This study shows that chondrocytes from autologous chondrocyte implantation donors can be efficiently reprogrammed into induced pluripotent stem cells using a nonintegrating method based on mRNA delivery. Results suggest that RNA-based technology eliminates the risk of genomic integrations or aberrations, an important step toward a clinical-grade cell source for regenerative medicine such as treatment of cartilage defects and osteoarthritis.
Keywords: Induced pluripotency, Induced pluripotent stem cells, mRNA, Reprogramming, Chondrogenesis, Chondrocytes
Abstract
Human induced pluripotent stem cells (iPSCs) are potential cell sources for regenerative medicine; however, clinical applications of iPSCs are restricted because of undesired genomic modifications associated with most reprogramming protocols. We show, for the first time, that chondrocytes from autologous chondrocyte implantation (ACI) donors can be efficiently reprogrammed into iPSCs using a nonintegrating method based on mRNA delivery, resulting in footprint-free iPSCs (no genome-sequence modifications), devoid of viral factors or remaining reprogramming molecules. The search for universal allogeneic cell sources for the ACI regenerative treatment has been difficult because making chondrocytes with high matrix-forming capacity from pluripotent human embryonic stem cells has proven challenging and human mesenchymal stem cells have a predisposition to form hypertrophic cartilage and bone. We show that chondrocyte-derived iPSCs can be redifferentiated in vitro into cartilage matrix-producing cells better than fibroblast-derived iPSCs and on par with the donor chondrocytes, suggesting the existence of a differentiation bias toward the somatic cell origin and making chondrocyte-derived iPSCs a promising candidate universal cell source for ACI. Whole-genome single nucleotide polymorphism array and karyotyping were used to verify the genomic integrity and stability of the established iPSC lines. Our results suggest that RNA-based technology eliminates the risk of genomic integrations or aberrations, an important step toward a clinical-grade cell source for regenerative medicine such as treatment of cartilage defects and osteoarthritis.
Introduction
Osteoarthritis (OA) disease and cartilage injuries induce pain and joint dysfunction in patients and cause a severe burden to society and health care systems worldwide. Methods for regenerating cartilage tissue are expected to improve therapies for such disease and injuries. The autologous chondrocyte implantation (ACI) technology [1] has emerged over the past decade as the first disease-modifying treatment with excellent long-term clinical results in patients with isolated cartilage injuries [2, 3]. Although widely used, there are some inherent problems with the use of autologous chondrocytes: these cells show restricted proliferative capacity, rapidly lose their functional properties in culture, the initial biopsy is in itself an additional injury to the joint surface, and cell quality varies. Furthermore, during ACI the patient is subjected to two surgical procedures—the harvesting procedure, and, later on, the transplantation of the expanded chondrocytes—although an arthroscopic implantation is possible with matrix-assisted implantation and has reduced the rehabilitation period. Because cartilage tissue is immune privileged due to lack of vascularization, a universal donor cell line, such as human embryonic stem cells (hESCs), human mesenchymal stem cells (hMSCs), or human induced pluripotent stem cells (iPSCs), could putatively be used to replace the autologous cells. In vivo assays have shown that hESCs and hMSCs can form cartilaginous tissues; unfortunately, hMSCs have a default differentiation toward hypertrophic cartilage and bone [4]. Moreover, the ethical issues associated with hESCs, and the limited and age-related decline of proliferative capacity of hMSCs [5] are additional disadvantages of these cell types. Furthermore, it is relatively easy to make stem cells match an expression profile of mature chondrocytes, but making chondrocytes with high matrix-forming capacity from hESCs and hMSCs has proved challenging.
Induced pluripotent stem cells (iPSCs) can be derived from somatic cells by expression of various transcription factors and were achieved in murine fibroblasts in 2006 [6] and later on in human fibroblasts [7, 8]. These cells exhibit pluripotency, proliferative capacity, and gene expression similar to hESCs and have been reported to generate cartilaginous tissue in teratomas in vivo [7–9] but are not subjected to the same ethical issues as hESCs. Furthermore, iPSCs have been shown to more easily differentiate along lineages related to the cell type of origin [10], probably because of a residual epigenetic memory [11, 12], implying that the cellular origin of the iPSC facilitates successful directed differentiation. Viral integration into the genome has been an impediment of the iPSC method for clinical use, and the search for ways to induce pluripotency without causing genetic change has become the focus of intense research [13–23]. Toward this end, we chose to use modified synthetic messenger RNAs [22] to reprogram surplus chondrocytes from anonymized donors undergoing ACI [1]. Established iPSC lines originating from chondrocytes could offer an invaluable cell source for cell replacement therapy as well as in vitro modeling of the OA disease, provided that such lines can be produced without any genomic aberrations and that these cells are able to efficiently differentiate into matrix-producing chondrocytes without any remaining pluripotent abilities that risk tumor formation.
In the current study, we reprogrammed human chondrocytes and human foreskin fibroblast cells (BJ fibroblasts), established chondrocyte-derived iPSC (c-iPSC) and fibroblast-derived iPSC (f-iPSC) lines, and compared their capacities for differentiation into cartilage. We developed a novel differentiation protocol, based on previous protocols [24, 25], involving differentiation in three-dimensional (3D) pellets into chondrogenic lineage via generation of a putative chondrogenic progenitor cell population. Using this protocol, we found one c-iPSC line that readily differentiated into cartilage comparable to that of mature chondrocytes. Our c-iPSC lines, particularly the line with high in vitro cartilage differentiation potential, could facilitate studies of the OA disease and cartilage differentiation as well as lead the way to development of an iPSC-derived clinical-grade cell source for cell-replacement therapies for cartilage injuries and diseases.
Materials and Methods
Cells and Cell Culture
Surplus chondrocytes were obtained from anonymized donors (Table 1) undergoing autologous chondrocyte implantation after informed consent (ethical approval number: S040-01). The treated cartilage defects had International Cartilage Repair Society grades of 3–4, and the biopsy for cell isolation was taken from a nonbearing area on the upper side of the medial femoral condyle (International Cartilage Repair Society grade 0). Chondrocytes were isolated, as described earlier [1], and were expanded in chondrocyte medium (Table 2) [4, 26].
Table 1.
Starting cells used for reprogramming
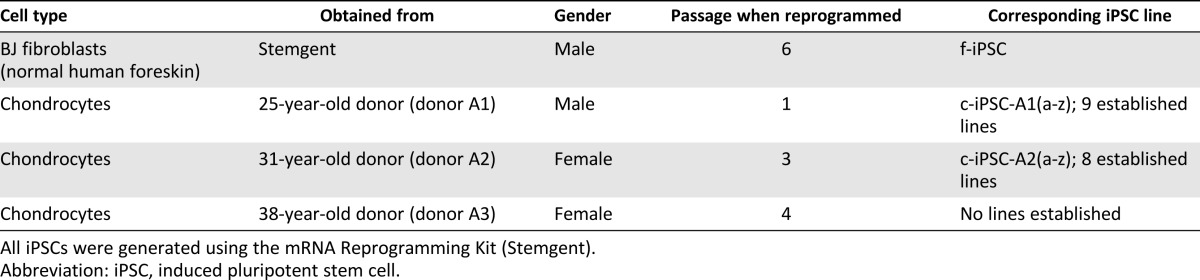
Table 2.
Reagents for cell culture
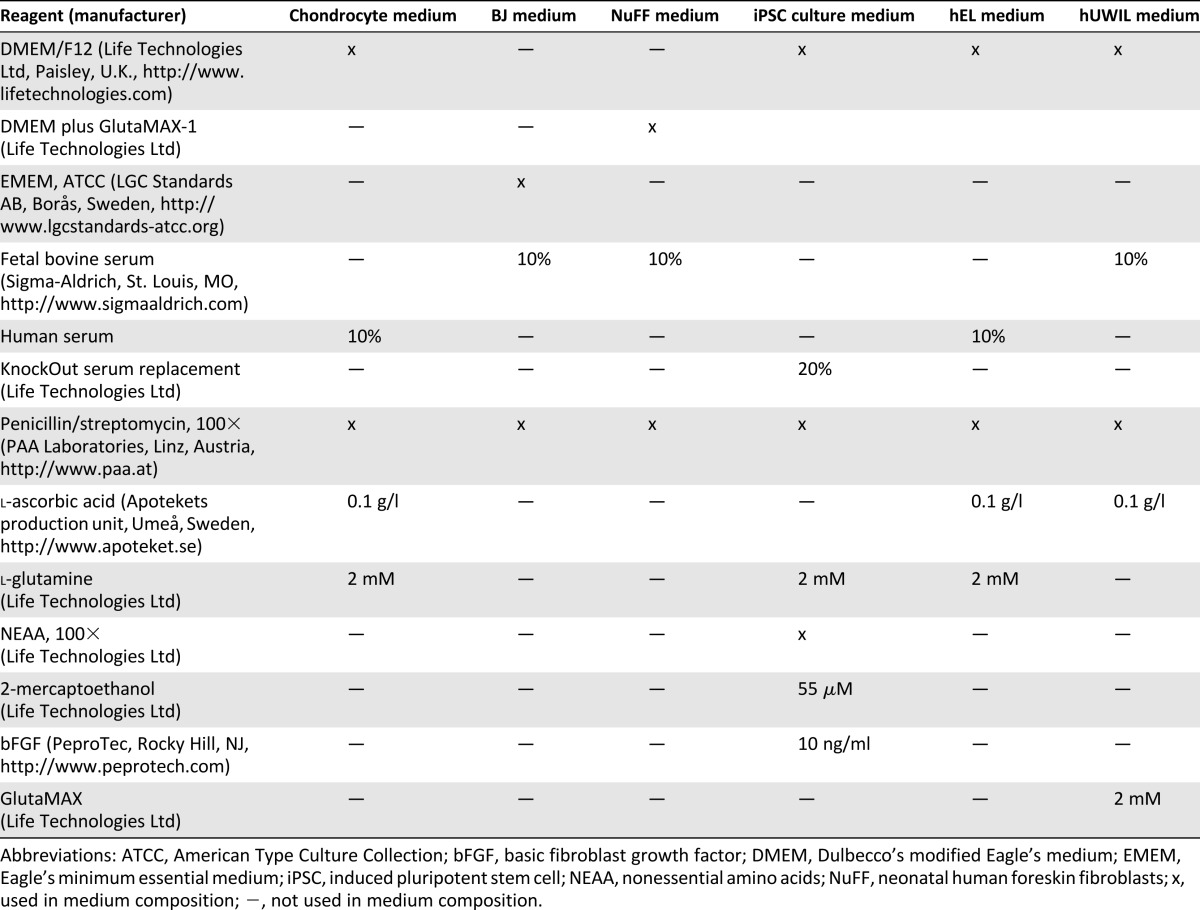
Human BJ fibroblasts were obtained from Stemgent (Stemgent, Cambridge, MA, https://www.stemgent.com) and cultured in BJ medium (Table 2) in a humidified atmosphere at 37°C and 7% carbon dioxide (CO2; Steri-Cult 200 incubator; Forma Scientific, Marietta, OH, http://www.formascientific.org).
Mitotically inactivated neonatal human foreskin fibroblasts (NuFF) were obtained from GlobalStem (AMS Biotechnology, Abingdon, U.K., http://www.amsbio.com), plated in NuFF medium (Table 2), and used as feeders during the reprogramming procedure and first passages.
The iPSCs (both c-iPSCs and f-iPSCs) were maintained in human iPSC culture medium (Table 2) at 37°C and 5% CO2 (Steri-Cult 200 incubator; Forma Scientific). The iPSCs were mechanically passaged by microdissection every 4–5 days for the first couple of passages and enzymatically passaged using collagenase type IV, 200 U/ml (Life Technologies, Paisley, U.K., http://www.lifetechnologies.com) in later passages. At the day of passaging, 10 μM Stemolecule Y27632 (Stemgent) was added to the medium.
Irradiated (25 Gy) human diploid embryonic lung fibroblast (hEL) cells [27] were expanded in hEL medium (Table 2) at 37°C in 7% CO2 (Steri-Cult 200 incubator; Forma Scientific), and used as feeders during the early passages.
The human feeder cell line hUWIL (Cellectis Bioresearch, Gothenburg, Sweden, http://www.cellectis-bioresearch.com) was expanded in hUWIL medium (Table 2) at 37°C in 5% CO2 (Steri-Cult 200 incubator; Forma Scientific) and was mitotically inactivated by irradiation (25 Gy) before plating and used as feeders during the early passages.
The hESCs (SA121) were obtained from Cellectis Bioresearch.
Three feeder systems (NuFF, hEL, and hUWIL) and two feeder-free systems (Matrigel and DEF-CS [Cellectis Bioresearch]) were tested during establishment and maintenance of our iPSC lines. All systems were able to keep our iPSCs pluripotent with regard to expression of pluripotency markers.
mRNA Reprogramming
mRNA reprogramming was conducted using the Stemgent mRNA Reprogramming Kit according to the manufacturer’s instructions, with some minor modifications. The Oct4 variant used in this protocol is Oct4A (NM_002701; the isoform responsible for the pluripotency properties of ESCs). The entire reprogramming cycle was conducted in Pluriton Reprogramming Medium (Stemgent) under hypoxic conditions in a humidified atmosphere at 37°C, 5% CO2 and 5% oxygen (Heraeus BBD6220; Thermo Scientific, Waltham, MA, http://www.thermo.com). Seeding density for chondrocytes was optimized to 1 × 103/cm2 in a previous experiment. Clonal iPSC lines were established by picking hESC-like colonies after daily mRNA transfections for 21 days (chondrocytes) and 17 days (BJ fibroblasts).
Immunohistochemical Analysis
Live staining of unfixed cells was performed using Stemgent StainAlive TRA-1-60 Antibody (DyLight 488) according to the manufacturer’s instructions. Undifferentiated human iPSCs and plated embryoid bodies (EBs) were fixed in Histofix (5% paraformaldehyde [PFA]; HistoLab Products AB, Västra Frölunda, Sweden, http://www.histolab.se), permeabilized using 0.1% Triton X-100 in phosphate-buffered saline (PBS), and blocked using Triton Block solution (0.1M glycine, 2% bovine serum albumin, 0.1% Triton X-100). Primary antibody incubations were performed in blocking solution at 4°C overnight. Primary antibodies used were rabbit-anti-Oct4 with 1:400 dilution (C30A3; Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com), rabbit-anti-Nanog with 1:800 dilution (D73G4; Cell Signaling Technology), mouse-anti-SSEA4 with 1:100 dilution (eBioMc81370; eBioscience, Ltd, Hatfield, U.K., http://www.ebioscience.com/), rabbit-anti βΙΙΙ-tubulin with 1:200 dilution (D71G9; Cell Signaling Technology), mouse-anti-actin smooth muscle with 1:200 dilution (CBL171; Millipore, Billerica, MA, http://www.millipore.com/), and rabbit-anti FoxA2/HNF3β with 1:400 dilution (D56D6; Cell Signaling Technology). Chondrogenic pellet mass culture sections were deparaffinized, rehydrated, and treated with 8,000 U/ml of hyaluronidase (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) in PBS for 60 minutes at 37°C. Sections were blocked with 3% bovine serum albumin (Sigma-Aldrich) in PBS and incubated in primary mouse-anti-human collagen II antibody with 1:150 dilution (MP Biomedicals Europe, Illkirch, France, http://www.mpbio.com/) at 4°C overnight. Incubations with secondary antibodies were performed for 2 hours at room temperature in darkness using goat-anti-mouse Alexa Fluor 546 with 1:400 dilution (A21133; Life Technologies) or goat-anti-rabbit Alexa Fluor 546 with 1:400 dilution (A11071; Life Technologies). Samples were mounted using ProLong Gold Antifade with 4′,6-diamidino-2-phenylindole (Life Technologies) to visualize nuclei. Alkaline phosphatase (ALP) activity was determined using the Stemgent Alkaline Phosphatase Staining Kit II according to the manufacturer’s instructions. All samples were observed in a Nikone Eclipse Ti fluorescence microscope (Nikon, Tokyo, Japan, http://www.nikon.com).
Western Blot Analysis
Cells (1 × 106) in feeder-free cultures (DEF-CS) were collected by centrifugation, and cell pellets were washed once with PBS. Each pellet was suspended in 300 μl RIPA buffer (50 mM Tris, 150 mM sodium chloride, 1% NP40, 1% deoxycholate, and 0.1% sodium dodecyl sulfate) with protease inhibitors (Complete ethylenediaminetetraacetic acid [EDTA] free; Roche Applied Science, Penzberg, Germany, http://www.roche-applied-science.com/) and incubated on ice for 10 minutes. Samples were cleared by centrifugation, and protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific Inc., Rockford, IL, http://www.thermofisher.com/) according to the manufacturer’s instructions. Samples were mixed with one-quarter volume NuPAGE LDS Sample Buffer (4×) (Life Technologies) complemented with protease inhibitors (Complete EDTA free; Roche Applied Science, http://www.roche-applied-science.com/) and 100 mM dithiothreitol (DTT; final concentration) and incubated at 70°C for 10 minutes. Equal amounts of total protein (10 μg) were added to each well of a denaturing NuPAGE Novex 4%–12% Bis-Tris gel (1.5 mm, 15 well; Life Technologies). Proteins were blotted to an Amersham Hybond ECL membrane (GE Healthcare, Life Sciences, Piscataway, NJ, http://www.gelifesciences.com/), and membranes were blocked in 5% milk in PBSt (PBS plus 0.1% [w/w] Tween 20; Bio-Rad Laboratories, Hercules, CA, http://www.bio-rad.com) and probed with relevant antibodies in 5% milk in PBSt (primary antibody overnight at 4°C, secondary antibody 90 minutes at room temperature). Antibodies used were rabbit-anti-Oct-4A with 1:1,000 dilution (Oct-4A [C30A3]; Cell Signaling Technologies), rabbit-anti-Nanog with 1:2,000 dilution (Nanog [D73G4] XP; Cell Signaling Technologies), and rabbit-anti-GAPDH with 1:5,000 dilution (sc-25778; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, http://www.scbt.com). Anti-rabbit-HRP with 1:3,000 dilution (Cell Signaling Technologies) was used as secondary antibody. Membranes were washed 3 × 10 minutes with PBSt between and after antibody incubations. Membranes were developed using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific Inc.), and signals were visualized using a CCD camera (ChemiDoc XRS+ System; Bio-Rad Laboratories).
In Vitro Differentiation
For in vitro differentiation or EB formation, undifferentiated iPSC colonies were excised using the Stem Cell Cutting Tool (Vitrolife, Gothenburg, Sweden, http://www.vitrolife.com) and transferred to a low-adhesion cell culture dish containing EB medium (KnockOut Dulbecco’s modified Eagle's medium, 20% fetal bovine serum; Sigma-Aldrich), 2mM L-glutamine, nonessential amino acids, 0.1 mM β-mercaptoetanol (Life Technologies), and penicillin/streptomycin (PAA Laboratories, Linz, Austria, http://www.paa.at) to form EBs. The EBs were cultured in suspension for 6 days, plated in gelatin-coated culture dishes, and cultured for two additional weeks in EB medium in a humidified atmosphere at 37°C and 5% CO2 (Steri-Cult 200 incubator). The medium was renewed every 2–3 days. Some beating EBs were fixed in Histofix (5% PFA; HistoLab Products AB) for 24 hours, dehydrated by treatment with increasing concentrations of ethanol, and embedded in paraffin. Sections (5 μm) were placed onto microscope slides, deparaffinized, and stained using hematoxylin and eosin or monoclonal anti-actin (α-Sarcomeric) antibody with 1:400 dilution (#A2172; Sigma-Aldrich) and goat-anti-mouse Alexa Fluor 568 (#A21043; Molecular Probes, Eugene, OR, http://probes.invitrogen.com; Life Technologies), as described in the previous section.
In Vitro Teratoma Formation in Agarose Culture System
In vitro teratoma formation has been described previously [28]. After 8 weeks, EBs were fixed in Histofix (5% PFA; HistoLab Products AB) for 24 hours, dehydrated by treatment with increasing concentrations of ethanol, and embedded in paraffin. Sections (5 μm) were placed onto microscope slides, deparaffinized, and stained with hematoxylin and eosin. All sections were observed in a light microscope (Nikon, Tokyo, Japan, http://www.nikon.com).
RNA Extraction and Quantitative Reverse Transcriptase Polymerase Chain Reaction
Monolayer cultures were harvested using RLT lysis buffer (Qiagen, Hilden, Germany, http://www.qiagen.com) containing DTT (40 mM). Frozen micromass cultures were homogenized using 5 mm stainless steel beads (Qiagen) and a TissueLyser (Qiagen) for 2 minutes at 25 Hz. RLT lysis buffer containing DTT (40 mM) was added to the homogenized tissues and further mixed for 2 minutes at 25 Hz. Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the Animal Cells Using Spin protocol. DNase I (Qiagen) was used to remove contaminating genomic DNA from the isolated RNA. RNA concentrations were measured with NanoDrop 1000 (ThermoScientific), with a 260/280-nm ratio between 1.9 and 2.1 considered good purity.
Quantitative Reverse Transcriptase Polymerase Chain Reaction
For cDNA synthesis and quantitative reverse transcriptase polymerase chain reaction (PCR), all reagents, instruments, and software were purchased from Applied Biosystems (Life Technologies). The cDNA was prepared from total RNA using the High-Capacity cDNA Reverse Transcriptase Kit with random hexamers and RNase Inhibitor on a 2720 Thermal Cycler. Four samples with small amounts of cDNA were preamplified without introducing amplification bias into the samples using the TaqMan PreAmp Master Mix. All samples were analyzed in duplicate on the 7900HT instrument using the TaqMan Gene Expression Master Mix. The following human TaqMan gene expression assays were used: OCT4 Hs01895061_u1, LIN28 Hs00702808_s1, NANOG Hs02387400_g1, SOX2 Hs01053049_s1, SOX9 Hs00165814_m1, COL1A1 Hs00164004_m1, COL2A1 type A Hs00156568_m1, COL2A1 type B Hs01064869_m1, ACAN Hs00153936_m1, COL10A1 Hs00166657_m1, CDH1 Hs01023894_m1, MIXL1 Hs00430824_g1, GSC Hs00418279_m1, PDGFRB Hs01019589_m1, SOX6 Hs00264525_m1, and Runx2 Hs00231692_m1. CREBBP Hs00231733_m1 was used as a reference gene [29]. All samples were tested for genomic DNA contamination using a –RT step or ValidPrime Kit on the cDNA (TATAA Biocenter, Gothenburg, Sweden, http://www.tataa.com). Fold change for each sample was calculated using the 2-ΔΔCT method [30], and the expression was calculated relative to a calibrator.
Statistical Analysis
Linear real-time PCR Ct values were used to calculate the correlation between different cell types and determine the Pearson’s correlation coefficient (r). A 95% confidence interval (CI) was calculated on r.
Datasets were analyzed for normal distribution with a nonparametric one-sample Kolmogorov-Smirnov test. Normally distributed data were analyzed by analysis of variance and Tukey's Honestly Significant Difference post hoc test. The sample size was n = 3 per group unless otherwise stated.
Chondrogenic Differentiation in Monolayer
For adaptation of iPSCs to feeder-free expansion, DEF-CS (Cellectis Bioresearch) was used according to the manufacturer’s instructions. At passage 5, cells were passaged by TrypLE Select (Life Technologies) and seeded at 100,000 cells per square centimeter in 24-well plates in DEF-CS. After 24 hours, medium was changed to basal medium supplemented with appropriate growth factors, as previously described [24] (supplemental online Table 1). The directed differentiation protocol was followed, with the following modifications: Cells were seeded in the DEF-CS system in lieu of fibronectin and only passaged once, during the differentiation cycle. Cells were harvested at 4, 9, and 14 days for analysis.
Chondrogenic Differentiation in 3D
The 3D pellet environment mimics the condensation phase during embryonic development of the limbs, during which high cell density improves intercellular communication and which is essential for chondrogenesis to occur [31, 32]. This knowledge was used to develop a novel differentiation protocol for chondrogenic differentiation. The 3D pellet mass for the predifferentiation stage was formed by releasing iPSCs on feeders with Trypsin-EDTA (Life Technologies), and 16 pellets of each iPSC line were analyzed. Cells were diluted to 2 × 106 iPSCs per milliliter in defined chondrogenic medium (Dulbecco’s modified Eagle's medium high glucose; PAA Laboratories) supplemented with 5.0 μg/ml linoleic acid (Sigma-Aldrich), 1× ITS-G premix (6.25 μg/ml insulin, 6.25 μg/ml transferrin, 6.25 ng/ml selenious acid; Life Technologies), 1.0 mg/ml human serum albumin (Equitech-Bio, Kerrville, TX, http://www.equitech-bio.com/), 10 ng/ml transforming growth factor β 1 (R&D Systems, Abingdon, U.K., http://www.rndsystems.com/), 10−7 M dexamethasone (Sigma-Aldrich), 14 μg/ml L-ascorbic acid (Apotekets production unit, Umeå, Sweden, http://www.apoteket.se), and penicillin/streptomycin (PAA Laboratories) (supplemental online Table 2). Cell suspension (200 μl) was placed into a conical polypropylene tube with 0.5 ml of defined medium, centrifuged at 400g for 5 minutes, and maintained at 37°C in 5% CO2. Medium was changed three times a week. Relevant control cultures with only irradiated feeders were kept throughout the differentiation protocol.
After 14 days of predifferentiation, the 3D pellet mass cultures were digested with collagenase type II (Worthington Biochemicals, Lakewood, NJ, http://www.worthington-biochem.com) to release cells from surrounding matrix. Released cells were washed once in chondrocyte medium (Table 2) and plated in six-well plates and expanded in chondrocyte medium. After four passages, these predifferentiated c-iPSCs were harvested for mRNA extraction, as described, for determination of chondrogenic state or cultured in a second 3D pellet mass culture. This monolayer expansion ensured the disappearance of irradiated feeder cells that were present in the predifferentiation stage. For pellet mass culture, predifferentiated c-iPSCs and primary donor chondrocytes, as control, were released with Trypsin-EDTA and resuspended in defined chondrogenic medium at 500,000 cells per milliliter. Cell suspension (500 μl) was placed into conical polypropylene tubes, centrifuged at 400g for 5 minutes, and maintained at 37°C in 5% CO2. The medium was changed three times per week. After 2 weeks, donor chondrocyte pellets were harvested for histology or mRNA extraction. Predifferentiated c-iPSC pellets were cultured for 2 weeks and 5 weeks, the longer time point allowing for progenitors to mature properly. The 3D pellet mass cultures were fixed in Histofix (5% PFA; Histolab Products AB), dehydrated with ethanol, and embedded in paraffin. Sections (5 μm) were placed onto microscope slides (Superfrost Plus; Menzel-Gläser, Braunschweig, Germany, http://www.menzel.de/), deparaffinized and stained with Alcian blue van Gieson, and then were observed with a light microscope (Nikon). The 3D pellet mass cultures were snap frozen in liquid nitrogen after 2 weeks and 5 weeks in culture and stored at −80°C before mRNA extraction, as described.
Karyotype Analysis and Single Nucleotide Polymorphism Array Analyses
Karyotype analysis was conducted using standard protocols for the chromosomal Giemsa banding at the Department for Clinical Chemistry at Sahlgrenska University Hospital, using protocols standardized for clinical samples. For each sample, 25 karyotypes were examined.
For single nucleotide polymorphism (SNP) array, DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. Array analyses were performed at the Department of Clinical Chemistry at Sahlgrenska University Hospital and at the Bioinformatics and Expression Analysis core facility at Novum, using the Affymetrics Genome-Wide Human SNP 6.0 Array. Data were analyzed using Nexus discovery (6.0) (BioDiscovery, Hawthorne, CA, http://www.biodiscovery.com).
Results
Efficient Generation of c-iPSCs Using mRNA Reprogramming
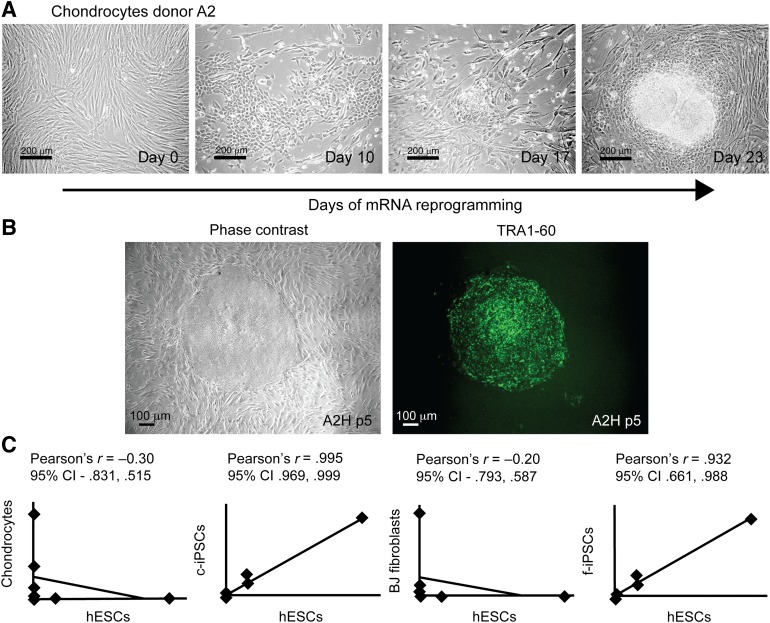
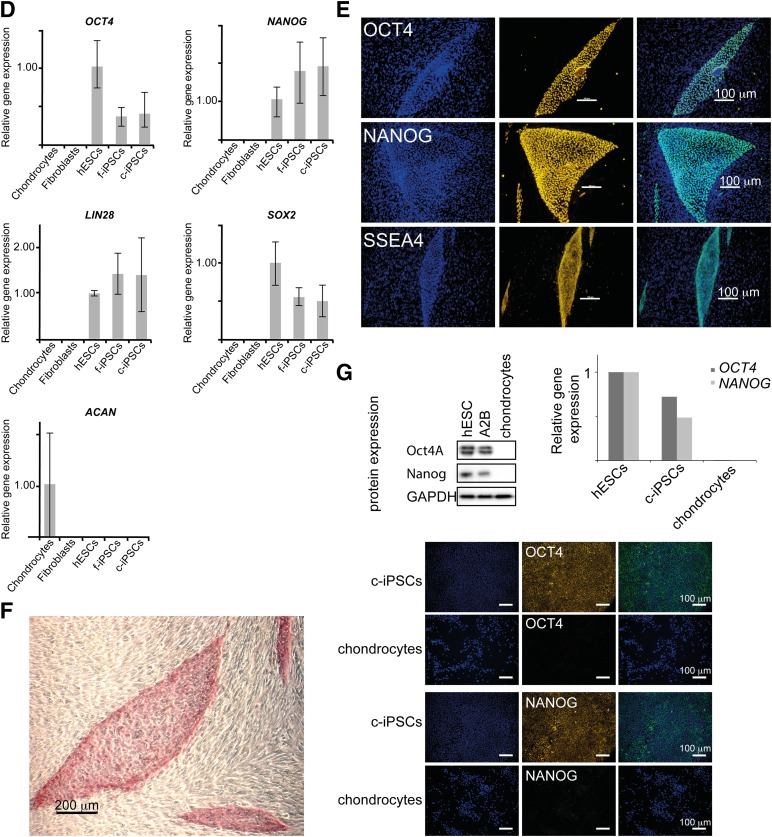
To create a stem cell model for chondrogenic differentiation and OA disease, we generated c-iPSC lines using surplus chondrocytes from three anonymized donors undergoing ACI treatment (Table 1). Prior to reprogramming, donor cells were analyzed for expression of pluripotency markers, cartilage markers, and matrix-degrading enzymes (Fig. 1D). Previously, iPSC lines have been shown to acquire genomic and epigenomic abnormalities during viral reprogramming and expansion [33–41]; consequently, we used nonintegrating mRNA reprogramming technology [22] to reduce risks of such abnormalities. Clonal iPSC lines were readily established from BJ fibroblasts (control, f-iPSC) and human chondrocytes transfected with synthetic mRNAs encoding OCT4, SOX2, KLF4, c-MYC, and LIN28 daily for 17 days and 21 days, respectively (Fig. 1A). On average, nine c-iPSC lines were successfully established out of every 10,000 chondrocytes (efficiency of ∼0.1% based on the number of established lines), and more than 200 hESC-like colonies were present among the control reprogrammed BJ fibroblasts (efficiency of ∼2% based on the number of colonies). All expanded c-iPSC lines stained positive for alkaline phosphatase and showed expression of pluripotency markers (OCT4, SSEA4, TRA-1-60, NANOG) visualized by immunofluorescent staining and quantitative Reverse Transcriptase PCR (Fig. 1B–1F). To verify protein quantities of OCT4 and NANOG in relation to mRNA levels, Western blots and additional quantitative Reverse Transcriptase PCR and immunohistochemical analyses were performed using feeder-free expanded c-iPSCs (line A2B), hESCs, and donor chondrocytes (Fig. 1G).
Figure 1.
Efficient generation of c-iPSCs using mRNA reprogramming. (A): Phase contrast images taken during reprogramming showing morphology changes. At day 0, mainly the feeders are visible (seeding density: chondrocytes 1 k/cm2, feeders 20 k/cm2). Small hESC-like colonies appeared at day 17 (chondrocytes). Scale bars = 200 μm. (B): Phase contrast image and immunofluorescence live staining showing expression of pluripotency marker TRA-1-60 in c-iPSC clone A2H at passage 5. Scale bar = 100 μm. (C): Scatter plots showing gene expression profiles of chondrocytes, BJ fibroblasts, hESCs, c-iPSCs, and f-iPSCs. Each data point represents expression of a single gene (eight genes in total). Excellent correlation in expression profiles between hESCs and iPSCs, and divergence from the profile of the cell type of origin, indicates successful reprogramming. (D): Induction of pluripotency genes OCT4, NANOG, LIN28, and SOX2 in c-iPSCs and f-iPSCs at early passage (passage 2–4, 11 clones and 9 clones, respectively) and extinction of ACAN in c-iPSCs (passage 2–4, 11 clones). Expression levels are normalized to CREBBP. Data are means with error bars showing standard deviation (chondrocytes, n = 3; hESCs, n = 3; f-iPCs, n = 9; c-iPSCs, n = 11). (E): Immunofluorescence staining showing expression of pluripotency markers (OCT4, NANOG, SSEA4) in c-iPSC clones (passage 14). DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain to show total cell number. Scale bars = 100 μm. (F): Alkaline phosphatase staining of c-iPSC clone. Scale bar = 200 μm. (G): Western blot, quantitative reverse transcriptase polymerase chain reaction data and immunofluorescent staining showing expression of pluripotency markers OCT4 and NANOG in hESCs and c-iPSCs (line A2B) after establishment in feeder-free culture on both protein and mRNA levels. GAPDH was used in the Western blot analysis to show equal loading. No expression of pluripotency markers was detected in the chondrocyte cultures. Scale bars = 100 μm. Abbreviations: BJ fibroblasts, human foreskin fibroblast cells; CI, confidence interval; c-iPSCs, chondrocyte-derived induced pluripotent stem cells; f-iPSCs, fibroblast-derived induced pluripotent stem cells; hESCs, human embryonic stem cells.
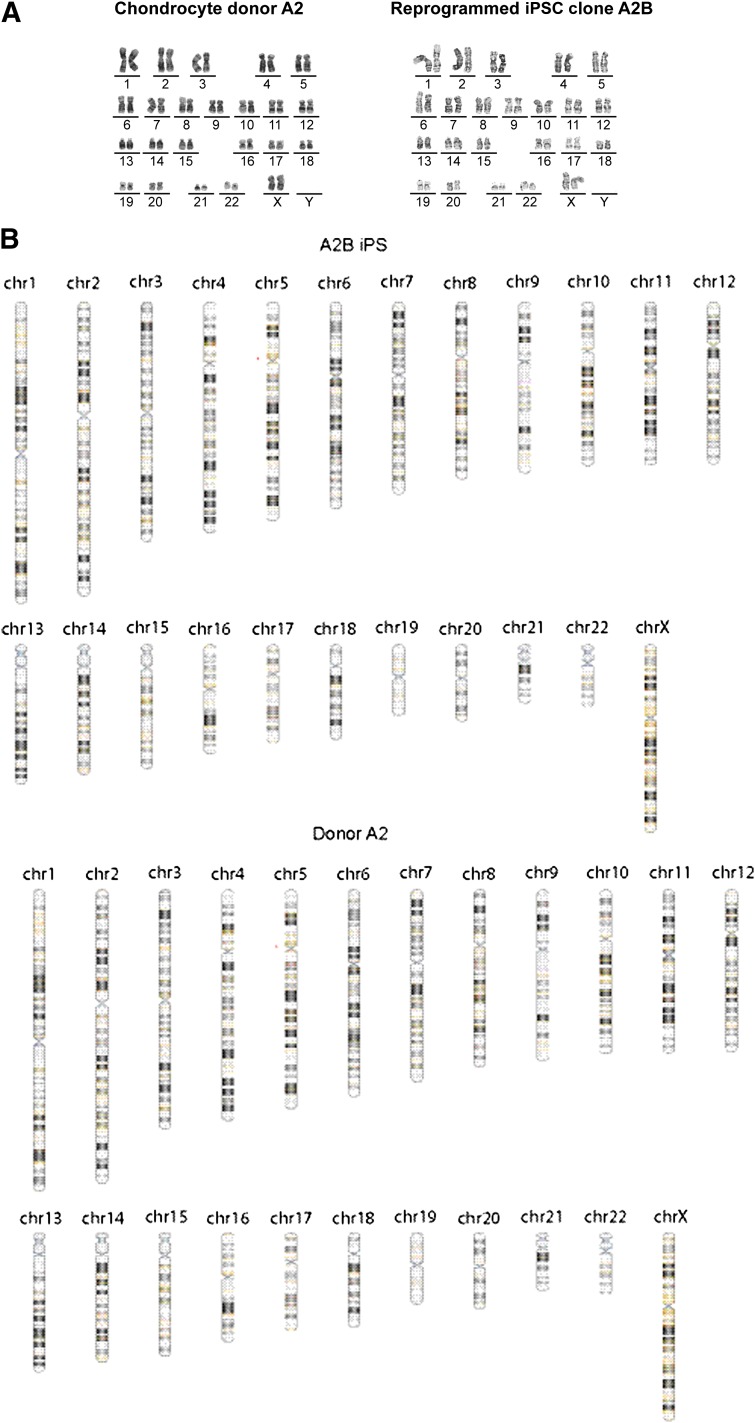
To ascertain the genomic stability of the reprogrammed lines, Giemsa-banded chromosome analysis (karyotyping) and whole-genome detection of copy number variants using a genomewide SNP array were performed on three selected c-iPSC lines and one f-iPSC line as well as the donor cell populations (karyotyping: donor A1, donor A2, c-iPSC lines; A1K, A2H, A2B, and f-iPSC and SNP array: donor A2, BJ fibroblasts, A1K, A1R, A2B, A2H, and f-iPSCs). All analyses revealed a normal karyotype (46 XY or 46 XX) (Fig. 2) without any acquired genomic copy number aberrations in all lines analyzed.
Figure 2.
mRNA reprogramming generates footprint-free iPSCs. (A): Cytogenetic analysis of donor chondrocytes and chondrocyte-derived iPSC line A2B showing normal karyotype. (B): Whole-genome single nucleotide polymorphism array analysis showing normal single nucleotide polymorphism patterns in chondrocyte-derived iPSC line A2B and original donor A2 (red arrows indicate losses). Abbreviation: iPSC, induced pluripotent stem cell.
mRNA-Induced c-iPSCs Can Differentiate to All Three Germ Layers In Vitro
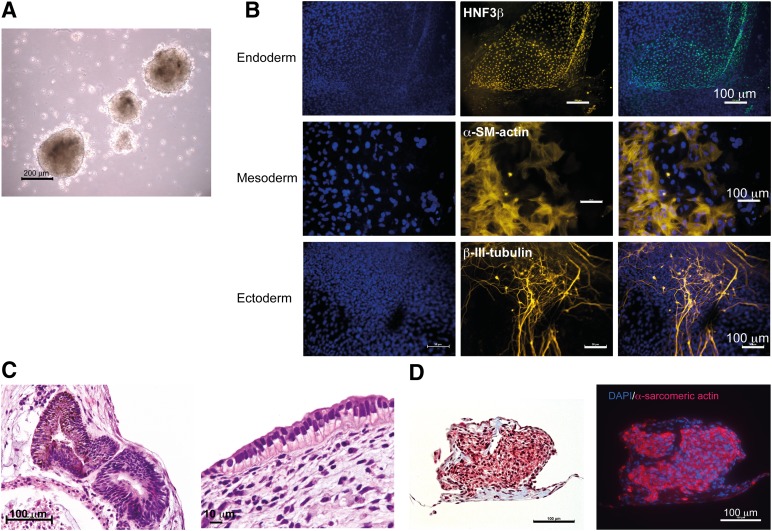
To examine the in vitro differentiation capacity of the c-iPSCs, we performed EB-mediated differentiation (Fig. 3A). After 1 week in suspension culture, EBs were plated on gelatin-coated plates. Outgrowths of heterogeneous populations of differentiating cells began to appear soon after plating. Derivatives of cells from all three embryonic germ layers were identified by morphology and expression of cell-type specific markers using immunofluorescence staining: mesoderm (α-smooth muscle actin), endoderm (HNF3β), and ectoderm (βΙΙΙ-tubulin) (Fig. 3B). Notably, within 2 weeks of plating, spontaneously beating cardiomyocytes were observed for EBs from clones A1K, A1R, A2B, A2H, and f-iPSCs.
Figure 3.
mRNA-induced chondrocyte-derived iPSCs have the capacity to differentiate to all three germ layers in vitro. (A): Embryoid bodies in suspension. Scale bar = 200 μm. (B): Immunofluorescence staining showing expression of HNF3β (endoderm), α-SM-actin, and βIII tubulin (ectoderm). DNA was counterstained with DAPI nuclear stain, to show total cell number. Scale bars = 100 μm (endoderm), 50 μm (mesoderm and ectoderm). (C): Histological examination of teratomas showing representative tissues originating from ectoderm (neural rosettes) and endoderm (epithelial cells). Scale bars = 100 μm (left), 10 μm (right). (D): Histological examination and immunofluorescent staining of sections from a beating embryoid body showing striated muscle tissue, representative of the mesoderm lineage. The left image shows hematoxylin and eosin staining, and the right image shows staining for α-Sarcomeric actin; DNA was counterstained with DAPI nuclear stain to show total cell number. Scale bars = 100 μm. Abbreviations: α-SM-actin, α-smooth muscle actin; DAPI, 4′,6-diamidino-2-phenylindole.
To mimic teratoma formation in mice and to facilitate long-term differentiation, a second set of EBs were cultured for 8 weeks in agarose as a semisolid matrix [28]. Histological examination of teratomas revealed representative tissues originating from all three embryonic germ layers, including neural epithelium (ectoderm), columnar epithelium (endoderm), and spontaneously beating cardiomyocytes positively stained for α-Sarcomeric actin (mesoderm) (Fig. 3C, 3D).
Taken together, these data demonstrate derivation of truly pluripotent, footprint-free human iPSC lines from chondrocytes that closely recapitulate the functional and molecular properties of hESCs.
Directed Chondrogenic Differentiation in Monolayer
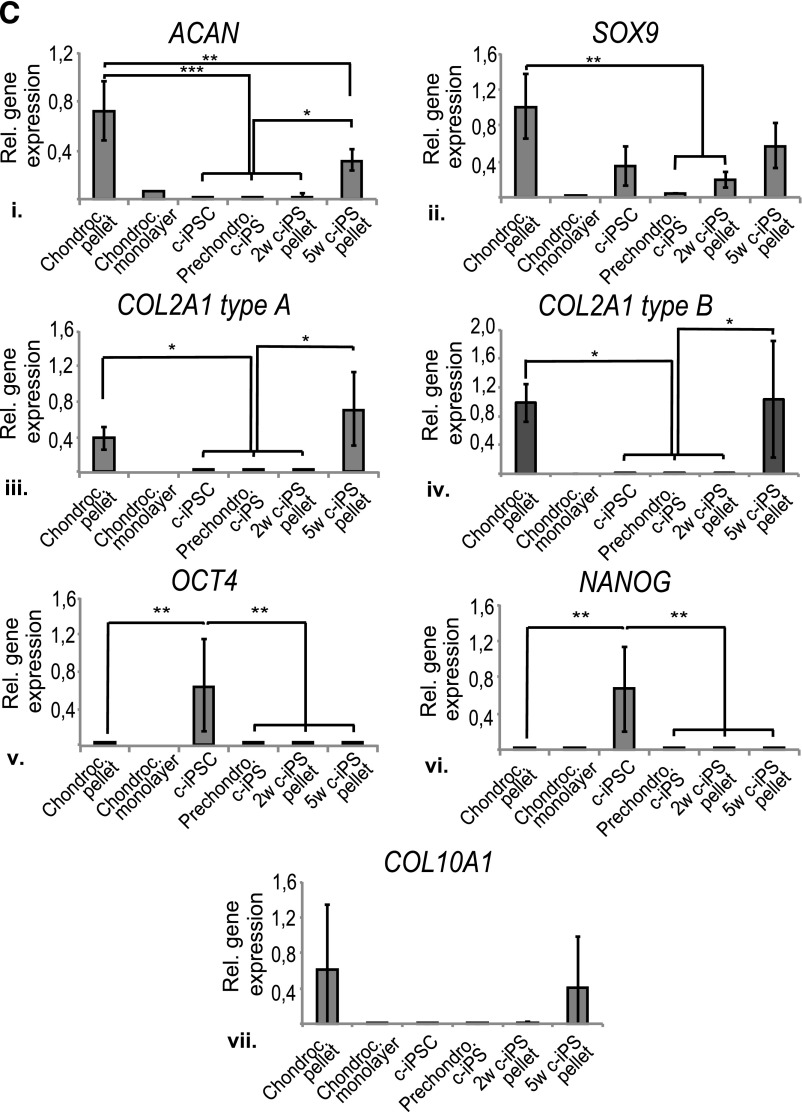
Excellent chondrogenic differentiation capacity is the hallmark of a potential universal donor cell line for ACI. This ability is also necessary for modeling cartilage development and OA disease progression. The c-iPSCs and f-iPSCs were differentiated in monolayer using a protocol initially developed for hESCs by Oldershaw et al. [24], exposing the iPSCs in a defined, stage-specific manner to activin A, WNT3A, FGF2, BMP4, follistatin, GDF5, and neurotrophin 4 (supplemental online Table 1). The protocol is based on known pathways during development from pluripotent cells, through primitive-streak mesendoderm to mesoderm intermediates to chondrocytes, and involves differentiation in three stages (Fig. 4A). At each stage, the cells were analyzed for gene expression of pluripotency and primitive-streak mesendodermal, mesodermal, and chondrocyte lineage markers (Fig. 4B). All four lines (three c-iPSC and one f-iPSC) went through the chondrogenic progression, from a stem cell phenotype to a chondrogenic phenotype. Markers of pluripotency (OCT4, NANOG, and SOX2) were significantly decreased from the pluripotency stage to the final stage (Fig. 4Bi–4Biii). From the pluripotency stage to stages 1 and 2, markers of primitive-streak mesendoderm (CDH1, MIXL1, GSC) were upregulated and then downregulated in the final stage (Fig. 4Biv–4Bvi), showing development of the cell lines through the mesodermal lineage. Simultaneously, mesodermal and chondrogenic markers (PDGFRB, SOX6, SOX9, ACAN, COL2A1 types A and B) were significantly upregulated (Fig. 4Bvii–4Bxii), showing progression to the chondrogenic lineage. Type II collagen (COL2A1) is a major component of cartilage providing structural integrity to the tissue. Differential splicing of the primary gene transcript results in two splice variants that are differentially expressed during cartilage development; chondroprogenitor cells predominantly express the type A splice variant, whereas differentiated chondrocytes mainly express the type B splice variant [42]. The dynamics between the COL2A1 splice variants A and B observed from stage 1 to stage 3 indicate a maturation process from immature chondroprogenitors in stage 1 toward mature chondrocytes in stage 3 (Fig. 4Bxi–4Bxii). No hypertrophic differentiation could be seen at the end of stage 3, as shown by the decrease in COL10A1 expression (Fig. 4Bxiii).
Figure 4.
Monolayer differentiation of c-iPSCs to chondrogenic cells. (A): Morphology of induced pluripotent stem cells (iPSCs) at the four stages of the monolayer chondrogenic differentiation protocol. Pluripotent iPSCs adapted to feeder-free expansion in defined medium. Cells show high nucleus-to-cytoplasm ratio with prominent nucleoli A2B (Ai), A2H (Aii), A1K (Aiii), and BJ (f-iPSC) (Aiv). At the end of stage 1, the four lines showed different responses to stimuli, with A1K (Avii), A2H (Avi), and BJ (Avii) lines forming colony-like structures that the A2B line (Av) did not. During stage 2, all c-iPSCs A2B (Aix), A2H (Ax), and A1K (Axi) formed dense clusters appearing bright in phase contrast images, whereas the BJ line (Axii) did not. At the end of stage 3, the four lines showed different morphologies. The A2B (Axiii) and BJ (Axvi) lines expanded in ML, although the BJ line at reduced capacity. The A1K (Axiv) and A2H (Axv) lines formed three-dimensional cell aggregates similar to embryoid body structures. (B): Relative gene expression patterns of the differentiating iPSCs. Each graph shows the expressions of f-iPSC, c-iPSC A1K, c-iPSC A2H, and c-iPSC A2B at the pluripotency stage and at stages 1–3 of the differentiation protocol: genes associated with pluripotency (Bi–Biii), genes associated with primitive-streak mesendoderm (Biv–Bvi), the gene associated with mesoderm (Bvii), genes expressed in chondrocytes (Bviii–Bxii), and the marker for hypertrophic differentiation (Bxiii). Each bar represents one cell line (n = 3), with error bars showing standard deviation and expressions normalized to the highest expression for each separate cell line. Statistical significance was determined for all lines collectively at each time point (n = 12). ∗, p < .05, ∗∗, p < .01, ∗∗∗, p < .001. Abbreviations: c-iPSC, chondrocyte-derived induced pluripotent stem cell; f-iPSC, fibroblast-derived induced pluripotent stem cell; FF-iPS, induced pluripotent stem cell adapted to feeder-free expansion; Rel., relative.
In summary, we demonstrate that the gene expression profile of our iPSCs during directed chondrogenic differentiation in monolayer (Fig. 4B) follows the previously published profile of hESCs [24].
c-iPSCs Form Articular Cartilage Matrix in 3D Pellet Mass Cultures
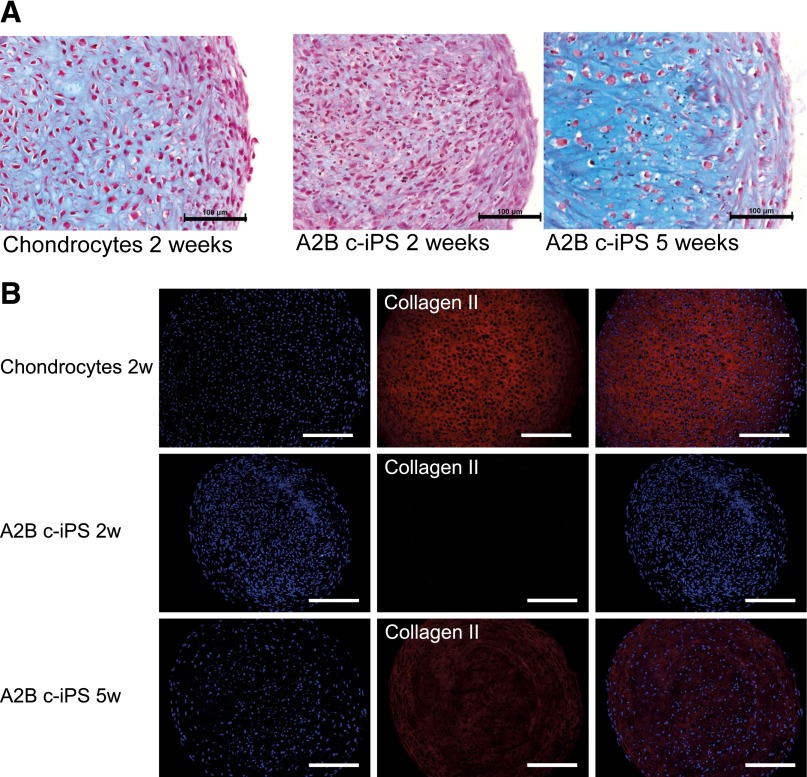
To further investigate the cartilage differentiation capacity of our c-iPSCs and their ability to form cartilage extracellular matrix, a novel differentiation protocol was developed involving a 3D pellet mass predifferentiation stage (2 weeks) followed by monolayer expansion of chondrogenic progenitors. These progenitors were subsequently differentiated in a second chondrogenic 3D pellet mass culture (for 2 weeks and 5 weeks, respectively) to study their chondrogenic capacity. Three of the four c-iPSC clones, but not the f-iPSC clone, could be expanded in monolayer after the 2-week predifferentiation stage. These three chondrogenic progenitor lines showed minor or no expression of the pluripotency markers OCT4 and NANOG (represented by A2B in Fig. 5C). All three showed prechondrogenic characteristics, expressing both ACAN and SOX9 (represented by A2B in Fig. 5C). One line in particular (A2B) showed a significantly higher expression of ACAN than the others. During the 5-week 3D pellet mass differentiation, the prediffentiatied c-iPSCs showed an increasing deposition of extracellular matrix, as seen by Alcian blue van Gieson staining (Fig. 5A). The c-iPSC line A2B in particular formed intensely stained 3D pellet masses, comparable to pellets from donor chondrocytes, thus linking higher ACAN expression during predifferentiation to superior in vitro cartilage differentiation capacity. During the last 3 weeks of differentiation, collagen II deposition also increased from no deposition at 2 weeks to a rich staining at 5 weeks (Fig. 5B). Expression of chondrogenic markers ACAN, SOX9, and COL2A1 types A and B increased throughout the differentiation process, finally reaching the same expression levels as donor chondrocytes in primary culture before reprogramming. Expression of pluripotency markers OCT4 and NANOG was extinct during the differentiation process (Fig. 5C). Relevant control cultures with only irradiated feeders were kept, and these cells did not show any chondrogenic differentiation capacity.
Figure 5.
c-iPSCs form articular cartilage matrix in three-dimensional (3D) pellet mass cultures. (A): Histological examination of 3D pellet mass cultures stained with Alcian blue van Gieson showing chondrocytes from donor A2 before reprogramming at 2 weeks of differentiation as control and A2B c-iPSCs at 2 weeks and 5 weeks of differentiation. Blue color stains proteoglycans in the extracellular matrix. Scale bar = 100 μm. (B): Histological sections of the differentiated A2B c-iPSCs in pellet mass culture, stained with 4′,6-diamidino-2-phenylindole (DAPI) for cell nuclei and collagen II antibody at 2 weeks and 5 weeks in culture. Sections of pellet mass culture of chondrocytes from donor A2 are shown as control. Scale bar = 200 μm. Staining shows increased collagen II content in the pellets with time in culture. (C): Relative gene expression patterns of chondrogenic markers ACAN (Ci), SOX9 (Cii), COL2A1 type A (Ciii), COL2A1 type B (Civ); pluripotency markers OCT4 (Cv) and NANOG (Cvi); and hypertrophic marker COL10A1 (Cvii). From left to right, the donor chondrocytes in 3D pellet mass cultures before reprogramming, donor chondrocytes in monolayer just before reprogramming, A2B c-iPSCs, prechondrogenic cells, A2B c-iPSCs differentiated for 2 weeks, and A2B c-iPSCs differentiated for 5 weeks. Results shown as mean (n = 3; chondrocyte monolayer, n = 1) with error bars showing standard deviation. Expressions normalized to highest expression for each separate gene. ∗, p < .05, ∗∗, p < .01, ∗∗∗, p < .001. Abbreviations: 2w, 2 weeks; 5w, 5 weeks; c-iPS, chondrocyte-derived induced pluripotent stem cells; hEL, human diploid embryonic lung fibroblast cells; Rel., relative.
Taken together, these data show that at least one of our c-iPSC lines has the cartilage differentiation capacity needed for modeling cartilage development and OA disease and that the expression of ACAN is a putative marker for chondrogenic potential, possibly taking us one step closer to a clinical-grade cell source for regenerative medicine.
Discussion
Cartilage regeneration with ACI has been widely accepted since the first clinical report [1] and is clinically approved by both the U.S. Food and Drug Administration and the European Medicines Agency; however, current ACI technology is labor intensive and costly because it is based on personalized treatment. Replacing autologous chondrocytes with differentiated autologous iPSCs would neither be less laborious nor more cost effective; however, a universal chondroprogenitor donor cell line that could be mass produced as an off-the-shelf product could be a potential future treatment. Because of the immunoprivileged nature of cartilage tissue, a universal donor cell product is a potential replacement for the autologous cell source used today. Toward this end, we generated c-iPSCs using a nonintegrative method, the modified mRNA reprogramming method [22], and subsequently compared their differentiation capacity toward the chondrogenic lineage with BJ fibroblast-derived iPSCs.
In the present study, we reprogrammed chondrocytes from two donors (one male and one female) with cartilage injuries. We successfully established c-iPSC clonal lines with normal karyotype without any insertions or deletions acquired during the reprogramming process. We demonstrated that all lines analyzed have the capacity to differentiate along the chondrogenic lineage, matching the expression profile of mature chondrocytes, and that one c-iPSC clone in particular has the capacity to differentiate into chondrocytes with high cartilage matrix-forming capacity. Our results show that chondrocytes can be reprogrammed into iPSCs but that BJ fibroblasts are reprogrammed faster and more efficiently, probably because of the juvenescence of this cell source, in comparison with the somatic chondrocytes obtained from adult donors. Furthermore, the rapid turnover of BJ fibroblasts in comparison with donor chondrocytes could be an additional reason for the difference in the observed reprogramming efficiency. Although the efficiency is lower for primary chondrocytes than for BJ fibroblasts, the mRNA protocol is still more efficient than integrating viral methods [7, 8] as well as the recently reported nonintegrating episomal plasmid vector-based reprogramming technique [23]. The efficiency of the protocol, however, is not a marker of quality of the generated iPSC line; in reality, all we need is one high-quality iPSC line from each patient.
The mRNA procedure probably enables delivery of a precise amount of mRNA molecules into the cells, resulting in more consistent expression of reprogramming factors in each transfected cell in comparison with other reprogramming methods. The precise level of OCT4 has been shown to be crucial for murine ESCs [43, 44], and altering this level by as little as a factor or two causes differentiation [43]. Also in the iPSC field, the OCT4 level and stoichiometry of reprogramming factors have been proven to be important, with three times more OCT4 as the most efficient [45], equal to the amount used in this study and by Warren et al. [22, 43, 45]. Moreover, the exogenous reprogramming mRNA molecules are degraded within a day following transfection, and prolonged pluripotency relies on a steady state of expression of endogenous induced pluripotency genes, limiting the number of partially reprogrammed clones. All analyzed c-iPSC lines are pluripotent, show phenotypical characteristics similar to hESCs, and can spontaneously differentiate into all three germ layers as well as nerves and beating cardiomyocyte clusters.
The criteria for the selection of a potential Good Manufacturing Practice-grade iPSC line for an off-the-shelf product would be an iPSC line without any genomic modifications and with a high chondrogenic differentiation capacity toward hyaline cartilage. Directed differentiation along the chondrogenic lineage of iPSCs from different donors and cell origins was compared to address the hypothesis of whether iPSCs derived from chondrocytes have better differentiation capacity toward cartilage than cells of other origins. In the search for predictive markers of chondrogenic differentiation capacity, the Oldershaw protocol originally developed for hESCs [24] was chosen. This protocol is based on known embryonic developmental pathways and was successful for both f-iPSCs and c-iPSCs: All lines went through the chondrogenic progression from a stem cell phenotype to a chondrogenic phenotype. When analyzing the total pattern, it is difficult to single out a gene that might select a clone with better chondrogenic potential, which is not surprising because all lines successfully went through the chondrogenic differentiation equally well. Using our novel 3D differentiation protocol, one c-iPSC clone in particular (i.e., A2B) was singled out as having superior in vitro cartilage differentiation capacity. In their undifferentiated state, all four clones (two from each donor) expressed similarly high levels of the pluripotency markers OCT4 and NANOG and similarly low levels of the chondrogenic markers ACAN, SOX9, and COL2A1 types A and B. It was only after prechondrogenic differentiation that the clones started to differ considerably in expression. Only three of the investigated clones survived the expansion after this predifferentiation stage, and expression of pluripotency markers was reduced to original levels before reprogramming in all three clones—an important safety issue because remaining pluripotency could give rise to teratomas in vivo. The chondrogenic markers SOX9 and COL2A1 types A and B were similar for all surviving lines; however, higher ACAN expression singled out A2B, proposing ACAN as a putative predictive marker of chondrogenic capacity. The search for additional markers is ongoing, and the downregulation of disease markers of OA such as matrix metalloproteinases and cytokines as well as markers of pluripotency have to be taken into account.
The use of the novel 3D differentiation protocol resulted in COL10A1 expression in the cells, indicating that the differentiation process was that of the mesenchyme toward endochondral ossification [46], possibly because of epigenetic memory of an osteoarthritic phenotype. Notably, the iPSCs expressed COL10A1 to the same extent as the donor chondrocytes, indicating that they are on a similar differentiation path as the chondrocytes, approaching hypertrophy. Notably, in the more intricate monolayer differentiation protocol specifically designed by Oldershaw et al. for chondrogenic differentiation without hypertrophy, COL10A1 expression was strongly reduced.
The varying chondrogenic differentiation capacity between the c-iPSC clones suggests that the reprogramming process is not equal and that some clones better retain a memory of their heritage. This could be attributed to a putatively suboptimal reprogramming cocktail producing c-iPSCs that can more easily recover their somatic gene profile but could also be a result of the heterogeneity of cells in primary culture that were reprogrammed. Either way, this supports the idea of a remaining epigenetic memory after reprogramming that has been shown with other reprogramming methods previously [10–12, 47–49]. A conclusive experiment would be to collect and reprogram cells from different tissues (e.g., blood, bone, skin, cartilage) of the same individual to truly verify the putative tissue-dependent differentiation bias.
The risk of altered epigenetic landscapes that cause developmental plasticity is a safety concern that could hold back clinical application of iPSCs; however, an iPSC or progenitor line that has superior differentiation potential along a specific lineage could also deliver important clinical benefits. For iPSCs in cartilage tissue renewal to progress further into the clinic, there is a pressing need for safe and efficient means to redirect cell fate. This is apparent when one considers that iPSCs are only a starting point for patient-specific therapies, and specification of clinically useful cell types is still required to produce autologous tissues for transplantation or for disease modeling. Importantly, we have demonstrated that modified mRNA-based technology enables highly efficient reprogramming and that epigenetic memory persists so that cartilage-specified cell lines can be produced, like our line A2B, without compromising genomic integrity.
These findings in combination with the varying differentiation capacity of the c-iPSCs, show that to find a new potential cell source for an off-the-shelf chondrocyte product, careful genomic analyses as well as extended differentiation studies would be required to identify the perfect candidate. Taken together, we believe that our c-iPSCs hold great promise in investigating OA diseases and drug screening as well as being potential cell sources for future clinical ACI products.
Conclusion
Our study reports, for the first time, reprogramming of human chondrocytes by synthetic mRNA. Mechanistically, we show that our derived iPSC clones in a chemically defined media supplemented with exogenous growth factors at different time points significantly upregulated chondrogenic markers (PDGFRB, SOX6, SOX9, ACAN, COL2A1 types A and B), showing progression toward a chondrogenic expression profile regardless of donor origin.
One of our derived iPSC clones differentiates in vitro into cartilage matrix producing mature chondrocytes, comparable to the differentiation capacity of patient-derived chondrocytes, a stem cell characteristic that has been sought after in the field of regenerative medicine for years. This iPSC clone, in comparison with the original donor, exhibits no detectable genomic gains or losses (analyzed by karyotyping and whole-genome SNP array), a quality that is important for mechanistic studies and crucial for future clinical applications. This paper improves understanding of nonintegrating cellular reprogramming of cells from adult individuals as well as chondrogenic differentiation of pluripotent stem cells.
Supplementary Material
Acknowledgments
The study was funded by Swedish Research Council Grant 2009-4849, the ALF research grant (Avtalet om Läkarutbildning och medicinsk Forskning) research grant from the Sahlgrenska University Hospital, and the Inga Britt and Arne Lundberg Research Foundation. We thank Sonja Ranjbar and the departments of Cytogenetics and Clinical Genetics for excellent technical assistance and Dr. Lars Palmqvist for single nucleotide polymorphism array result interpretation. We also would like to thank the Bioinformatics and Expression Analysis core facility at Novum, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska Hospital. We also wish to acknowledge Jenny Johannisson, Tina Nilsson, Dr. Raimund Strehl, and Dr. Peter Sartipy at Cellectis Bioresearch for valuable discussions.
Author Contributions
C. Boreström, S.S., and L.E.: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; N.B.: collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; C.B.: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; C.E. and J.H.: provision of study material or patients, final approval of manuscript; A.L.: conception and design, financial support, provision of study material or patients, manuscript writing, final approval of manuscript
Disclosure of Potential Conflicts of Interest
C.E. has compensated employment with Cellectis AB. J.H. had compensated employment with Cellectis AB/Cellartis AB.
References
- 1.Brittberg M, Lindahl A, Nilsson A, et al. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N Engl J Med. 1994;331:889–895. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- 2.Vasiliadis HS, Danielson B, Ljungberg M, et al. Autologous chondrocyte implantation in cartilage lesions of the knee: Long-term evaluation with magnetic resonance imaging and delayed gadolinium-enhanced magnetic resonance imaging technique. Am J Sports Med. 2010;38:943–949. doi: 10.1177/0363546509358266. [DOI] [PubMed] [Google Scholar]
- 3.Peterson L, Vasiliadis HS, Brittberg M, et al. Autologous chondrocyte implantation: A long-term follow-up. Am J Sports Med. 2010;38:1117–1124. doi: 10.1177/0363546509357915. [DOI] [PubMed] [Google Scholar]
- 4.Tallheden T, Dennis JE, Lennon DP, et al. Phenotypic plasticity of human articular chondrocytes. J Bone Joint Surg Am. 2003;85-A(Suppl 2):93–100. doi: 10.2106/00004623-200300002-00012. [DOI] [PubMed] [Google Scholar]
- 5.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Maherali N, Ahfeldt T, Rigamonti A, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K, Zhao R, Doi A, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polo JM, Liu S, Figueroa ME, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez F, Barragan Monasterio M, Tiscornia G, et al. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc Natl Acad Sci USA. 2009;106:8918–8922. doi: 10.1073/pnas.0901471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadtfeld M, Nagaya M, Utikal J, et al. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia F, Wilson KD, Sun N, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soldner F, Hockemeyer D, Beard C, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang CW, Lai YS, Pawlik KM, et al. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- 20.Yusa K, Rad R, Takeda J, et al. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woltjen K, Michael IP, Mohseni P, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 24.Oldershaw RA, Baxter MA, Lowe ET, et al. Directed differentiation of human embryonic stem cells toward chondrocytes. Nat Biotechnol. 2010;28:1187–1194. doi: 10.1038/nbt.1683. [DOI] [PubMed] [Google Scholar]
- 25.Koyama N, Miura M, Nakao K, et al. Human induced pluripotent stem cells differentiated into chondrogenic lineage via generation of mesenchymal progenitor cells. Stem Cells Dev. 2013;22:102–113. doi: 10.1089/scd.2012.0127. [DOI] [PubMed] [Google Scholar]
- 26.Tallheden T, van der Lee J, Brantsing C, et al. Human serum for culture of articular chondrocytes. Cell Transplant. 2005;14:469–479. doi: 10.3727/000000005783982909. [DOI] [PubMed] [Google Scholar]
- 27.Bigdeli N, Andersson M, Strehl R, et al. Adaptation of human embryonic stem cells to feeder-free and matrix-free culture conditions directly on plastic surfaces. J Biotechnol. 2008;133:146–153. doi: 10.1016/j.jbiotec.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 28.Stenberg J, Elovsson M, Strehl R, et al. Sustained embryoid body formation and culture in a non-laborious three dimensional culture system for human embryonic stem cells. Cytotechnology. 2011;63:227–237. doi: 10.1007/s10616-011-9344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Synnergren J, Giesler TL, Adak S, et al. Differentiating human embryonic stem cells express a unique housekeeping gene signature. Stem Cells. 2007;25:473–480. doi: 10.1634/stemcells.2006-0247. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.DeLise AM, Fischer L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage. 2000;8:309–334. doi: 10.1053/joca.1999.0306. [DOI] [PubMed] [Google Scholar]
- 32.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 33.Taapken SM, Nisler BS, Newton MA, et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- 34.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 36.Martins-Taylor K, Nisler BS, Taapken SM, et al. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol. 2011;29:488–491. doi: 10.1038/nbt.1890. [DOI] [PubMed] [Google Scholar]
- 37.Mayshar Y, Ben-David U, Lavon N, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 38.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elliott AM, Elliott KA, Kammesheidt A. High resolution array-CGH characterization of human stem cells using a stem cell focused microarray. Mol Biotechnol. 2010;46:234–242. doi: 10.1007/s12033-010-9294-1. [DOI] [PubMed] [Google Scholar]
- 40.Laurent LC, Ulitsky I, Slavin I, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji J, Ng SH, Sharma V, et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 42.Ryan MC, Sandell LJ. Differential expression of a cysteine-rich domain in the amino-terminal propeptide of type II (cartilage) procollagen by alternative splicing of mRNA. J Biol Chem. 1990;265:10334–10339. [PubMed] [Google Scholar]
- 43.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 44.Radzisheuskaya A, Chia GB, dos Santos RL, et al. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papapetrou EP, Tomishima MJ, Chambers SM, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelttari K, Winter A, Steck E, et al. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum. 2006;54:3254–3266. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- 47.Miura K, Okada Y, Aoi T, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 48.Hu BY, Weick JP, Yu J, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.