This study generated an induced pluripotent stem cell (iPSC) model of hypoplastic left heart syndrome (HLHS) malformation and characterized the properties of cardiac myocytes (CMs) differentiated from these and control-iPSC lines. CMs derived from an HLHS patient demonstrate a number of marker expression and functional differences to human embryonic stem cells/control iPSC-derived CMs, thus providing some evidence that cardiomyocyte-specific factors may influence the risk of HLHS.
Keywords: Hypoplastic left heart syndrome, Induced pluripotent stem cells, Cardiac myocytes, Cardiac development, Pluripotent stem cell differentiation
Abstract
Hypoplastic left heart syndrome (HLHS) is a serious congenital cardiovascular malformation resulting in hypoplasia or atresia of the left ventricle, ascending aorta, and aortic and mitral valves. Diminished flow through the left side of the heart is clearly a key contributor to the condition, but any myocardial susceptibility component is as yet undefined. Using recent advances in the field of induced pluripotent stem cells (iPSCs), we have been able to generate an iPSC model of HLHS malformation and characterize the properties of cardiac myocytes (CMs) differentiated from these and control-iPSC lines. Differentiation of HLHS-iPSCs to cardiac lineages revealed changes in the expression of key cardiac markers and a lower ability to give rise to beating clusters when compared with control-iPSCs and human embryonic stem cells (hESCs). HLHS-iPSC-derived CMs show a lower level of myofibrillar organization, persistence of a fetal gene expression pattern, and changes in commitment to ventricular versus atrial lineages, and they display different calcium transient patterns and electrophysiological responses to caffeine and β-adrenergic antagonists when compared with hESC- and control-iPSC-derived CMs, suggesting that alternative mechanisms to release calcium from intracellular stores such as the inositol trisphosphate receptor may exist in HLHS in addition to the ryanodine receptor thought to function in control-iPSC-derived CMs. Together our findings demonstrate that CMs derived from an HLHS patient demonstrate a number of marker expression and functional differences to hESC/control iPSC-derived CMs, thus providing some evidence that cardiomyocyte-specific factors may influence the risk of HLHS.
Introduction
Hypoplastic left heart syndrome (HLHS) is characterized by underdevelopment of the left-sided cardiac structures, variably including hypoplasia or atresia of the left ventricle, ascending aorta, and aortic and mitral valves. HLHS is the commonest cause of heart transplantation in infancy and is the cardiovascular malformation most frequently resulting in childhood death (without surgical intervention, HLHS is invariably fatal). There is a substantial familial predisposition to HLHS [1–3], although it is not usually a Mendelian condition and first-degree relatives of probands with HLHS have a high incidence (11%, versus 1%–2% in the general population) of bicuspid aortic valve. Despite this, only a minority of neonates with severe obstruction of left ventricular outflow have HLHS. Diminished flow through the left side of the heart [1, 2] and disruption of genetic networks specific to left ventricular chamber development [4–7] have been suggested to cause HLHS. In particular it has been hypothesized that disruption of genetic networks specific to the left ventricular chamber, which depend on the differential expression of such genes as Tbx5 and Lrx1, is also contributory to the pathogenesis of the condition, and loss-of-function mutations in the gene HAND1, which is expressed in left-sided cardiac structures including the LV myocardium, are present in a proportion of patients with HLHS. One report has noted that HLHS patient cardiac myocytes show inappropriate expression of platelet endothelial cell adhesion molecule-1 (PECAM-1) (CD31) and indications that the HLHS left ventricle has a gene expression pattern reminiscent of a heart failure or fetal gene expression pattern [8]. As yet, however, there is relatively little evidence in favor of the role of cardiac myocyte (CM)-specific factors in HLHS risk. We have derived induced pluripotent stem cell (iPSC) lines from one HLHS patient who demonstrated mild facial dysmorphism, bilateral hip dislocation, hypoplastic fingernails, mild microcephaly, aortic atresia, and mitral atresia who subsequently died at 10 days of age. CMs differentiated from these lines show substantial differences to those obtained from human embryonic stem cells (hESCs) [9] and control-iPSC lines derived from unaffected neonatal human dermal fibroblast (NHDF), supporting the hypothesis that patient-specific myocardial developmental factors make a substantial contribution to the development of HLHS.
Materials and Methods
Cell Source
Dermal fibroblasts derived from the skin biopsy of one HLHS patient (GM12601) were obtained from the Coriell Institute for Medical Research. The patient was clinically diagnosed with hypoplastic left heart syndrome characterized by aortic and mitral atresia. Associated malformations included mild facial dysmorphism, bilateral hip dislocation, hypoplastic fingernails, and mild microcephaly. The patient died 10 days after birth.
Derivation, culture, and characterization of human iPSC lines were performed using a polycistronic lentiviral system (Allele Biotech, San Diego CA, http://www.allelebiotech.com) [10]. More details are given in the supplemental online data.
Cardiac differentiation of human embryonic stem cell (ESC)/iPSC lines and characterization of cardiac progenitors were carried out as described in [11], with full details presented in the supplemental online data.
DNA fingerprinting and karyotype analysis were performed as described in [12] (more details are available in the supplemental online data).
Flow Cytometric Analysis
Human iPSCs were cultured under feeder-free conditions (Matrigel [BD Biosciences, San Diego, CA, http://www.bdbiosciences.com] and murine embryonic feeder cell-conditioned media) for two passages. Single-cell dissociation was achieved by adding Accutase (Life Technologies, Rockville, MD, http://www.lifetech.com) for 5 minutes and incubating at room temperature. A total of 200,000–500,000 single cells were resuspended in phosphate-buffered saline (PBS) supplemented with 5% fetal calf serum (FCS) and added to a well of a 96-well plate (U-shaped). The plate was spun at 200g for 5 minutes at 4°C, and the supernatant was discarded by flicking the whole plate with adherent cells remaining attached to the bottom. Isotype control or conjugated TRA-1-60 (Millipore, Billerica, MA, http://www.millipore.com) and NANOG (Cell Signaling Technology, Beverly, MA, http://www.cellsignal.com) were added at a dilution of 1:50, followed by incubation on ice for 1 hour. Cells were rinsed three times with PBS as before and finally resuspended in 0.5 ml of PBS plus 5% FCS for fluorescence-activated cell sorting analysis using an LSR II analyzer (BD Biosciences). At least 10,000 cells were analyzed for each experiment. A similar procedure was followed for the NANOG staining, with the exception of one additional step of cell permeabilization and fixation with 2% paraformaldehyde at 37°C for 10 minutes before antibody addition.
Transmission electron microscopy, measurement of cytosolic [Ca2+], and multielectrode array recordings were used to further characterize hESC/iPSC-derived CMs (full details are given in the supplemental online data).
DNA Sequencing of Cardiac-Specific Genes
Coding regions of NKX2.5, HAND1, and HAND2 genes, including exon-intron boundaries, were amplified by polymerase chain reaction from genomic DNA of control- and HLHS patient-derived fibroblasts and iPSCs and sequenced (full details are given in the supplemental online data).
Results
Two iPSC clones were derived from one HLHS patient and one unaffected control. Both HLHS clones behaved very similarly, so the data are presented as the average of the two clones wherever possible. iPSC derivation efficiency was lower for HLHS (0.0002%) than control-iPSCs (0.3%). A recent paper that was published while this report was under review has shown the prevalence of a senescent phenotype in HLHS-derived CMs and endothelial cells [13]. If this was to be true for fibroblasts derived from HLHS patients, it could explain the low efficiency of iPSC derivation, as suggested by Marión et al. [14].
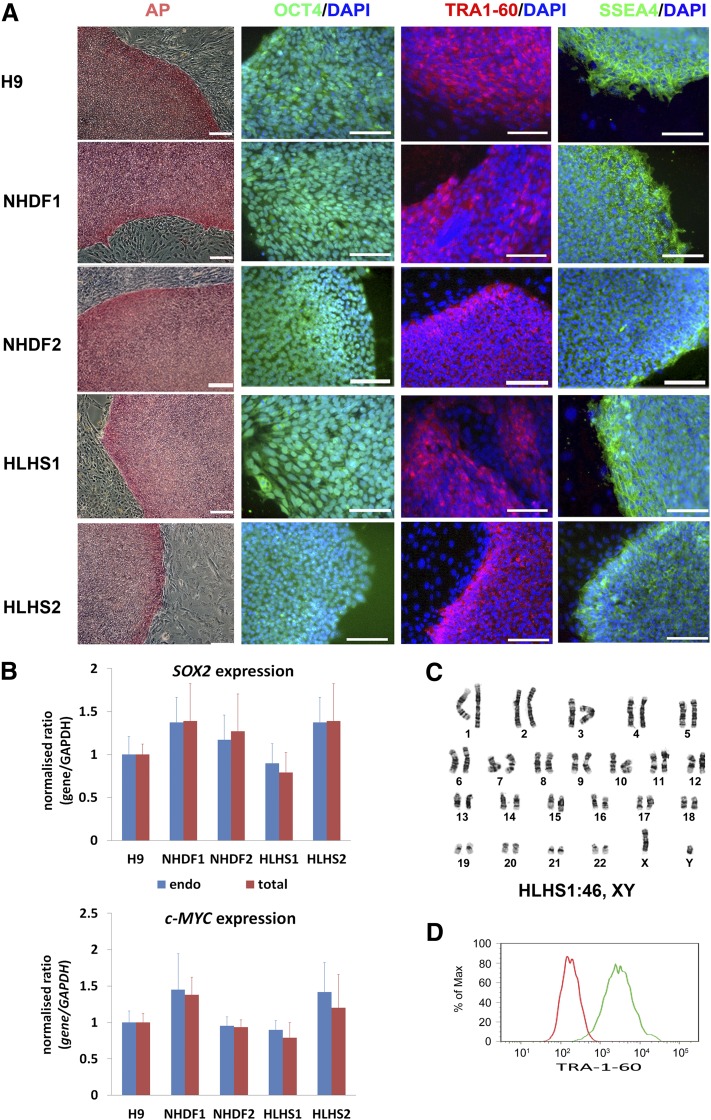
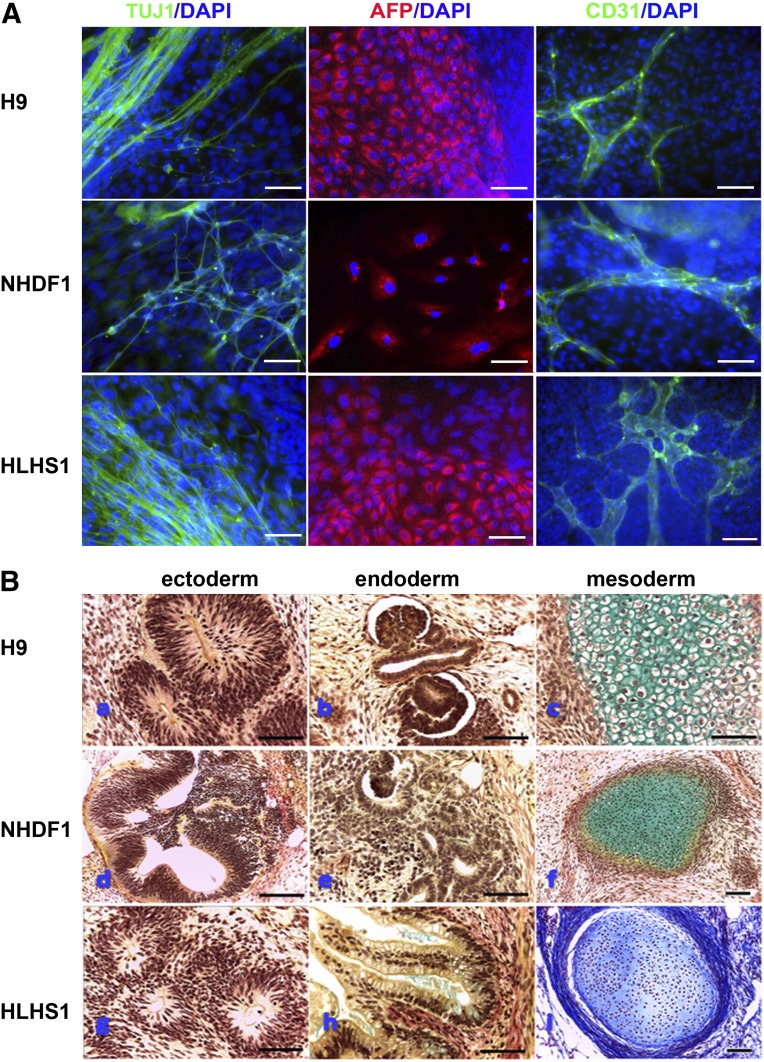
All resulting iPSCs were highly similar to hESCs (H9) (Fig. 1A). Exogenous transgenes were silenced in all pluripotent iPSC lines (Fig. 1B). All HLHS iPSC lines were karyotypically normal (Fig. 1C) and isogenic with the parent fibroblasts (supplemental online Fig. 1). Flow cytometric analysis for NANOG and TRA-1-60 expression (Fig. 1D) was performed in all iPSCs, and only the iPSC lines that showed expression of those markers in more than 90% of the cells were taken forward for further analysis. Pluripotency was further confirmed by multilineage differentiation as embryoid bodies (EBs), followed by expansion in monolayer culture. Immunohistochemical staining for β-III-tubulin, α-fetoprotein, and CD31 indicated the presence of differentiated cell types derived from ectoderm, endoderm, and mesoderm in all samples (Fig. 2A). Subcutaneous injection of HLHS- and control-iPSCs into immunocompromised NOD/SCID mice gave rise to teratomas containing tissues derived from all three embryonic germ layers (Fig. 2B). These results demonstrated that fully reprogrammed iPSCs can be derived from HLHS patients.
Figure 1.
Molecular characterization of human induced pluripotent stem cell (iPSC) lines derived from an HLHS patient and an unaffected control. (A): Staining of NHDF-derived control- and HLHS-iPSC lines and H9 human embryonic stem cells with pluripotency markers and AP. DAPI staining of the nuclei is shown in blue. This is a representative example of at least three independent experiments. Scale bars = 100 μm. (B): Quantitative reverse transcription-polymerase chain reaction analysis for the expression of total and endogenous c-MYC and SOX2 relative to GAPDH with H9 as calibrator (set to 1). Data are presented as mean ± SEM, n = 3. (C): A representative example of karyotypic analysis of HLHS-iPSC1 showing a normal 46 XY karyotype. (D): Flow cytometric analysis for expression of TRA-1-60. The red line represents the cell population stained with the isotype control, and the green line represents the control sample stained with TRA-1-60 antibody. This is a representative example of at least three independent replicates with the NHDF1 iPSC clone. Abbreviations: AP, alkaline phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HLHS, hypoplastic left heart syndrome; NHDF, neonatal human dermal fibroblast.
Figure 2.
In vitro and in vivo differentiation capacity of human embryonic stem cell (hESC), NHDF-derived control-induced pluripotent stem cell (iPSC), and HLHS-iPSC lines. (A): In vitro differentiation capacity to all three germ layers demonstrated by immunocytochemical staining with an anti-AFP antibody (endoderm), anti-CD31 antibody (mesoderm), and anti-βIII-tubulin antibody TUJ1 (ectoderm). DAPI staining of nuclei is shown in blue. Shown is a representative example of at least three independent experiments. Scale bar = 100 μm. (B): In vivo differentiation of hESC line H9 (upper panel), NHDF1-iPSCs (middle panel), and HLHS1-iPSCs (lower panel). All three pluripotent stem cell lines produced teratomas containing structures representative of each germ layer, notably neuroepithelium (Ba, Bd, Bg) (ectoderm), kidney (Bb, Be)/gut (Bh) wall (both endoderm), and cartilage (Bc, Bf, Bi) (mesoderm). Histological staining: Masson’s trichrome for iPSC-HLHS mesoderm and Weigert’s stain for all other sections. Scale bars = 150 µm (NHDF1-iPSCs and iPSC-HLHS) and 75 µm (all others). Abbreviations: AFP, α-fetoprotein; DAPI, 4′,6-diamidino-2-phenylindole; HLHS, hypoplastic left heart syndrome; NHDF, neonatal human dermal fibroblast.
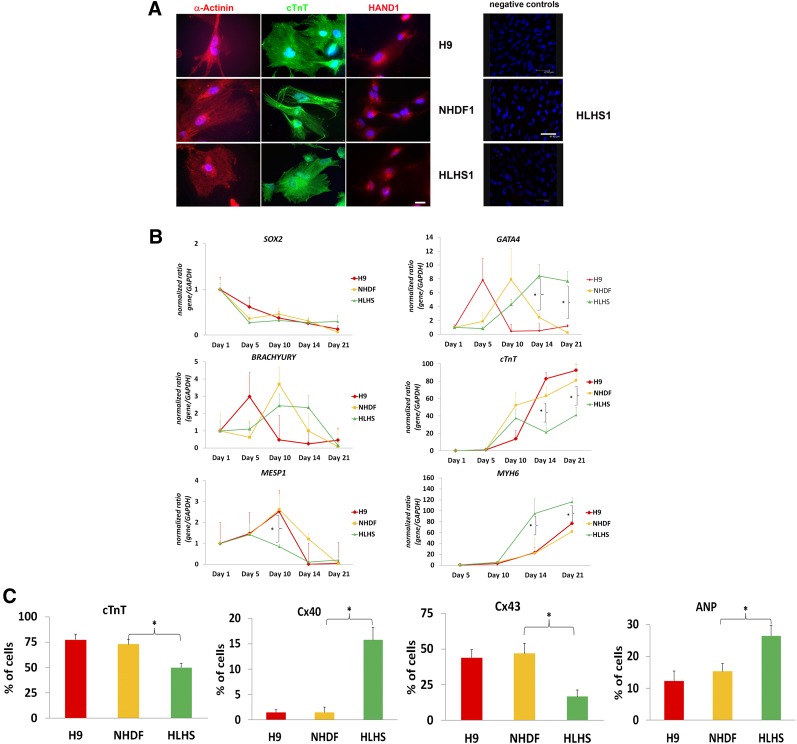
We differentiated hESC, HLHS-, and control-iPSC lines into CMs using protocols published by Kattmann et al. [11]. We were particularly interested in investigating the differences between HLHS and patient CMs as they emerged during human embryonic development, and for this reason we focused our studies on the first 21 days of iPSC and ESC differentiation that has been reported by a large number of studies to be sufficient for cardiac commitment and generation of contracting CMs. Contracting clusters containing cells expressing cardiac markers such as α-actinin, cardiac troponin (cTnT), and the early cardiac development transcription factor HAND1 [15] could be readily detected by immunohistochemistry from day 10 of differentiation (Fig. 3A). Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis during differentiation suggested that HLHS-iPSCs, control-iPSCs, and hESCs downregulated pluripotency markers as shown by SOX2 (Fig. 3B) and NANOG expression (supplemental online Fig. 2A) and formed mesoderm indicated by expression of BRACHYURY (Fig. 3B). We noticed a lower expression of a key marker of cardiac mesoderm, MESP1, at differentiation day 10 (Fig. 3B) in addition to delayed and persistent expression of the cardiac progenitor marker, GATA4, and reduced expression of a more mature CM marker, cardiac troponin (cTnT), at days 14 and 21 of differentiation for the HLHS sample. Although analysis of variance two-factor analysis indicated no significant differences between H9, NHDF, and HLHS samples during the differentiation time course for the expression of above-mentioned cardiac markers, t test analysis performed at specific and key time points indicated that the changes observed between HLHS and each of the two controls (NHDF and H9) were significant (p < .05). Quantitative RT-PCR analysis also indicates higher than expected expression of PECAM-1 (CD31) and the embryonic atrial myosin essential light chain (ALC-1) in HLHS-derived CMs compared with hESC and control iPSC lines (supplemental online Fig. 3), which supports the observations made by Bohlmeyer et al. [8] about persistence of fetal gene expression in the heart of HLHS patients. It is interesting to note higher expression of MYH6 (fast isoform of the cardiac myosin heavy chain preferentially expressed in developing atria; Fig. 3B) and decreased expression of MYH7 (myosin heavy chain β predominantly expressed in ventricles; supplemental online Fig. 2B). This was corroborated by flow cytometric analysis of beating clusters, which show reduced cTnT expression and CX43 (found mainly in ventricular myocardium) in parallel to increased expression of CX40 (enriched in atrial CMs) and ANP (atrial natriuretic peptide; Fig. 3C).
Figure 3.
Differentiation of NHDF-derived control-induced pluripotent stem cells (iPSCs), HLHS-iPSCs, and H9 human embryonic stem cells (hESCs) to cardiac myocytes (CMs) in vitro. (A): Staining with cardiac markers α-actinin, cTnT, and HAND1. Note a mixed expression pattern of HAND1 in nucleoli and nucleus, as previously described in the literature. 4′,6-Diamidino-2-phenylindole staining of nuclei is shown in blue. Shown is a representative example of at least three independent experiments. Scale bar = 100 μm (left panel) and 47.62 μm (negative controls). (B): Quantitative reverse transcription-polymerase chain reaction analysis showing changes in cardiac marker expression during the differentiation of iPSC and hESC lines. Human embryonic heart cDNA was used as calibrator for MESP1, GATA4, cTnT, and MYH6, and hESC cDNA was used as calibrator for SOX2 and BRACHYURY. The calibrator value was set to the value of 1, and all other data were calculated with respect to that. Data are presented as mean ± SEM, n = 6. ∗, Significant differences (p < .05) in expression between HLHS and each of the controls: H9 and NHDF as revealed by t test at the particular time point of differentiation. (C): Flow cytometric data analysis of cardiac markers demonstrating changes in expression profiles of HLHS-iPSC-derived CMs when compared with control-derived cells at day 21 of differentiation. Data are presented as mean ± SEM, n = 6. Analysis of variance single-factor analysis, followed by Bonferroni post hoc test, indicated significant differences between HLHS and each of the controls (H9 and NHDF) but no differences between NHDF and H9. For simplicity, only p < .05 (∗) is indicated for the comparison between NHDF and HLHS iPSC lines. Abbreviations: ANP, atrial natriuretic peptide; cTnT, cardiac troponin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HAND1, heart- and neural crest derivatives-expressed protein 1; HLHS, hypoplastic left heart syndrome; NHDF, neonatal human dermal fibroblast.
Discussion
A number of publications to date have suggested variations between iPSC clones derived from the same patients as well as iPSC lines derived from different patients in the ability to give rise to cardiomyocytes, thus necessitating inclusion of iPSC lines from multiple HLHS patients. This is difficult for congenital diseases such as HLHS, in which improvement in fetal imagining has actually led to decreased numbers of babies with HLHS. During this study, we have not observed significant differences between iPSC clones derived from either the unaffected control or the HLHS patients. Furthermore, we have not observed significant differences between the H9 human ESC lines and the unaffected control, which lead us to speculate that differences observed between control and HLHS patient are the result of the HLHS phenotype rather than intra- and interline variability. It is also encouraging to notice that another group reported similar differences in gene expression profile and ventricular/atrial commitment of cardiomyocytes using independently derived iPSC clones from two other HLHS patients [16]. Together these findings suggest that HLHS may be characterized by impaired differentiation of cardiac lineages affecting multiple steps, such as cardiac mesoderm formation, maturation of cardiac progenitors to fully mature CMs, and commitment to atrial or ventricular phenotype.
Greatly reduced expression of CX43 has been observed in HLHS cardiac myocytes by other workers [17]. CX43 functions in the electromechanical transduction and signaling between cardiomyocytes to facilitate maturation, alignment, and proliferation during development of the myocardium. The reduced CX43 expression shown by HLHS iPSC-derived CMs supports the possibility that myocardial development may be compromised by failure to correctly align cardiac myocytes. Alternatively, the higher expression of CX40 may indicate that greater numbers of ventricular conduction system (VCS) cardiomyocytes arise during differentiation of HLHS iPSCs. CX40 is a known marker of this latter cell type [18–20], and our observed data support the possibility of increased numbers of VCS cardiomyocytes. Sequencing of coding regions of NKX2.5, HAND1, and HAND2 revealed no mutations in HLHS patient sample, excluding those genes as causal factors (data not shown).
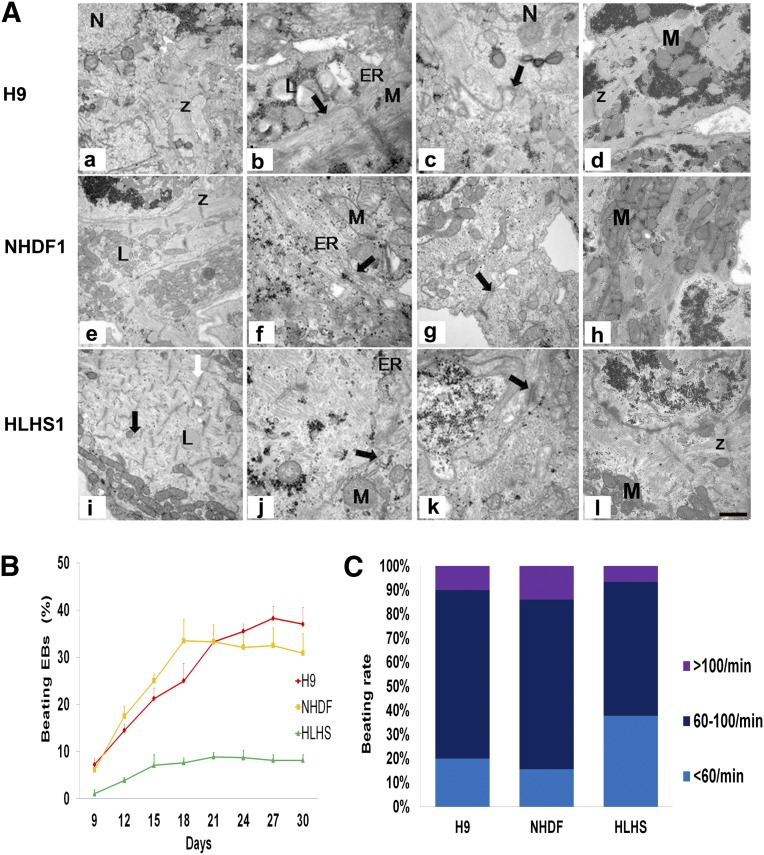
Transmission electron microscopy of hESC- and iPSC-derived CMs revealed that most CMs are mononucleated; however, whereas densely packed parallel myofibrils organized into apparently mature sarcomeres with Z-band formation were observed for hESC- and control-iPSC-derived CMs (Fig. 4Aa, 4Ae), a more random arrangement of myofibrils was observed in HLHS-iPSC-derived CMs despite the presence of abundant Z-bands (Fig. 4Ai). Moreover, CMs from hESCs and control-iPSCs show conspicuous rough endoplasmic reticulum and sarcoplasmic reticulum (Fig. 4Ab, 4Ac, 4Af, 4Ag, indicated by arrows), but these structures are less apparent in HLHS-iPSC-derived CMs (Fig. 4Aj, 4Ak).
Figure 4.
Transmission electron microscopy analysis of cardiac myocytes (CMs) derived from control- and HLHS-induced pluripotent stem cell (iPSC)-derived cells and H9 human embryonic stem cell line at day 21 of differentiation. (A): Left column shows highly organized, densely packed, paralleled myofilaments in H9 and NHD1-CMs with obvious Z-bands (Aa, Ae); less organized sarcomeres (indicated by black arrows [Ai]) were observed in the HLHS-derived CMs. Shown is a representative example of at least three independent experiments. (B): Analysis of beating embryoid bodies (EBs) derived from H9, control iPSC (NHDF), and HLHS patient-specific iPSC lines. Schematic presentation of the percentage of beating EBs throughout the differentiation window (300 EBs were analyzed for each time point). Data are presented as mean ± SEM, n = 6. Analysis of variance, two factors with replication, followed by Bonferroni post hoc test, indicated significant differences between HLHS and NHDF (p = .006) and between HLHS and H9 (p = .005). (C): Analysis of beating rate at day 21 of differentiation time course (100 EBs were analyzed in each experiment, n = 6). Abbreviations: ER, endoplasmic reticulum; HLHS, hypoplastic left heart syndrome; L, lipids; N, nuclei; NHDF, neonatal human dermal fibroblast; M, mitochondria; Z, Z-bands.
Numbers and beating rates of contractile areas from HLHS-iPSC EBs are substantially lower than those from hESCs or control-iPSCs (Fig. 4B, 4C), prompting detailed examination of CM calcium release characteristics. Spontaneous rhythmic calcium transients were detected in H9, unaffected control, and HLHS-iPSC-derived CMs (supplemental online Fig. 4A). Interestingly, HLHS-iPSC-derived CMs show an accelerated rate of Ca2+ transient decay compared with control-iPSC-derived CMs (supplemental online Fig. 4B). In fully differentiated cardiac muscle, the sarcoplasmic reticulum is the major source of Ca2+ required for contraction [21, 22]. HLHS-iPSC-derived CMs generate calcium transients in the presence of caffeine, whereas control-iPSC-derived CMs do not (supplemental online Fig. 4C), implying ryanodine receptor dysfunction in HLHS-iPSC-derived CMs. The contractile areas are still able to beat, so an alternative source of calcium such as the inositol trisphosphate system may be operative. This is indicated by upregulation of IP3R expression in HLHS-derived CMs compared with control CMs (supplemental online Fig. 5). Multielectrode array studies of contracting areas also support sarcoplasmic reticulum dysfunction. Treatment of CMs with the β1/β2 adrenergic receptor agonist isoproterenol normally increases the contraction frequency by enhancing Ca2+ ATPase activity in the sarcoplasmic reticulum [23]; however, the beat frequency of HLHS-iPSC-derived CMs increases by only 9% after isoproterenol treatment compared with 66% for hESC-derived CMs (supplemental online Fig. 6). Therefore, it is possible that for HLHS, inositol trisphosphate plays an important role in generating calcium transients. This is also an indicator of immature cardiac differentiation. Set against these data is the observation that control-iPSC-derived CMs demonstrated only a 20% increase in beat frequency; however, the inability of HLHS-iPSC-derived CMs to increase their beat frequency is significant.
Conclusion
We have presented data describing the derivation of two iPSC lines from an HLHS patient and demonstrated that CMs derived from these lines show developmental and/or functional defects that could compromise their ability to contribute to cardiogenesis in vivo. Our observations of such intrinsic cellular defects correlate with the findings of other groups, such as the possibility that gene expression programs more typical of a fetal cardiac phenotype may persist in HLHS-iPSC-derived CMs. Further studies are needed to determine whether these defects are causative of HLHS, not least of which will be the need to derive more patient-specific iPSCs to address questions of patient genetic background outside the factors responsible for the HLHS phenotype. Differentiation of HLHS-iPSCs to CMs is a valuable tool in this investigation because it permits examination of not only the more mature stages of CM development but also allows us to access several of the cardiac mesoderm cell types that precede CM formation, which is not possible from the HLHS patient tissues alone. Comparison of the transcriptomic and functional parameters of such cells will be a useful mechanism through which we may identify candidate genes whose mutations may contribute to the HLHS phenotype.
Supplementary Material
Acknowledgments
We thank Ian Dimmick and Dr. Owen Hughes for help with the flow cytometric analysis, Dr. Mauro Santibanez-Coref for help with the statistical analysis, Complement Genomics Ltd. for carrying out the DNA fingerprinting analysis, Addgene for provision of the Cre construct, Dr. Kathryn White for help with transmission electron microscopy, and Coriell Cell Repositories for providing the patient fibroblasts. This work was supported by the Newcastle Health Charity funds and the British Heart Foundation (A.W.T.). B.K. holds a British Heart Foundation Personal Chair.
Author Contributions
Y.J.: collection and assembly of data, manuscript writing; S.H., K.T., J.C., T.B., L.T., R.H., A.W.T., G.K., C.G.E., and S.P.: collection and assembly of data; J.Y.A.-A., E.S., and M.S.: collection and assembly of data, critical reading of manuscript; M.L., B.K., and L.A.: conception and design, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.L. has uncompensated research funding.
References
- 1.Harh JY, Paul MH, Gallen WJ, et al. Experimental production of hypoplastic left heart syndrome in the chick embryo. Am J Cardiol. 1973;31:51–56. doi: 10.1016/0002-9149(73)90810-2. [DOI] [PubMed] [Google Scholar]
- 2.deAlmeida A, McQuinn T, Sedmera D. Increased ventricular preload is compensated by myocyte proliferation in normal and hypoplastic fetal chick left ventricle. Circ Res. 2007;100:1363–1370. doi: 10.1161/01.RES.0000266606.88463.cb. [DOI] [PubMed] [Google Scholar]
- 3.D’Alto M, Russo MG, Pacileo G, et al. Left ventricular remodelling in outflow tract obstructive lesions during fetal life. J Cardiovasc Med. 2006;7:726–730. doi: 10.2459/01.JCM.0000247318.27041.1e. [DOI] [PubMed] [Google Scholar]
- 4.Hinton RB, Jr, Martin LJ, Tabangin ME, et al. Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol. 2007;50:1590–1595. doi: 10.1016/j.jacc.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Hinton RB, Martin LJ, Rame-Gowda S, et al. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J Am Coll Cardiol. 2009;53:1065–1071. doi: 10.1016/j.jacc.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reamon-Buettner SM, Ciribilli Y, Inga A, et al. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet. 2008;17:1397–1405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- 7.Iascone M, Ciccone R, Galletti L, et al. Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clin Genet. 2012;81:542–554. doi: 10.1111/j.1399-0004.2011.01674.x. [DOI] [PubMed] [Google Scholar]
- 8.Bohlmeyer TJ, Helmke S, Ge S, et al. Hypoplastic left heart syndrome myocytes are differentiated but possess a unique phenotype. Cardiovasc Pathol. 2003;12:23–31. doi: 10.1016/s1054-8807(02)00127-8. [DOI] [PubMed] [Google Scholar]
- 9.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y, Cowley SA, Siler U, et al. Derivation and functional analysis of patient-specific induced pluripotent stem cells as an in vitro model of chronic granulomatous disease. Stem Cells. 2012;30:599–611. doi: 10.1002/stem.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kattman SJ, Witty AD, Gagliardi M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Stojkovic M, Lako M, Stojkovic P, et al. Derivation of human embryonic stem cells from day-8 blastocysts recovered after three-step in vitro culture. Stem Cells. 2004;22:790–797. doi: 10.1634/stemcells.22-5-790. [DOI] [PubMed] [Google Scholar]
- 13.Gaber N, Gagliardi M, Patel P, et al. Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation. Am J Pathol. 2013;183:720–734. doi: 10.1016/j.ajpath.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Marión RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincentz JW, Barnes RM, Firulli AB. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91:485–494. doi: 10.1002/bdra.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bosman A, Blue G, Winlaw D et al. Investigating the genetic causation of hypoplastic left heart syndrome using induced pluripotent stem cells. Poster T-3147. International Society for Stem Cell Research 11th Annual Meeting; June 2013; Boston, MA. [Google Scholar]
- 17.Mahtab EA, Gittenberger-de Groot AC, Vicente-Steijn R, et al. Disturbed myocardial connexin 43 and N-cadherin expressions in hypoplastic left heart syndrome and borderline left ventricle. J Thorac Cardiovasc Surg. 2012;144:1315–1322. doi: 10.1016/j.jtcvs.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Miquerol L, Bellon A, Moreno N, et al. Resolving cell lineage contributions to the ventricular conduction system with a Cx40-GFP allele: A dual contribution of the first and second heart fields. Dev Dyn. 2013;242:665–677. doi: 10.1002/dvdy.23964. [DOI] [PubMed] [Google Scholar]
- 19.Harris BS, Spruill L, Edmonson AM, et al. Differentiation of cardiac Purkinje fibers requires precise spatiotemporal regulation of Nkx2-5 expression. Dev Dyn. 2006;235:38–49. doi: 10.1002/dvdy.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desplantez T, McCain ML, Beauchamp P, et al. Connexin43 ablation in foetal atrial myocytes decreases electrical coupling, partner connexins, and sodium current. Cardiovasc Res. 2012;94:58–65. doi: 10.1093/cvr/cvs025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trafford AW, Díaz ME, Negretti N, et al. Enhanced Ca2+ current and decreased Ca2+ efflux restore sarcoplasmic reticulum Ca2+ content after depletion. Circ Res. 1997;81:477–484. doi: 10.1161/01.res.81.4.477. [DOI] [PubMed] [Google Scholar]
- 22.Walden AP, Dibb KM, Trafford AW. Differences in intracellular calcium homeostasis between atrial and ventricular myocytes. J Mol Cell Cardiol. 2009;46:463–473. doi: 10.1016/j.yjmcc.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Ginsburg KS, Bers DM. Modulation of excitation-contraction coupling by isoproterenol in cardiomyocytes with controlled SR Ca2+ load and Ca2+ current trigger. J Physiol. 2004;556:463–480. doi: 10.1113/jphysiol.2003.055384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.