluted in hybridization buffer (6.15 g of dextran sulfate, 20 ml of formamide, 4 ml of 20× standard saline citrate (SSC), and 18 ml of water, brought to pH 7 with HCl [all from Sigma-Aldrich]) and denatured at 85°C for 4 minutes. Hybridization of the slides were performed at 37°C O/N. Thereafter, the slides were washed for 2 minutes in 0.4× SSC (Life Technologies) with 0.3% Igepal-CA630 (Sigma-Aldrich), 1 minute in 2× SSC with 0.1% Igepal-CA630, and 5 minutes in 2× SSC. For chromogranin A (CHGA) staining, FISH was performed first, followed by immunostaining. DAPI was used as nuclear counterstain in all the cases. Images of the sections were taken using the AxioImager-Z1 (Carl Zeiss, Jena, Germany, http://www.zeiss.com).
Statistical Analyses
Data values obtained on the differentiation were subject to the two-tailed Student’s t test. Values of p < .05 (∗), p < .01 (∗∗), and p < .001 (∗∗∗) were considered statistically significant.
Results
Generation of NGN3eGFP-hESC Lines
ZFNs were generated recognizing unique sites adjacent to the stop codon of the NGN3 gene. A gene targeting vector containing two isogenic homology regions of up to 800 bp each that flanked the targeting locus was created. Upon correct targeting, the NGN3 translation stop codon was deleted and the protein was tagged with a polycistronic cassette containing the enhanced green fluorescent protein (eGFP) and puromycin resistance cassette (PuroR) linked by T2A and P2A sequences (Fig. 1A), which led to the production of three individual proteins once NGN3 was expressed. An EF1α-hygromycinR/thymidine kinase (Hyg/TK) fusion cassette allowed the selection of recombinant clones.
Figure 1.
Characterization of the NGN3eGFP human embryonic stem cell (hESC) lines. (A): Schematic overview of the wild-type allele, targeting vector and targeted NGN3 alleles. Red lines: probes used in Southern blot; white boxes: coding sequences; black lines: Zinc finger nuclease genomic recognition site; dark gray boxes: 3′ untranslated region; light gray boxes: T2A and P2A sequences; eGFP and PURO-pA: PuromycinR gene and polyadenylation signal; white arrows, EF1α promoter; Hyg/TK, hygromycinR-thymidine kinase fusion gene. Polymerase chain reaction genotyping primers are indicated in blue (amplification of the integration) and red (amplification of random integrants). (B): Southern blot of NGN3eGFP-hESC lines. Genomic DNA was digested with AvrII and hybridized with external (5′) and internal (3′) probes. (C): Immunostaining of the NGN3eGFP-hESC lines for pluripotency markers OCT4, NANOG, SOX2, SSEA4, and TRA-1-60. Scale bars = 100 μm. (D): Hematoxylin and eosin staining of teratoma sections generated from the NGN3eGFP-hESC lines representing all three lineages. Scale bars = 25 μm. Abbreviations: EF1α, human elongation factor 1α; huEF1α, human elongation factor 1α; wt, wild-type.
After nucleofection of the ZFNs and the targeting vector in H9- and H1-hESCs, antibiotic selection was performed and 18 and 5 hygromycin-resistant colonies, respectively, were manually picked. Of these, 12 of 18 H9 colonies and 4 of 5 H1 colonies were correctly targeted without random integrations (supplemental online Fig. 1), and the genotypes of three clones of each line were further characterized by Southern blot (Fig. 1B). Genome editing based on homologous recombination with ZFNs is known to cause off-target mutations, which could affect ESC characteristics. Therefore, the genetically modified cell lines were assessed for maintenance of pluripotency and genome integrity. The targeted lines expressed the pluripotency markers OCT4, NANOG, SOX2, TRA-1-60, and SSEA4 (Fig. 1C) and formed teratomas wherein cells of the three germ layers could be detected (Fig. 1D). Genome integrity was assessed by both array comparative genomic hybridization (aCGH), due to the higher sensitivity for detecting copy number variations than traditional cytogenetic techniques, and conventional karyotyping. aCGH demonstrated that no genetic abnormalities were present after ZFN treatment and that the karyotype and ploidy of both NGN3eGFP-hESC lines were normal (supplemental online Table 1; supplemental online Fig. 2).
Differentiation of hESCs Toward Endocrine Pancreas
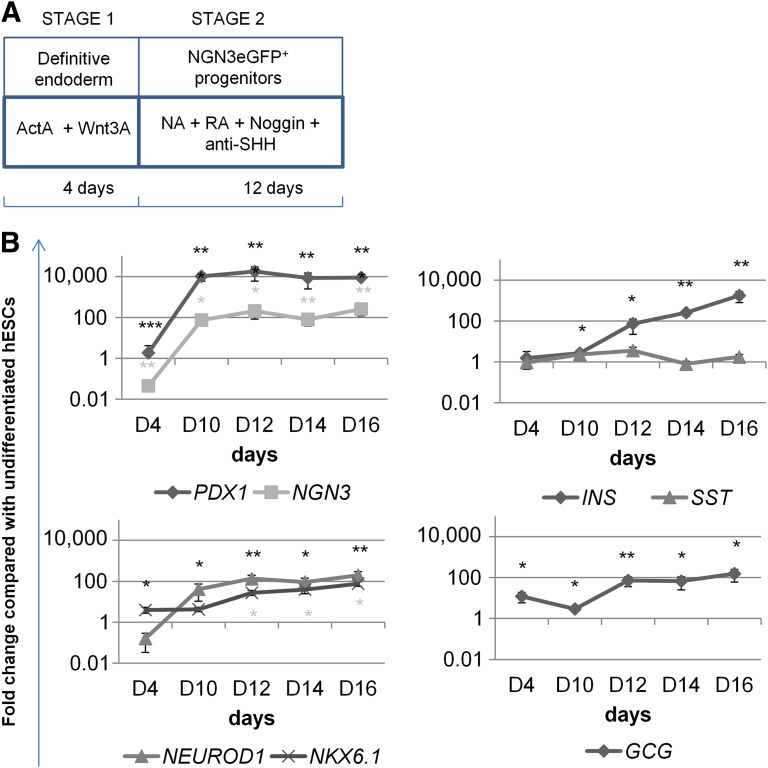
We used the protocol depicted in Fig. 2A to differentiate hESCs toward endocrine pancreas. Activin A and Wnt3a were used for induction of definitive endoderm [1, 16, 17]. We next used a mix of anti-SHH antibody, to inhibit SHH signaling [18], RA to direct ESCs toward endocrine pancreas by inducing PDX1 expression [19], noggin to inhibit BMP signaling [17, 19], and NA, which is known to induce differentiation and maturation of undifferentiated fetal pancreatic cells [20]. We evaluated the differentiation process using WT H9 cells by qRT-PCR on days 0, 4, 10, 12, 14, and 16. Transcript levels for the pluripotency genes OCT4 and NANOG decreased from day 4 onward and were no longer expressed following day 10. Transcripts of the primitive streak marker genes (EOMES and MIXL1) were maximally expressed on day 4, and transcripts for the definitive endoderm genes (CXCR4, FOXA2 and SOX17) increased from day 4 to days 10–12 but decreased afterward (supplemental online Fig. 3). From day 4 onward, we could detect transcripts for the pancreatic endocrine gene PDX1, followed by an increase in NGN3, NEUROD1, and NKX6.1 expression levels. INS and GCG transcript levels increased moderately and SST was not expressed as compared with undifferentiated cells, which did not express any of these pancreatic genes (Fig. 2B), but expression levels remained ∼9,000-, ∼2,000-, and ∼80,000-fold lower, respectively, than in human pancreas (data not shown).
Figure 2.
Generation of NGN3eGFP+ cells from the NGN3eGFP-hESC lines. (A): The differentiation scheme depicts the mix of factors used to commit hESCs toward endocrine pancreas. (B): Transcript levels for different pancreatic genes during endocrine pancreas differentiation using H9 cells. All results shown are from three or more independent experiments. Data are represented as mean ± SEM. Asterisks indicate statistical significance as determined by t test. Values of p < .05 (∗), p < .01 (∗∗), and p < .001 (∗∗∗) were considered statistically significant. Abbreviations: D, day; hESC, human embryonic stem cell; NA, nicotinamide; RA, retinoic acid; SHH, Sonic hedgehog.
The transgenic cell lines were further assessed for their in vitro differentiation potential by directing them toward endocrine pancreatic-like cells as described above. No significant differences in the transcript levels of these genes were observed between WT-H9 and NGN3eGFP-H9 of day 16 progeny, whereas some pancreatic genes in NGN3eGFP-H1 differentiated progeny expressed levels lower compared with WT-H1 cells (supplemental online Fig. 4a). This observation may be explained either by the experimental variation commonly seen during hESC differentiation or by a moderate loss of differentiation potential of the transgenic lines after the gene editing process. In any case, estimation of the relevance of this finding is difficult, as direct comparison of in vitro differentiation potential to the specific lineage studied after gene targeting has not previously been assessed in hESC lineage tracing studies [21–28]. When this protocol was used to differentiate the WT-H1 line, gene expression of pancreatic endocrine genes was also different from WT-H9 on day 16 (supplemental online Fig. 4b), reflecting the common observation that different human ESC lines have different abilities to differentiate to specific lineages [1, 17, 19, 29].
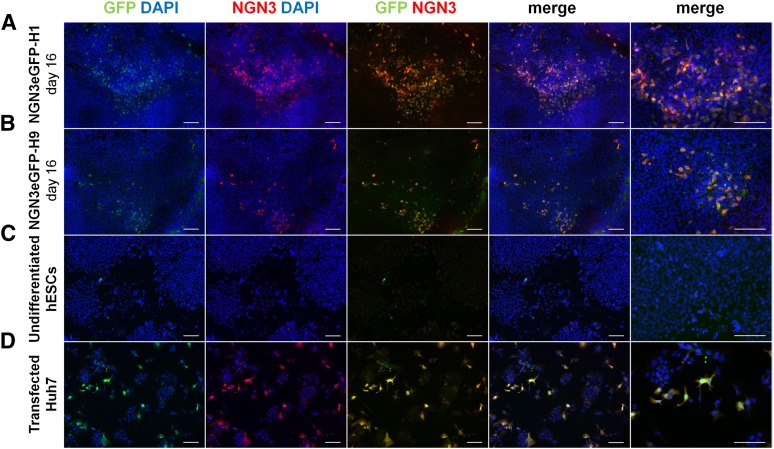
As robust NGN3 expression could be detected between day 12 and day 16, we choose day 12 for the initiation of the puromycin selection and day 16 to isolate committed NGN3+ cells from the targeted lines. In order to determine the faithfulness of the knockin add-on, we analyzed GFP and NGN3 expression on nonselected day 16 progeny of NGN3eGFP-H9 and -H1 expressing cells. The specificity of both antibodies was first tested on positive control Huh7.5 cells (Fig. 3D; supplemental online Fig. 5d) to eventually demonstrate that the GFP signal colocalizes consistently with the NGN3 expression in the transgenic lines (Fig. 3A–3C; supplemental online Fig. 5a–5c). Some studies have suggested that retention of the cassette used for the selection of recombinant clones may interfere with the faithfulness of the lineage specific reporter cassette [24, 30]. However, in day 16 progeny of NGN3eGFP-H1 and -H9 cell lines, all GFP+ cells were NGN3+ and all NGN3+ cells were GFP+, demonstrating that the reporter lines faithfully express GFP from the endogenous NGN3 locus and ruling out the need to remove the selectable cassette.
Figure 3.
NGN3 and GFP immunocytochemistry of NGN3eGFP-hESCs-derived day 16 progeny. Day 16 NGN3eGFP-hESCs-derived cells (unsorted and nonselected) (A, B), undifferentiated hESCs (C), and NGN3-GFP transfected Huh7.5 cells (D) were stained with anti-NGN3 to confirm the colocalization of NGN3 and GFP. Scale bars = 100 μm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; hESC, human embryonic stem cell.
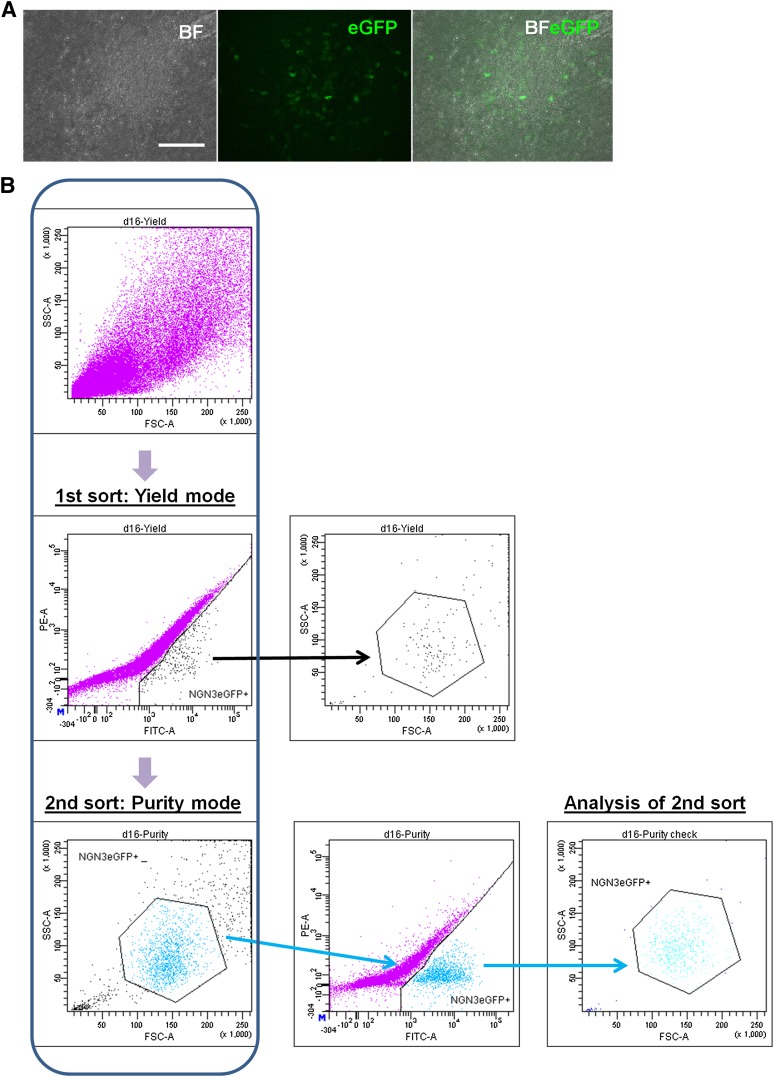
The differentiating hESCs were treated with 600 ng/ml puromycin from day 12 to day 16 to pre-enrich the NGN3eGFP+ fraction (ranging from 0.3% to 2%) before FACS (Fig. 4A). To ensure rapid selection of highly purified cells, two sequential sorts were performed (Fig. 4B): first, a low purity stringency sort that recovered all putative positive cells and pre-enriched the NGN3+ cell population up to 25%, followed by a high purity stringency sort. The final purity of the FACS-selected NGN3eGFP+ cells was 90.9% ± 2.5% (n = 3); the remaining ∼10% can be mostly accounted for by cell debris, as determined by their low forward scatter and side scatter (Fig. 4B). The NGN3eGFP+ cells expressed NGN3 mRNA at levels 63-fold higher than unsorted cells (supplemental online Fig. 6a).
Figure 4.
Isolation of NGN3eGFP+ cells from the NGN3eGFP human embryonic stem cell (hESC) lines. (A): Bright field, eGFP, and overlay images of puromycin-selected differentiation cultures. Scale bar = 100 μm. (B): Fluorescence-activated cell sorting plots showing the sorting strategy to enrich for NGN3eGFP+ cells. The NGN3eGFP+ cells were first sorted in yield mode by gating on the GFP+ cells, followed by purity sort according to both their forward and side scatter profiles and GFP signal. After the purity sort, cells were reanalyzed to check for purity. Abbreviations: BF, bright field; BFeGFP, overlay of bright field and eGFP; d, day; FITC, fluorescein isothiocyanate; FSC, forward scatter; SSC, side scatter.
Characterization of the NGN3eGFP+ Cells From Differentiating hESCs
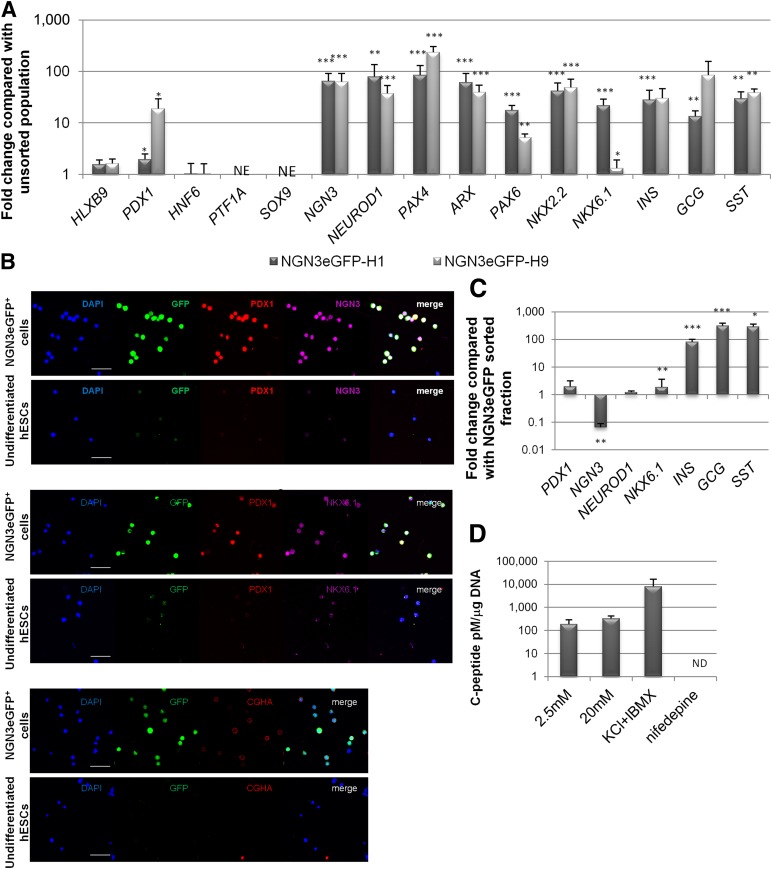
To characterize the FACS-purified NGN3eGFP+ cells, we used qRT-PCR and immunostaining. Aside from the 63-fold increased transcript levels of NGN3, other pancreatic marker genes (PDX1, NEUROD1, PAX4, ARX, PAX6, NKX2.2, NKX6.1, INS, SST, and GCG) were significantly enriched compared with the unsorted fraction (Fig. 5A). Although we could detect INS, SST, and GCG transcripts in NGN3eGFP+ progenitors, immunostaining demonstrated that protein expression was very faint (supplemental online Fig. 6b). A similar mRNA expression profile has been described by Xu et al. for the most committed subpopulation of Ngn3-expressing cells (eGFP+-high side scatter [eGFP/HSSC]) isolated from adult mouse pancreas after partial duct ligation [31]. Like mouse eGFP-HSSCs, human NGN3eGFP+ cells did not express SOX9 or PTF1a transcripts. PTF1a is not required for NGN3 specification [32], whereas SOX9 is expressed in early multipotential progenitors of the pancreas during development [33], indicating that the human NGN3eGFP+ cells may already have passed through that stage. HLXB9 and HNF6 transcripts were present in NGN3eGFP+ cells but at levels similar to those in the unsorted population (Fig. 5A), likely because of the upstream and more widespread expression of these genes [34]. All eGFP+ cells stained positive for NGN3 (confirming again the faithfulness of the knockin), PDX1, and NKX6.1, further confirming their progenitor state. In addition, NGN3eGFP+ cells stained positive for CHGA (Fig. 5B) but were negative for CD142 (data not shown). CHGA was recently shown to be absent in pancreatic endodermal progenitors but present on endocrine cells derived from hESCs [6].
Figure 5.
Characterization of isolated NGN3eGFP+ cells and in vitro maturation in response to insulin-like growth factor II (IGF-II) and NA. (A, B): Freshly isolated NGN3eGFP+ cells: fold change of pancreatic markers compared with unsorted cell population (A) and immunostaining for GFP, PDX1, NGN3, NKX6.1, and CHGA (B). Scale bars = 25 μm (top two rows), 50 μm (middle two rows), and 25 μm (bottom two rows). (C): Sorted NGN3eGFP+ cells cultured for 24 days with IGF-II and NA. Shown are relative mRNA expression levels of pancreatic markers compared with the initial NGN3eGFP sorted fraction. (D): C-peptide release of NGN3eGFP+-derived clusters was induced with 2.5 mM, 20 mM glucose, 2.5 mM d-glucose with 1 mM IBMX, and 30 mM KCl or 20 mM d-glucose with 50 μM nifedepine. All results shown are from three or more independent experiments. Data are represented as mean ± SEM. Asterisks indicate statistical significance as determined by t test. Values of p < .05 (∗),p < .01 (∗∗), and p < .001 (∗∗∗) were considered statistically significant. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; hESC, human embryonic stem cell; IBMX, 3-isobutyl-1-methylxanthine; ND, not detected; NE, nonexpressed.
NGN3eGFP+ Cells Undergo Further Maturation In Vitro
To induce further differentiation of the NGN3eGFP+ cells to pancreatic endocrine hormone-producing cells, we plated NGN3eGFP+ cells in low-attachment plates to allow cell cluster formation. The clusters were maintained for an additional 24 days with IGF-II and NA, a combination of factors previously shown to support in vitro maturation of progenitors to INS+ cells [35]. Transcript levels for NGN3 were, as expected with maturation, significantly down regulated, whereas expression levels of NKX6.1 and the endocrine hormones INS, GCG, and SST were significantly increased (Fig. 5C). We assessed the glucose responsiveness of the NGN3eGFP+-derived clusters. Exposure to 20 mM d-glucose did not induce secretion of C-peptide. However, stimulation with KCl and the phosphodiesterase inhibitor IBMX induced C-peptide release. On the other hand, nifedipine, a calcium antagonist, blocked the basal C-peptide release seen with 20 mM glucose-stimulation (Fig. 5D). These findings are in accordance with other published reports and may reflect the significantly lower levels of INS expression in the in vitro NGN3eGFP+ clusters compared with islets of Langerhans [1, 35, 36].
NGN3eGFP+ Cells Have the Capacity to Differentiate Toward Endocrine Hormone-Expressing Cells In Vivo
Approximately 200,000 NGN3eGFP+ cells were transplanted immediately after sorting under the kidney capsule of normoglycemic immunodeficient mice to assess whether further maturation occurs in vivo. Mice were sacrificed after 8–9 weeks, and kidneys were harvested. Grafts were present in all transplanted mice, and human cells could be identified by FISH with a specific human DNA CotI probe (supplemental online Fig. 7). Human cells positive for CHGA were detected (Fig. 6A), as well as INS, GCG, and SST (Fig. 6B–6D). Some of the CHGA+ cells stained positive for INS, GCG, or SST (Fig. 6C, 6D, 6F, 6G). Although some authors have noticed INS+/GCG+ and INS+/SST+ cells in hESC-derived grafts [1, 3], no double hormone positive cells were found in our grafts. Likewise, no NGN3eGFP+ cells were detected (Fig. 6E). Immunostaining for exocrine marker Amylase and the ductal marker CK19 was negative. As NGN3-expressing cells can also give rise to glial precursor cells [37], we tested whether the NGN3eGFP+-derived cells stained positive for GFAP, but we did not find positive cells. The grafted cells were also negative for the liver marker Albumin; the intestinal markers CDX2, MUCIN-2, and LYSOZYME; the neuronal marker β-tubulin III; and the mesodermal marker α-smooth muscle actin (data not shown). Teratomas were not detected.
Figure 6.
NGN3eGFP+ cells transplanted under the kidney capsule give rise to endocrine-positive progeny. (A–E): Fluorescence in situ hybridization with labeled CotI DNA and/or immunostaining for CHGA, INS, GCG, SST, and GFP. Human CHGA+ cells are indicated by green arrows (A). (B): Human INS+ and GCG+ cells (green and purple arrows, respectively). Single-positive INS (green arrows), GCG (purple arrows), and SST (magenta arrows) cells coexpressed CHGA (C, D). NGN3eGFP+ cells were not detected in the graft (E). (F, G): Magnification of CotI, CHGA, and SST (E) and CotI, CHGA, and INS (F). Scale bars = 50 μm (A–C, E), 100 μm (D), and 20 μm (F, G). Abbreviations: CHGA, chromogranin A; DAPI, 4′,6-diamidino-2-phenylindole.
Discussion
Many attempts have been made to generate pancreatic β cells from pluripotent stem cells. An important hurdle that investigators generally face is the fact that most of these directed differentiations generate mixed progeny, and therefore, the identification of the specific cell population that contributes to the generation of β cells is unclear [1–3]. Here we used ZFN-mediated homologous recombination to knock in an eGFP-PuroR cassette in the NGN3 gene in hESCs. Most gene targeting studies in hESCs have disrupted one copy of the gene of interest by introducing a selection cassette in the initial coding exons [24, 26]. Abrogation of the expression of one allele may cause haploinsufficiency of the gene of interest, which may be important for key transcription factors, as has been shown for Oct4 [38]. To retain both NGN3 alleles intact, we choose to insert a polycistronic 2A-linked reporter-selectable cassette at the C terminus of NGN3, deleting the stop codon. This allowed the selection of pure NGN3+ cells from mixed hESC progeny. To our knowledge, this is the first detailed study reporting a knockin add-on in a nonexpressed gene in hESCs, which allows for the successful isolation of a specific cell lineage.
We selected NGN3eGFP+ cells from hESC progeny instructed to differentiate to endoderm with Activin A and Wnt3, followed by differentiation toward endocrine pancreas. The NGN3eGFP+-sorted cells could be further differentiated in vitro to endocrine cells when cultured with presence of IGF-II and NA, although no glucose responsiveness could be detected. This is not unexpected, as expression levels for INS, GCG, and SST remained 10- to 100-fold lower than in human islets. We also further demonstrated that pure NGN3eGFP+ cells, when transplanted immediately after sorting under the kidney capsule of immunodeficient mice, generated CHGA+ progeny that costained mainly single-positive for INS, GCG, or SST, whereas neither ductal nor exocrine pancreatic cells were identified. The fact that we could identify relatively few cells under the kidney capsule may be attributable to the low number of transplanted cells [2, 3, 39] or the need for a more appropriate environment for further differentiation, which might be provided by cotransplantation of pancreatic embryonic tissue [6]. Under the current differentiation protocol, the number of NGN3eGFP+ cells was low, precluding transplantation of more cells. However, the availability of the NGN3eGFP-hESC reporter line should now enable the development of improved differentiation protocols.
Lineage tracing studies in mouse demonstrated that Ngn3+ cells from both embryonic and adult pancreas tissue are endocrine progenitors that give rise to all endocrine cells and suggested that regeneration may recapitulate developmental pathways [40]. Xu et al. also demonstrated that Ngn3eGFP+ cells isolated from regenerating pancreas in the adult mouse contain eGFP+ low side scatter (eGFP/LSSC) cells that are similar to NGN3eGFP+ cells from embryonic day 13.5, further supporting the previous study [31]. Aside from the eGFP/LSSC population, another Ngn3+ subpopulation (eGFP/HSSC), more granulated and believed to be more committed, was also detected and showed an expression profile comparable to that of the human ESC-derived NGN3eGFP+ cells described here. Using differentiating hESCs, Kelly et al. demonstrated that CD142+/PDX1+/NKX6.1+/CHGA− cells give rise to all three pancreatic cell types and represent pancreatic endoderm [6]. On the basis of these murine and human studies, we believe that the NGN3eGFP+ cells isolated in the current study, which are also PDX1+ and NKX6.1+, do not express CD142 or PTF1A but express CHGA, mature in vivo to islet cells, and are pancreatic endocrine progenitors. Thus, our findings provide new insight in the development of pancreatic endocrine fate specification in human.
The NGN3eGFP-hESC lines will be a useful tool for optimization of hESC differentiation protocols, as they will allow high-throughput screening for small molecules, which can help increase the number of NGN3eGFP+ cells during differentiation [4, 5], as well as for molecules that may stimulate NGN3+ cell differentiation toward β cells. In addition, the purified NGN3eGFP+ cell fraction should allow the identification of cell-surface markers that could then be used to detect NGN3+ cells in fetal and postnatal tissues [31, 40]. Finally, it will be of interest to test whether the NGN3eGFP-hESC line could be used to isolate glial, intestine, or gut progenitors from hESC cultures directed toward the neuroectodermal or intestinal lineage.
Conclusion
We demonstrated that NGN3eGFP+ cells prospectively isolated from differentiating hESCs can develop further toward single-hormone expressing cells in vivo, providing proof of principle for the purification of a human pancreatic endocrine progenitor. We also demonstrated that the NGN3eGFP knockin add-on strategy is a reliable and powerful tool for lineage tracing purposes that can also be used in other areas of human stem cell research.
Supplementary Material
Acknowledgments
We thank V. Van Duppen for his expertise in cell sorting and J. Laureys for his excellent transplantation skills. Prof. T. Voet was very kind in suggesting the use of the DNA CotI probe. K. Eggermont and V. Vanslembrouck contributed significantly to this work with valuable advice and help, and W. Vandendries and P. Berckmans were very helpful in the technical work. The array comparative genomic hybridization analysis was verified by Prof. H. Van Esch from the Center of Human Genetics at KU Leuven (Leuven, Belgium), and the karyotype analysis and chromosome counts were performed by Dr. C. Staessen from the Unit of Human Reproduction at the Vrije Universiteit Brussel (Brussels, Belgium). Q.C., K.V., and J.V. were supported by the Agentschap voor Innovatie door Wetenschap en Technologie (IWT); P.B. was supported by the European Molecular Biology Organization and the European Foundation for the Study of Diabetes/Juvenile Diabetes Research Foundation; R.S. was supported by the Dutch Diabetes Foundation; H.H. was supported by Fonds Wetenschappelijk Onderzoek Vlaanderen (FWO) G034613N; L.O. was supported by Gobierno de Aragon, Instituto Aragonés de Ciencias de la Salud, and IWT; and C.M.V. was supported by KU Leuven (EIW-B4855-EF/05/11, ETH-C1900-PF, EME-C2161-GOA/11/012), an FWO Odysseus award, FWO G.0601.07, and Fonds Jouw Gezondheid.
Author Contributions
Q.C.: conception and design, financial support, administrative support, provision of study material, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; P.B.: financial support, provision of study material, collection and/or assembly of data, manuscript writing, final approval of manuscript; R.S.: provision of study material, collection and/or assembly of data, manuscript writing, final approval of manuscript; K.V., J.V., C.G., M.D.-R., and S.R.: provision of study material, collection and/or assembly of data, final approval of manuscript; H.H.: financial support, provision of study material, collection and/or assembly of data, final approval of manuscript; L.O.: conception and design, financial support,, provision of study material, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; C.M.V.: conception and design, financial support, provision of study material, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure
The authors indicate no potential conflicts of interest.
References
- 1.D’Amour KA, Bang AG, Eliazer S, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 2.Kroon E, Martinson LA, Kadoya K, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 3.Rezania A, Bruin JE, Riedel MJ, et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61:2016–2029. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Borowiak M, Fox JL, et al. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 5.Borowiak M, Maehr R, Chen S, et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly OG, Chan MY, Martinson LA, et al. Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat Biotechnol. 2011;29:750–756. doi: 10.1038/nbt.1931. [DOI] [PubMed] [Google Scholar]
- 7.Piper K, Brickwood S, Turnpenny LW, et al. Beta cell differentiation during early human pancreas development. J Endocrinol. 2004;181:11–23. doi: 10.1677/joe.0.1810011. [DOI] [PubMed] [Google Scholar]
- 8.Polak M, Bouchareb-Banaei L, Scharfmann R, et al. Early pattern of differentiation in the human pancreas. Diabetes. 2000;49:225–232. doi: 10.2337/diabetes.49.2.225. [DOI] [PubMed] [Google Scholar]
- 9.Lyttle BM, Li J, Krishnamurthy M, et al. Transcription factor expression in the developing human fetal endocrine pancreas. Diabetologia. 2008;51:1169–1180. doi: 10.1007/s00125-008-1006-z. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar SA, Kobberup S, Wong R, et al. Global gene expression profiling and histochemical analysis of the developing human fetal pancreas. Diabetologia. 2008;51:285–297. doi: 10.1007/s00125-007-0880-0. [DOI] [PubMed] [Google Scholar]
- 11.Castaing M, Péault B, Basmaciogullari A, et al. Blood glucose normalization upon transplantation of human embryonic pancreas into beta-cell-deficient SCID mice. Diabetologia. 2001;44:2066–2076. doi: 10.1007/s001250100012. [DOI] [PubMed] [Google Scholar]
- 12.Castaing M, Duvillie B, Quemeneur E, et al. Ex vivo analysis of acinar and endocrine cell development in the human embryonic pancreas. Dev Dyn. 2005;234:339–345. doi: 10.1002/dvdy.20547. [DOI] [PubMed] [Google Scholar]
- 13.Xie R, Everett LJ, Lim HW, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apelqvist A, Li H, Sommer L, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 15.Gradwohl G, Dierich A, LeMeur M, et al. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo A, Shinozaki K, Shannon JM, et al. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 17.Nostro MC, Sarangi F, Ogawa S, et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- 19.Mfopou JK, Chen B, Mateizel I, et al. Noggin, retinoids, and fibroblast growth factor regulate hepatic or pancreatic fate of human embryonic stem cells. Gastroenterology. 2010;138:2233–2245. doi: 10.1053/j.gastro.2010.02.056. e1–e14. [DOI] [PubMed] [Google Scholar]
- 20.Otonkoski T, Beattie GM, Mally MI, et al. Nicotinamide is a potent inducer of endocrine differentiation in cultured human fetal pancreatic cells. J Clin Invest. 1993;92:1459–1466. doi: 10.1172/JCI116723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue H, Wu S, Papadeas ST, et al. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27:1836–1846. doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Rodriguez RT, Wang J, et al. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis RP, Ng ES, Costa M, et al. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- 26.Elliott DA, Braam SR, Koutsis K, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 27.Micallef SJ, Li X, Schiesser JV, et al. INS(GFP/w) human embryonic stem cells facilitate isolation of in vitro derived insulin-producing cells. Diabetologia. 2012;55:694–706. doi: 10.1007/s00125-011-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruby KM, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- 29.Osafune K, Caron L, Borowiak M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 30.Scarff KL, Ung KS, Sun J, et al. A retained selection cassette increases reporter gene expression without affecting tissue distribution in SPI3 knockout/GFP knock-in mice. Genesis. 2003;36:149–157. doi: 10.1002/gene.10210. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, D’Hoker J, Stangé G, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 32.Burlison JS, Long Q, Fujitani Y, et al. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seymour PA, Freude KK, Tran MN, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacquemin P, Durviaux SM, Jensen J, et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang J, Au M, Lu K, et al. Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells. 2007;25:1940–1953. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- 36.Jiang W, Shi Y, Zhao D, et al. In vitro derivation of functional insulin-producing cells from human embryonic stem cells. Cell Res. 2007;17:333–344. doi: 10.1038/cr.2007.28. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Wu Y, Qi Y, et al. Neurogenin3 participates in gliogenesis in the developing vertebrate spinal cord. Dev Biol. 2003;253:84–98. doi: 10.1006/dbio.2002.0868. [DOI] [PubMed] [Google Scholar]
- 38.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 39.Sneddon JB, Borowiak M, Melton DA. Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature. 2012;491:765–768. doi: 10.1038/nature11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.