Hutchinson-Gilford progeria syndrome is a rare congenital disease characterized by premature aging in children. The results of this systematic comparative study of the three main treatments currently administered or proposed to progeria-affected children reveal the complexity of the modes of action of different drugs and underscore the use of induced pluripotent stem cell derivatives as a critical and powerful tool for standardized, comparative pharmacological studies.
Keywords: Pluripotent stem cells, Progeria, Aging, Drug development, Mesenchymal stem cells
Abstract
Hutchinson-Gilford progeria syndrome is a rare congenital disease characterized by premature aging in children. Identification of the mutation and related molecular mechanisms has rapidly led to independent clinical trials testing different marketed drugs with a preclinically documented impact on those mechanisms. However, the extensive functional effects of those drugs remain essentially unexplored. We have undertaken a systematic comparative study of the three main treatments currently administered or proposed to progeria-affected children, namely, a farnesyltransferase inhibitor, the combination of an aminobisphosphonate and a statin (zoledronate and pravastatin), and the macrolide antibiotic rapamycin. This work was based on the assumption that mesodermal stem cells, which are derived from Hutchinson-Gilford progeria syndrome-induced pluripotent stem cells expressing major defects associated with the disease, may be instrumental to revealing such effects. Whereas all three treatments significantly improved misshapen cell nuclei typically associated with progeria, differences were observed in terms of functional improvement in prelamin A farnesylation, progerin expression, defective cell proliferation, premature osteogenic differentiation, and ATP production. Finally, we have evaluated the effect of the different drug combinations on this cellular model. This study revealed no additional benefit compared with single-drug treatments, whereas a cytostatic effect equivalent to that of a farnesyltransferase inhibitor alone was systematically observed. Altogether, these results reveal the complexity of the modes of action of different drugs, even when they have been selected on the basis of a similar mechanistic hypothesis, and underscore the use of induced pluripotent stem cell derivatives as a critical and powerful tool for standardized, comparative pharmacological studies.
Introduction
Since the discovery of the molecular mechanisms underlying Hutchinson-Gilford progeria syndrome (HGPS) (OMIM no. 176670), three independent clinical trials have been proposed to treat the patients affected with this accelerated aging disorder: lonafarnib (ClinicalTrials.gov identifier NCT00425607) [1], pravastatin and zoledronate (ClinicalTrials.gov identifier NCT00731016), and their combination (ClinicalTrials.gov identifier NCT00916747). Although some patients are currently treated with these drugs, little is known regarding each of their specific effects on different cell defects observed in progeria. In this study, we have taken advantage of the unique potential of induced pluripotent stem (iPS) cells to address that issue.
Progeria is a rare genetic disorder caused by a point mutation in the LMNA gene that leads to the production and accumulation of a truncated form of lamin A called “progerin” [2, 3]. This toxic form of lamin cannot be terminally matured and thus causes disruption of the nuclear structure, defects in DNA repair processes, and other molecular defects associated with premature aging [4]. The disease manifests itself in a set of symptoms that includes growth delay, loss of body fat, osteoporosis, and atherosclerosis leading to premature death [5, 6]. Based on the identification of the toxic mechanism leading progeria to be associated with the accumulation of misprocessed farnesylated progerin, two therapeutic strategies have been investigated. Farnesyltransferase inhibitors (FTIs) have been assayed following the restoration of some defects both in vitro and in progeroid animal models [7, 8]. The recently published results of this clinical trial indicate the partial clinical benefits of an FTI, lonafarnib, although several adverse effects have been reported [1]. In parallel, because of the previously described antiproliferative effect of FTIs on cancer cells [9], through the activation of an alternative prenylation pathway called “geranylgeranylation,” another clinical trial has been initiated. This second approach is based instead on the reduction of global progerin protein prenylation through the combination of a statin (pravastatin) and an aminobisphosphonate (zoledronate) [10]. Direct benefits of this combination, called “ZoPra,” were also sought through the reduction of atherosclerosis and bone turnover defects, two of the key hallmarks of progeria [10]. More recently, another strategy was proposed through the use of the macrolide antibiotic rapamycin (Rapa), demonstrating its effect in improving the cell nuclear morphology of HGPS fibroblasts through mammalian target of rapamycin (mTOR) inhibition and progerin clearance stimulation [11].
Before the discovery of iPS cells, biological resources allowing the study at a cellular level of such new treatments were limited to patients’ primary fibroblast cultures and genetically modified healthy cells overexpressing progerin [12–14]. Thanks to their intrinsic self-renewal and pluripotency properties, iPS cells constitute an inexhaustible and reproducible biological material that can be amplified, characterized, and banked. Accordingly, over the past 5 years, several studies highlighted that iPS cell derivatives were an interesting alternative tool with which to study a potentially unlimited number of genetic pathologies [15]. More recently, several groups, including ours, have confirmed the usefulness of these cells to decipher the molecular mechanisms involved in HGPS [16–19].
In the present study, we have taken advantage of this unique cell model to create a pharmacological study platform to systematically compare the effects of the drugs currently studied or proposed for children affected by progeria. For that purpose, mesodermal stem cells (MSCs) have been derived from iPS cells to monitor relevant molecular parameters such as prelamin A maturation inhibition and to quantify functional defects such as nuclear architecture, progerin expression, energy metabolism, cell proliferation, and osteogenic differentiation properties.
Materials and Methods
Fibroblast Reprogramming
Fibroblasts used in this study were isolated from patient biopsies performed in the Assistance Publique Hôpitaux de Marseille for the patient 13-8243 and provided by the Coriell cell repository (Coriell Institute for Medical Research, Camden, NJ, http://www.coriell.org) for the control DM4603. The fibroblasts were successfully reprogrammed to iPS cells using Yamanaka’s original method with OCT4, KLF4, SOX2, and c-Myc, transferred using retroviral vectors [20]. The iPS cell lines were amplified up to the 15th passage before differentiation. Molecular characterization of pluripotency and self-renewal capacities of these cells was described previously [19].
Pluripotent Stem Cell Culture and Differentiation
Control and HGPS iPS cells were grown on STO mouse fibroblasts, inactivated with 10 mg/ml mitomycin C seeded at 30,000/cm2 and grown as described previously [19]. For differentiation, iPS cells were differentiated into MSCs using directed protocols for differentiation previously published by our group [21].
Cell Culture and Drug Treatments
MSCs derived from HGPS iPS cells (HGPS MSCs) and control MSCs were cultured in KnockOut Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, http://www.invitrogen.com), supplemented with 20% of fetal bovine serum (SH30066.0.3; Thermo Fisher Scientific, Waltham, MA, http://www.thermoscientific.com), 1% nonessential amino acids (Invitrogen), 1% glutamine (Invitrogen), and 0.1% β-mercaptoethanol (Invitrogen). Six hours after seeding, MSCs were treated with 0.1% dimethyl sulfoxide, 1 μM pravastatin (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), 1 μM zoledronate (Zometa; Novartis International, Basel, Switzerland, http://www.novartis.com), 10 nM rapamycin (R0395, Sigma-Aldrich), or 1 μM FTI (tipifarnib, R115777; Selleck Chemicals, Houston, TX, http://www.selleckchem.com). Concentrations of drugs were defined based on the literature and their effect on nuclear shape abnormalities.
Osteogenic Differentiation
MSCs were seeded at 15,000 cells per cm2 in 24-well plates in MSC culture medium and treated as described previously. After 72 hours, the MSC medium was replaced by StemPro osteogenic induction medium (Invitrogen) in the presence or absence of the different drugs. The medium was replaced every 3–4 days. After 7 days of culture, cells were fixed with 95% ethanol and stained by adding a colorimetric substrate of alkaline phosphatase, 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (Sigma-Aldrich).
Cell Proliferation Assay
Cell proliferation analysis of HGPS MSCs was performed using the Click-iT 5-ethynyl-2′-deoxyuridine (EdU) flow cytometry assay (Life Technologies, Rockville, MD, http://www.lifetech.com), according to the manufacturer’s protocol. Cells were treated with 10 μM EdU for 2 hours and then fixed, permeabilized, and stained with propidium iodide. The proportion of cells in S phase was determined by magnetic affinity cell sorting flow cytometry (Miltenyi Biotec, Bergisch Gladbach, Germany, http://www.miltenyibiotec.com) using FlowJo software. The number of events analyzed for each experiment was 10,000. Three independent experiments were performed for each condition.
ATP Measurement
HGPS MSCs and MSCs derived from control iPS cell lines (wild-type [WT] MSCs) were seeded in 96-well plates (3917; Corning Life Sciences, Acton, MA, http://www.corning.com/lifesciences) (5000 cells per well) and treated with the different drugs. After 72 hours of treatment, the ATP content was measured using CellTiter-Glo (Promega, Madison, WI, http://www.promega.com) according to the manufacturer’s protocol. Luminescence was measured with an Analyst GT counter luminometer (Molecular Devices, Sunnyvale, CA, http://www.moleculardevices.com). In parallel, nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI) were counted in each well with an automated microscope called LEAP (Laser-Enabled Analysis and Processing) (Intrexon, Germantown, MD, http://www.dna.com). The results are given in the ratio between ATP measurement and the number of nuclei counted.
Array Scan Analysis
Prelamin A localization, progerin expression, and Ki-67 positive nuclei detection were analyzed with an ArrayScan VTI HCS Reader (Cellomics Inc., Pittsburgh, PA, http://www.thermoscientific.com). The first channel was used for nucleus identification (DAPI staining), and the second channel was used to identify prelamin A, progerin, and Ki-67. Information was collected with vHCS View software (Cellomics). Pictures were acquired at ×20 objective in high-resolution camera mode and were analyzed based on the “spot detection” bioapplication for prelamin A and progerin and the “colocalization” bioapplication for Ki-67. Each analysis was performed on three independent experiments by counting at least 500 nuclei per well in 12 separated wells.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde (15 minutes at room temperature) before permeabilization and blocking in phosphate-buffered saline (PBS) supplemented with 0.1% Triton X-100 and 1% bovine serum albumin (Sigma-Aldrich). Primary antibodies were incubated for 1 hour at room temperature in blocking buffer. Antibodies included mouse anti-lamins A/C (clone JOL2; Millipore, Billerica, MA, http://www.millipore.com), rabbit anti-prelamin A (ANTOO45; Diatheva, Fano, Italy, http://www.diatheva.com/), rabbit anti-progerin (provided by K.D.), and anti-Ki-67 (clone Ki-S5, MAB4190; Millipore). Cells were stained with the species-specific fluorophore-conjugated secondary antibody (Invitrogen) (1 hour at room temperature), and nuclei were visualized with DAPI.
Western Immunoblotting
Whole-cell lysates of MSCs were collected, separated by SDS-PAGE, and transferred onto PVDF membrane by the liquid transfer method. Blots were blocked in 10% skim milk (Bio-Rad, Hercules, CA, http://www.bio-rad.com) containing Tween 0.1% Tris-buffered saline one time for 1 hour at room temperature. The primary antibodies used were a mouse anti-lamin A/C 1:200 (JOL2; Millipore) and a β-actin 1:200,000 (Sigma-Aldrich). Membranes were incubated overnight at 4°C. Antigen-antibody binding was detected using horseradish peroxidase-conjugated species-specific secondary antibodies (GE-Healthcare, Little Chalfont, U.K., http://www.gehealthcare.com), followed by enhanced chemiluminescence Western blotting detection reagents (PerkinElmer Life and Analytical Sciences, Waltham, MA, http://www.perkinelmer.com).
Quantitative Polymerase Chain Reaction
Total RNAs were isolated using the RNeasy Mini extraction kit (Qiagen, Courtaboeuf, France, http://www.qiagen.com) according to the manufacturer’s protocol. An on-column DNase I digestion was performed to avoid genomic DNA amplification. RNA levels and quality were checked using the NanoDrop technology. A total of 500 ng of RNA was used for reverse transcription using the SuperScript III reverse transcription kit (Invitrogen). Quantitative polymerase chain reaction (PCR) analysis was performed using an ABI 7900 system (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com). Lamin expressions were performed using the TaqMan gene expression Master Mix (Roche, Indianapolis, IN, http://www.roche.com), following the manufacturer’s protocol. Quantification of gene expression was based on the ΔCt method and normalized on 18S expression (Assay HS_099999). PCR primers were previously described by Rodriguez and colleagues [22]. Primer sequences were lamin A (exons 11/12), 5′-TCTTCTGCCTCCAGTGTCACG-3′ and 5′-AGTTCTGGGGGCTCTGGGT-3′; lamin C (exons 9/10), 5′-CAACTCCACTGGGGAAGAAGTG-3′ and 5′-CGGCGGCTACCACTCAC-3′; and progerin (exons 11/12), 5′-ACTGCAGCAGCTCGGGG-3′ and 5′-TCTGGGGGCTCTGGGC-3′. TaqMan MGB probe sequences were lamin A (exon 11), 5′-ACTCGCAGCTACCG-3′; lamin C (exon10), 5′-ATGCGCAAGCTGGTG-3′; and progerin (exon 11), 5′-CGCTGAGTACAACCT-3′. Reporter and quencher dyes for the LMNA locus assays were 5′-6FAM and 3′ nonfluorescent quencher dye (Applied Biosystems). Osteogenic differentiation markers were quantified using SYBR Green PCR Master Mix (Applied Biosystems), following the manufacturer’s protocol. Primer sequences were collagen type 1A, CCCCTGGAAAGAATGGAGAT and CCATCCAAACCACTGAAACC; alkaline phosphatase (ALP), CCACGTCTTCACATTTGGTG and AGACTGCGCCTGGTAGTTGT; Ki-67, TGATGGTTGAGGTCGTTCCTTG and TCCTTTGGTGGGCACCTAAGAC; proliferating cell nuclear antigen (PCNA), TCCACTCTCTTCAACGGTGACA and TCGATCTTGGGAGCCAAGTAGTA. Quantification of gene expression was based on the ΔCt method and normalized on 18S expression, GATATGCTCATGTGGTGTTG, AATCTTCTTCAGTCGCTCCA.
Flow Cytometry
Cells were detached from culture plates using trypsin-ethylenediaminetetraacetic acid (Invitrogen) and fixed in 4% paraformaldehyde (15 minutes at room temperature). After a PBS wash, cells were stained with CD29-PE Mouse IgG1 1:6 (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com), CD44-PE Mouse IgG1 1:6 (BD Biosciences), CD73-PE Mouse IgG1 1:2 (BD Biosciences), CD105-PE Mouse IgG1 1:20 (Invitrogen), and CD166-PE mouse IgG1 1:2 (BD Biosciences), diluted in PBS containing 5% fetal bovine serum (30 minutes at room temperature). Negative controls were unstained cells and isotype-control stained cells (mouse IgG1-PE). Cells were analyzed on a magnetic affinity cell sorting flow cytometry (Miltenyi Biotec) using FlowJo software. The number of events analyzed for each experiment was 10,000. Three independent experiments were performed for each cell line.
Statistical analysis
Statistical analysis was performed by a one-way analysis of variance using the Dunnett’s comparison test. Values of p < .05 were considered significant (∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001).
Results
This study was performed using MSCs derived from HGPS iPS cells (Fig. 1A). All of the experiments described in this study were performed on the same iPS cell line to rigorously compare the effects of the different drugs in cells presenting exactly the same genetic background. Pluripotency and self-renewal of the iPS cell line used was described previously by our group [19]. HGPS iPS cells were differentiated into MSCs (Fig. 1A) presenting 99% homogeneous for mesodermal markers (Fig. 1B). Compared with WT MSCs, HGPS MSCs overexpress progerin at the mRNA level (Fig. 1C) and the protein level (Fig. 1D, 1E) and present functional pathological defects characteristic of the disease, including nuclear shape abnormalities (Fig. 2A, 2B), premature osteoblastic differentiation (Fig. 2C, 2D), and defects in cell proliferation (Fig. 2E-2G) but not in cell metabolism (Fig. 2H). All the further pharmacological experiments were carried out after the same number of cell divisions, in vitro following MSC differentiation, with four passages before replicative senescence, as determined in preliminary experiments. The experimental workflow (supplemental online Fig. 1) and analysis (supplemental online Fig. 2) are presented as supplemental online data. The treatment dosage was chosen on the basis of dose-response effects of the drugs on nuclear shape abnormalities (supplemental online Fig. 3), in agreement with the existing literature on HGPS fibroblasts for ZoPra (1 μM), the selected FTI (1 μM), and Rapa (680 nM). Whereas dosages of ZoPra and FTI were consistent with the literature, our results showed that Rapa was toxic for HGPS MSCs at this reported concentration but significantly efficient on nuclear shape integrity at a classic dosage of Rapa for in vitro studies (10 nM) [23].
Figure 1.
Derivation and characterization of MSCs from HGPS iPSCs. (A): Schematic representation of the pathological modeling strategy and the iPSC differentiation protocol. Scale bars = 50 μm. (B): Flow cytometry analysis of CD29, CD44, CD73, CD105, and CD166 expression in WT MSCs and HGPS MSCs. (C): Quantitative polymerase chain reaction analysis of progerin expression in WT MSCs and HGPS MSCs. Data are normalized on the 18S housekeeping gene. Each chart represents the mean ± SD of three independent experiments. (D): Progerin immunostaining in WT MSCs and HGPS MSCs. Scale bars = 50 μm. (E): Automated quantification of progerin immunopositive nuclei in WT MSCs and HGPS MSCs. The chart represents the dispersion of 12 independent experiments. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; FBS, fetal bovine serum; FGF2, fibroblast growth factor 2; HGPS MSC, MSCs derived from Hutchinson-Gilford progeria syndrome iPSCs; iPSC, induced pluripotent stem cell; KSR, knockout serum replacement; MSC, mesodermal stem cells; WT MSC, MSCs derived from control iPSC lines (wild-type).
Figure 2.
Molecular characterization of HGPS MSCs. (A): Lamin A/C staining (JOL2) in WT MSCs and HGPS MSCs. Scale bars = 25 μm. (B): Automated quantification of abnormal nuclei in WT MSCs and HGPS MSCs. The chart represents the dispersion of eight independent experiments. (C): ALP activity in WT MSCs and HGPS MSCs differentiated in osteoblastic lineage (7 days of differentiation). Scale bars = 50 μm. (D): Quantitative polymerase chain reaction analysis of ALP and collagen type 1A expression in WT MSCs and HGPS MSCs. Data are normalized on the 18S housekeeping gene. Each chart represents the mean ± SD of three independent experiments. (E): Cell cycle analysis after 5-ethynyl-2′-deoxyuridine incorporation in WT MSCs and HGPS MSCs. Values represent the mean ± SD of three independent experiments. (F): Automated quantification of Ki-67 immunopositive nuclei in WT MSCs and HGPS MSCs. The chart represents the dispersion of eight independent experiments. (G): Cumulative number of WT MSCs and HGPS MSCs during 24 days of cultures. (H): Measure of ATP content in WT MSCs and HGPS MSCs. The chart represents the dispersion of eight independent experiments. Abbreviations: ALP, alkaline phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; FBS, fetal bovine serum; HGPS MSC, mesodermal stem cells derived from Hutchinson-Gilford progeria syndrome induced pluripotent stem cells; PI, prodidium iodide; WT MSC, mesodermal stem cells derived from control induced pluripotent stem cell lines (wild-type).
Effect of Pharmacological Drugs on Nuclear Shape Integrity, Prelamin A Maturation, and Progerin Expression
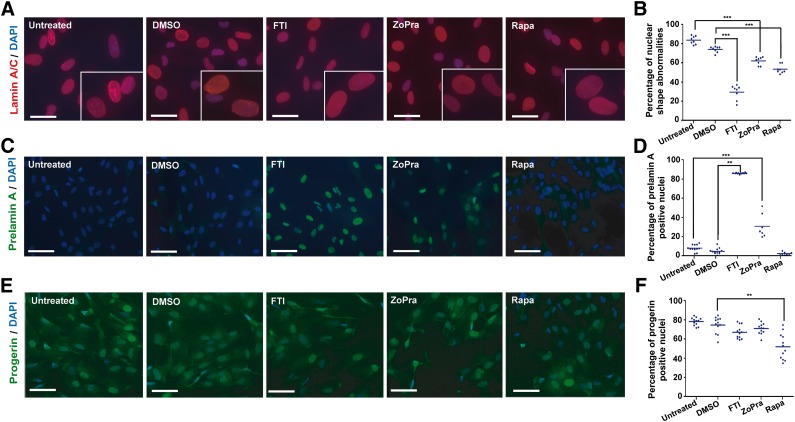
In vitro, misshapen nuclei are the main hallmark of the disease. WT MSCs exhibited homogeneous lamin A/C immunostaining, lining up regularly in the nuclear membrane. In contrast, HGPS MSCs displayed heterogeneous lamin A/C staining with nuclear lobulation, numerous endoplasmic reticulum folds, and macronuclei (Fig. 2A, 2B). When HGPS MSCs were treated for 72 hours, a significant decrease of the percentage of affected nuclei was observed in the presence of each of the three different treatments assayed, in comparison with untreated cells or dimethyl sulfoxide-treated cells, which display 83% and 74% misshapen nuclei, respectively (Fig. 3A, 3B). The quantitative analysis of the drugs’ effects on this parameter revealed improvement of nuclear shape integrity of 24% with ZoPra, 26% with Rapa, and 50% with FTI (Fig. 3B).
Figure 3.
Effect of the different pharmacological treatments on nuclear shape abnormalities, prelamin A maturation, and progerin expression. (A): Lamin A/C staining (JOL2) in mesodermal stem cells derived from Hutchinson-Gilford progeria syndrome induced pluripotent stem cells (HGPS MSCs) following 72 hours of treatment in a cumulative dose. Scale bars = 25 μm. (B): Quantification of misshapen nuclei in HGPS MSCs following 72 hours of treatment in a cumulative dose. Each plot represents the percentage of abnormal nuclei, and the horizontal bar represents the mean value of each condition. (C): Prelamin A immunostaining in HGPS MSCs following 72 hours of treatment in a cumulative dose. Scale bars = 30 μm. (D): Automated quantification of prelamin A-stained nuclei in HGPS MSCs following 72 hours of treatment in a cumulative dose. Each plot represents the percentage of prelamin A-positive nuclei, and the horizontal bar represents the mean value of each condition. (E): Progerin immunostaining in HGPS MSCs following 72 hours of treatment in a cumulative dose. Scale bars = 30 μm. (F): Automated quantification of progerin-stained nuclei in HGPS MSCs following 72 hours of treatment in a cumulative dose. Each plot represents the percentage of progerin-positive nuclei, and the horizontal bar represents the mean value of each condition. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; FTI, farnesyltransferase inhibitor; Rapa, rapamycin; ZoPra, zoledronate and pravastatin.
To evaluate the effect of these compounds on prelamin A maturation, an immunoassay allowing the quantification of the subcellular localization of this protein precursor was set up. HGPS MSCs exhibited no specific immunolabeling of prelamin A (Fig. 3C), whereas treatments affected it quite differently. Automated quantification of prelamin A immunostaining revealed high nuclear staining of the protein in cells treated with FTI, indicating an inhibition of the prelamin A maturation process. ZoPra was also efficient, although results were quantitatively less strong (35%) (Fig. 3C, 3D). Western blot analysis showed that expression of lamin A and C was not affected by either ZoPra or rapamycin (supplemental online Fig. 4A). FTI promoted an increase of prelamin A expression, whereas lamin A expression was decreased (supplemental online Fig. 4A). In clear contrast to the two other treatments, our results confirmed that rapamycin had no impact on prelamin A localization (Fig. 3C, 3D) but significantly decreased the percentage of progerin-expressing cells (30%) (Fig. 3E, 3F). The post-translational effect of these different drugs was finally confirmed by correlating these results to gene expression analysis, showing that lamin A/C and progerin mRNA levels were not affected by any of these drugs (supplemental online Fig. 4B, 4C).
Therapeutic Benefits of ZoPra, FTIs, and Rapa on Secondary Functional Parameters
The effects of the different drugs were then analyzed on three aging-related functional parameters, namely, premature osteoblastic differentiation, cell proliferation, and energy metabolism.
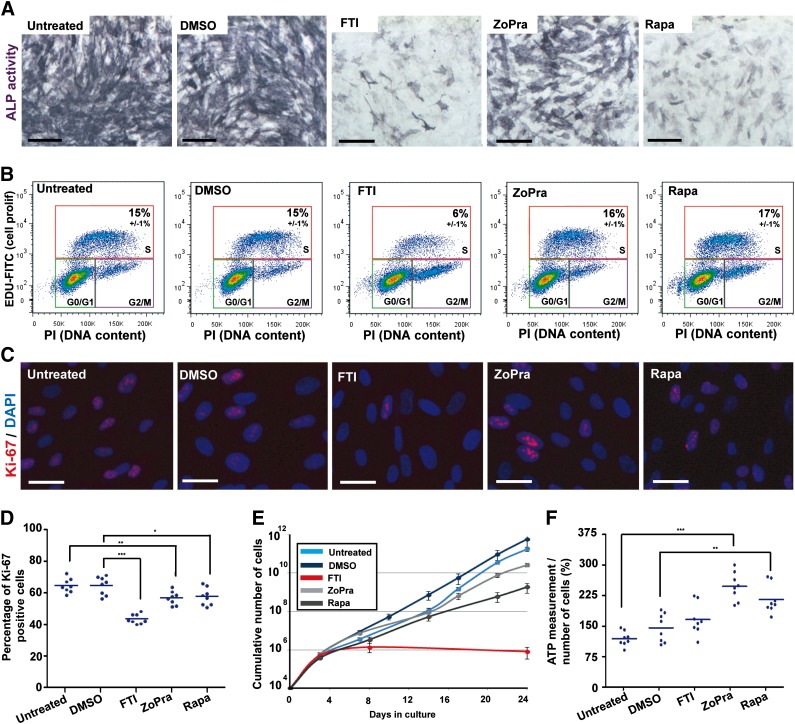
The capacity of MSCs to differentiate along the osteogenic lineage was monitored by measuring alkaline phosphatase activity after 7 days of differentiation in osteoblastic induction medium (OIM). Alkaline phosphatase activity quantification was performed using a colorimetric substrate and revealed a strong increase in the osteogenic differentiation of HGPS MSCs compared with WT MSCs (Fig. 2C). Premature osteogenic differentiation of HGPS MSCs was confirmed by quantitative PCR revealing an increased expression of two classical osteogenic markers, ALP and collagen type 1A (Fig. 2D). Whereas all of the drugs were efficient with nuclear shape abnormalities, major differences were identified between the effects of the treatments on this parameter. Accordingly, HGPS MSCs treated with either FTI or Rapa presented a high decrease in their premature osteogenic differentiation, whereas the effect of ZoPra was more limited (Fig. 4A).
Figure 4.
Effect of the different pharmacological treatments on osteogenic differentiation, cellular proliferation, and energy metabolism. (A): Alkaline phosphatase activity in mesodermal stem cells derived from Hutchinson-Gilford progeria syndrome induced pluripotent stem cells (HGPS MSCs) differentiated in osteoblastic lineage (7 days of differentiation) in the presence of the different treatments. Scale bars = 50 μm. (B): Cell cycle analysis after 5-ethynyl-2'-deoxyuridine incorporation in HGPS MSCs in the presence of the different treatments. Values represent the mean ± SD of three independent experiments. (C): Ki-67 immunostaining in HGPS MSCs following 72 hours of treatment. Scale bars = 25 μm. (D): Automated quantification of Ki-67 immunopositive nuclei in HGPS MSCs following 72 hours of treatment in a cumulative dose. Each plot represents the percentage of Ki-67-positive nuclei, and the horizontal bar represents the mean value of each condition. (E): The cumulative number of HGPS MSCs during 24 days of culture in the presence of the different drugs. (F): The measure of ATP content in HGPS MSCs following 72 hours of treatment. Each plot represents the values of ATP content per cell, and the horizontal bar represents the mean value of each condition. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; FTI, farnesyltransferase inhibitor; PI, prodidium iodide; Rapa, rapamycin; ZoPra, zoledronate and pravastatin.
As described previously, HGPS MSCs demonstrated a loss of proliferative capacity, as revealed by a decrease of the proportion of cycling cells incorporating EdU (Fig. 2E) and Ki-67-positive cells compared with WT MSCs (Fig. 2F). This led over time to a progressive reduction in the number of HGPS cells produced, with ∼7 times the difference to WT MSCs at 24 days (Fig. 2G). Measures of EdU incorporation after pharmacological treatments showed a decrease in the proportion of HGPS MSCs in S phase after 72 hours of treatment with FTI but not with either Rapa or ZoPra (Fig. 4B). Automated quantification of Ki-67 staining revealed more than 33% reduction of cycling cells in the presence of FTI, whereas the two other treatments were much less deleterious, with less than 15% reduction in the proportion of cycling cells (Fig. 4B–4D). These results were confirmed by quantitative PCR revealing a decrease of PCNA and Ki-67 expression after FTI treatment (supplemental online Fig. 5). Over the long term, when treatments were continuously applied over 24 days, ZoPra induced a mild reduction in the rate of cell proliferation (total number of cells reduced by 7 times compared with untreated HGPS MSCs), whereas Rapa had a moderate effect (reduction by 300 times), and FTI exhibited a strong cytostatic effect (Fig. 4E). FTI led to total proliferation arrest after 8 days of treatment. After 24 days, a final reduction in the number of cells was 700,000 times smaller compared with untreated HGPS cells.
Finally, the analysis of energy metabolism was monitored through the quantification of ATP production using CellTracker Green labeling. Although comparison between WT MSCs and HGPS MSCs revealed no statistically significant difference (Fig. 2H), pharmacological experiments highlighted differences among the treatments. In data normalized on the number of cells to avoid the bias introduced by the differential cell proliferation, ZoPra exhibited the most beneficial effect, with the amount of ATP almost doubling per cell (Fig. 4F). Rapa had a positive although much reduced effect, with a 30% increase in ATP content per cell, whereas FTI had no effect on ATP content per cell (Fig. 4E).
In Vitro Evaluation of the Different Drug Combinations on HGPS MSCs
In the light of these results, we have used this cellular model to evaluate the therapeutic potential of the different combinations of these three treatments. Quantification of nuclear shape abnormalities (Fig. 5A) and premature osteogenic differentiation (Fig. 5B) of HGPS MSCs revealed that all of the drug combination treatments could similarly restore these pathological defects. In parallel, measures of Ki-67-positive cells (Fig. 5C) and long-term culture experiments (Fig. 5D) revealed a strong cytostatic effect of treatments containing FTI in combination with either one or the two other treatments. In order to limit this deleterious effect of FTI in drug combinations, dose-response experiments were performed on the prelamin A maturation process (supplemental online Fig. 6A) and premature osteogenic differentiation (supplemental online Fig. 6B), showing a half maximal effective concentration of this FTI at 50 nM. Repeating single-drug and combination experiments with this lower efficient dose showed similar corrective effects on nuclear shape abnormalities but still associated with an antiproliferative effect on the cells (supplemental online Fig. 6C–6G).
Figure 5.
Molecular characterization of FTI, ZoPra, and Rapa drug combinations on mesodermal stem cells derived from Hutchinson-Gilford progeria syndrome induced pluripotent stem cells (HGPS MSCs). (A): Quantification of misshapen nuclei in HGPS MSCs following 72 hours of treatment with the different drug combinations. Values represent the mean ± SD of three independent experiments. (B): Alkaline phosphatase activity in HGPS MSCs differentiated in osteoblastic lineage (7 days of differentiation) in the presence of the different drug combinations. Scale bars = 50 μm. (C): Automated quantification of Ki-67 immunopositive nuclei in HGPS MSCs following 72 hours of treatment with the different drug combinations. Values represent the mean ± SD of three independent experiments. (D): The cumulative number of HGPS MSCs during 12 days of culture in the presence of the different drug combinations. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; FTI, farnesyltransferase inhibitor; Rapa, rapamycin; ZoPra, zoledronate and pravastatin.
Discussion
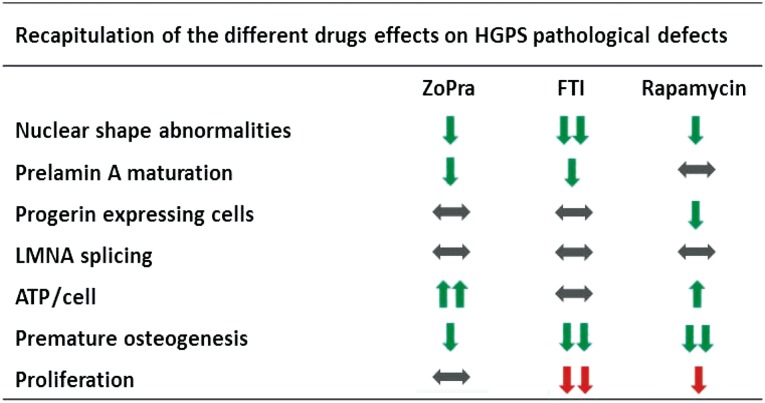
The main result of this study is the demonstration that the three treatments currently used or proposed in clinical trials for HGPS have somewhat different functional effects at the cellular level (Fig. 6). Altogether, in contrast to the common corrective effects of the three treatments on nuclear shape, this functional study revealed major differences in their therapeutic potential. This demonstration may have major consequences for the elaboration of new clinical trials, following those that have already been initiated. In addition, by demonstrating the capacity of iPS cells to enable quantified measurements in functional cellular assays, this study underscores the value of iPS cells as a platform for pharmacological studies, beyond the search for effective chemical compounds through drug screening.
Figure 6.
Recapitulation of the functional effects of ZoPra, FTI, and rapamycin on mesodermal stem cells derived from HGPS induced pluripotent stem cells. Abbreviations: HGPS, Hutchinson-Gilford progeria syndrome; FTI, farnesyltransferase inhibitor; ZoPra, zoledronate and pravastatin.
HGPS is caused by a point mutation (c.1824C > T) in exon 11 of LMNA, activating a cryptic splicing site and the production of the toxic protein progerin with two major defects: a truncation of the protein and the persistence of a farnesyl residue at its C-terminal end [24]. This latter defect has rapidly attracted attention because drugs that alter the farnesylation process were already available. FTIs that directly target the enzyme responsible for adding the farnesyl residue to the immature protein have elicited functional improvements, particularly with regard to the correction of nuclear shape abnormalities [7, 8]. In 2008, another approach was described to correct the same molecular defect by using a combination of two drugs that target the prenylation process in a broader way, with similar effects on misshapen nuclei [10]. More recently, another therapeutic route has been explored with the mTOR inhibitor rapamycin, known to stimulate autophagy [25]. Rapamycin treatment has been associated with the correction of misshapen nuclei in HGPS fibroblasts, suggesting that the increased clearance of progerin alone had corrective effects, in the likely absence of effects on the farnesylation process [11]. Quantitative results of the present study on prelamin A maturation and correction of the nuclear shape abnormalities were in keeping with the data underscoring the phenotypical rescue of treatments targeting the farnesylation process. Conversely, our results highlighted the fact that rapamycin’s therapeutic effect was independent from the maturation process of prelamin A. It is interesting to mention that rapamycin effects were recorded in the present study at a dose 68 times lower than in the original demonstration of the effects of rapamycin on HGPS cells [11]. Moreover, the robustness of this model has permitted study of the effect of the combination of these three drugs on nuclear shape abnormalities, showing neither synergistic nor additive effects of these combinations compared with single-drug treatments.
The decrease of cell proliferation and the premature senescence are well-characterized consequences of the HGPS mutation [26]. Interestingly, in our study, none of the treatments had a clear corrective effect on those parameters, suggesting that it is not affected by changes in the prelamin A maturation process. Moreover, it is interesting to mention that long-term analysis of the treatment effects revealed a potent cytostatic effect of FTIs, with cell proliferation arrest after 1 week. These deleterious effects were reminiscent of those demonstrated when FTIs were used in cancer therapy [27]. Even if extrapolation of in vitro results to patients stay elusive, these results raise the question of a potential disadvantage for therapeutic strategies based on the use of FTIs. At least these results highlight the interest in monitoring stem cell proliferation potential in patients treated in clinical trials using FTIs to avoid some undesirable side effects.
One of the main advantages of the iPS cell model is that it allows precise monitoring of stem cell differentiation known to be dysregulated in HGPS [28]. In light of these results and the loss of bone density observed in patients, we hypothesized that the premature differentiation of MSCs could be, at least partially, a component of the disease. Accordingly, our results revealed a premature osteogenic differentiation in vitro in HGPS iPS cell derivatives reminiscent of that previously reported in wild-type cells engineered to overexpress progerin [28]. Pharmacological experiments indicated that only ZoPra exhibited a slight corrective effect on that function, whereas rapamycin and FTI efficiently slowed it down both in the short and long terms.
Decreased energy metabolism with a major reduction in ATP production is a hallmark of cellular aging and has been classically associated with a limited replicative life span of cultured human HGPS fibroblasts [29]. Dysregulation of energy demand and/or inefficiency of oxidative phosphorylation could play a role in the curtailed replicative capacity of progeria cells. More recently, these data have been replicated, showing that the ATP content of progeria fibroblasts was 50% decreased compared with healthy controls, linking the effect to a decrease in proteasome activity [30]. At odds with these data, our study revealed no statistically significant difference between WT MSCs and HGPS MSCs when ATP production was normalized to the number of cells. This lack of alteration may be associated with the absence of cell senescence at the time of the analysis because the pharmacological treatment was always purposely performed ahead of that event. Nevertheless, the pharmacological study performed on HGPS MSCs revealed that ZoPra and, in a much more limited way, rapamycin, but not FTI, increased ATP production in a very significant way. Because correction of ATP concentrations has many consequences for major cell functions, it would be of interest to further explore the potential of treatments that would discretely target this mechanism in HGPS cells.
All the results presented in this study have been obtained with cells differentiated from iPS cell lines derived from HGPS patients. Such cell lines have already repeatedly been instrumental in replicating molecular mechanisms and major cellular defects associated with the disease [16, 17] as well as revealing physiological mechanisms that protect neural cells [19]. In this study, iPS cells have also allowed us to relate changes in those defects with different therapeutic tools, namely, to use them as a platform for pharmacological studies to detail functional outcomes that can be expected from the use of one or another treatment that targets causal pathological mechanisms. A number of studies have already made use of the in vitro replication of pathological hallmarks of a number of other genetic diseases to observe the effects of known drugs or to screen chemical libraries seeking yet unknown drugs. The present study is an extension of those experimental paradigms by demonstrating that iPS cell derivatives can be further exploited to search for associations between causative mechanisms and the functional effects of drugs. Such use of derivatives of iPS cells obtained from patients with a genetic disease may be extended to many other diseases for which potential treatments are proposed.
Conclusion
Since the discovery of the molecular mechanism leading to HGPS, several drugs have been described as candidates for drug repositioning. In this study, we have taken advantage of the unique potential of pluripotent stem cells to compare the functional effects of the three main treatments offered to patients, FTIs, ZoPra, and rapamycin. In addition to their comparison, this model has also permitted the evaluation of their combinations, showing the lack of additive effects of these drugs. Finally, beyond this functional analysis, this study opens up new therapeutic perspectives for the treatment of HGPS through the use of iPS cell derivatives as cellular platforms for drug screening approaches.
Supplementary Material
Acknowledgments
We thank Mélody Mazon, Kristell Lebozec, Karine Girault-Triboult, Pauline Poydenot, Pauline Georges, Johana Tournois, and Drs. Delphine Peric and Delphine Laustriat for their part in certain experiments; Yves Maury and Dr. Alexandra Benchoua for their expertise in high-content imaging; and Drs. Gisele Bonne, Walter Habeler, Alexandre Méjat, Pierre Cau, Odile Rigault, and Michèle Martin for helpful discussions. This work was supported by the Institut National de la Santé et de la Recherche Médicale, University Evry Val d’Essonne, Association Française contre les Myopathies, and Genopole.
Author Contributions
S.B.: collection and/or assembly of data, data analysis and interpretation, manuscript writing; A.-L.J., A.-L.E., A.L.C., C.N., V.C., C.M., and Y.L.: collection and/or assembly of data; K.D.: provision of study material or patients; A.d.S.-G. and N.L.: data analysis and interpretation, manuscript writing; M.P.: conception and design, data analysis and interpretation, manuscript writing; X.N.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Gordon LB, Kleinman ME, Miller DT, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2012;109:16666–16671. doi: 10.1073/pnas.1202529109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Sandre-Giovannoli A, Bernard R, Cau P, et al. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro CL, Cau P, Lévy N. Molecular bases of progeroid syndromes. Hum Mol Genet. 2006;15:R151–R161. doi: 10.1093/hmg/ddl214. [DOI] [PubMed] [Google Scholar]
- 5.Hennekam RC. Hutchinson-Gilford progeria syndrome: Review of the phenotype. Am J Med Genet A. 2006;140:2603–2624. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- 6.Merideth MA, Gordon LB, Clauss S, et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SH, Meta M, Qiao X, et al. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J Clin Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capell BC, Olive M, Erdos MR, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci USA. 2008;105:15902–15907. doi: 10.1073/pnas.0807840105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClintock D, Ratner D, Lokuge M, et al. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS One. 2007;2:e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela I, Pereira S, Ugalde AP, et al. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat Med. 2008;14:767–772. doi: 10.1038/nm1786. [DOI] [PubMed] [Google Scholar]
- 11.Cao K, Graziotto JJ, Blair CD, et al. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- 12.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paradisi M, McClintock D, Boguslavsky RL, et al. Dermal fibroblasts in Hutchinson-Gilford progeria syndrome with the lamin A G608G mutation have dysmorphic nuclei and are hypersensitive to heat stress. BMC Cell Biol. 2005;6:27. doi: 10.1186/1471-2121-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capell BC, Erdos MR, Madigan JP, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maury Y, Gauthier M, Peschanski M, et al. Human pluripotent stem cells: Opening key for pathological modeling [in French] Med Sci (Paris) 2011;27:443–446. doi: 10.1051/medsci/2011274023. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Lian Q, Zhu G, et al. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Liu GH, Barkho BZ, Ruiz S, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu GH, Suzuki K, Qu J, et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissan X, Blondel S, Navarro C, et al. Unique preservation of neural cells in Hutchinson-Gilford progeria syndrome is due to the expression of the neural-specific miR-9 microRNA. Cell Rep. 2012;2:1–9. doi: 10.1016/j.celrep.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Giraud-Triboult K, Rochon-Beaucourt C, Nissan X, et al. Combined mRNA and microRNA profiling reveals that miR-148a and miR-20b control human mesenchymal stem cell phenotype via EPAS1. Physiol Genomics. 2011;43:77–86. doi: 10.1152/physiolgenomics.00077.2010. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez S, Coppedè F, Sagelius H, et al. Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur J Hum Genet. 2009;17:928–937. doi: 10.1038/ejhg.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J, Meng Y, Kwiatkowski DJ, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120:103–114. doi: 10.1172/JCI37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira S, Bourgeois P, Navarro C, et al. HGPS and related premature aging disorders: From genomic identification to the first therapeutic approaches. Mech Ageing Dev. 2008;129:449–459. doi: 10.1016/j.mad.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 26.Graziotto JJ, Cao K, Collins FS, et al. Rapamycin activates autophagy in Hutchinson-Gilford progeria syndrome: Implications for normal aging and age-dependent neurodegenerative disorders. Autophagy. 2012;8:147–151. doi: 10.4161/auto.8.1.18331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efuet ET, Keyomarsi K. Farnesyl and geranylgeranyl transferase inhibitors induce G1 arrest by targeting the proteasome. Cancer Res. 2006;66:1040–1051. doi: 10.1158/0008-5472.CAN-05-3416. [DOI] [PubMed] [Google Scholar]
- 28.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein S, Ballantyne SR, Robson AL, et al. Energy metabolism in cultured human fibroblasts during aging in vitro. J Cell Physiol. 1982;112:419–424. doi: 10.1002/jcp.1041120316. [DOI] [PubMed] [Google Scholar]
- 30.Viteri G, Chung YW, Stadtman ER. Effect of progerin on the accumulation of oxidized proteins in fibroblasts from Hutchinson Gilford progeria patients. Mech Ageing Dev. 2010;131:2–8. doi: 10.1016/j.mad.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.