Abstract
Mycobacterium tuberculosis, the causative agent of tuberculosis in humans, is a bacterium with the unique ability to persist for years or decades as a latent infection. This latent state, during which bacteria have a markedly altered physiology and are thought to be dormant, is crucial for the bacteria to survive the stressful environments it encounters in the human host. Importantly, M. tuberculosis cells in the dormant state are generally refractory to antibiotics, most of which target cellular processes occurring in actively replicating bacteria. The molecular switches that enable M. tuberculosis to slow or stop its replication and become dormant remain unknown. However, the slow growth and dormant state that are hallmarks of latent tuberculosis infection have striking parallels to the “quasi-dormant” state of Escherichia coli cells caused by the toxin components of chromosomal toxin-antitoxin (TA) modules. An unusually large number of TA modules in M. tuberculosis, including nine in the mazEF family, may contribute to initiating this latent state or to adapting to stress conditions in the host. Toward filling the gap in our understanding of the physiological role of TA modules in M. tuberculosis, we are interested in identifying their molecular mechanisms to better understand how toxins impart growth control. Our recent publication1 uncovered a novel function of a MazF toxin in M. tuberculosis that had not been associated with any other MazF ortholog. This toxin, MazF-mt6, can disrupt protein synthesis by cleavage of 23S rRNA at a single location in an evolutionarily conserved five-base sequence in the ribosome active center.
Keywords: tuberculosis, ribosome, antitoxin, protein synthesis, endoribonuclease
Background
MazEF toxin-antitoxin (TA) systems are present in the genomes of many free-living bacteria.2 TA systems have been implicated in stress survival, persistence, and latent tuberculosis infection,3-5 in part because their general function is to facilitate reversible growth inhibition in response to stress. TA modules are autoregulated operons comprising adjacent genes that encode two small (~10 kDa) proteins, an intracellular toxin and its cognate antitoxin (Fig. 1). The cytotoxic activity of MazF can be triggered by stresses such as nutrient limitation, DNA damage, high temperature, oxidative stress, or exposure to various antibiotics.6-9 Although the MazF toxin and the MazE antitoxin are part of the same transcript, the amount of active, free MazF is variable because its activity can be inhibited upon binding to MazE. The dynamic interplay between the inactive MazEF complex and the active, free MazF toxin is due to the intrinsic instability of the antitoxin, which is readily degraded by cellular proteases (Fig. 1).6,7 Therefore, the activity of the toxin is dependent on the concentration of antitoxin, the concentration of the antitoxin is influenced by the milieu of cellular proteases, and the level of toxin and antitoxin synthesized is dependent on the transcriptional regulation of the operon, which is responsive to cellular signals that are not yet defined for M. tuberculosis TA systems.
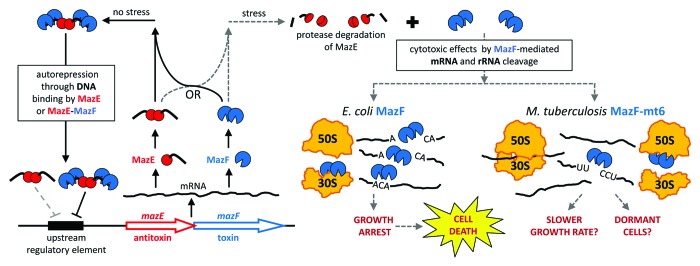
Figure 1. MazF toxins inhibit translation and arrest growth by cleaving mRNA and rRNA. All mazEF operons consist of two adjacent genes, one that encodes the intracellular toxin MazF and another that produces its inhibitor, antitoxin MazE.37 In the absence of stress, the MazE protein forms a stable complex with the MazF toxin to neutralize its toxicity.6,38 The MazEF heterohexamer complex (2 MazF: 2 MazE: 2 MazF)39 and to a lesser extent, the MazE dimer, repress the operon by binding to an upstream regulatory element that consists of a palindromic DNA sequence.10,40 The intrinsically unstable MazE antitoxin can be degraded by cellular proteases,6,7 which frees and activates the MazF toxin. The toxic activity of MazF is triggered by stresses such as nutrient limitation, DNA damage, high temperature, oxidative stress, or exposure to various antibiotics.6-9 MazF orthologs exert their cytotoxic effects by cleaving single-stranded RNA at unique and specific 3-, 5-, or 7-base sequences.1,11-23 Although MazF toxins were initially proposed to solely cleave mRNA as “mRNA interferases,”11-20E. coli MazF also cleaves 16S rRNA in the 30S ribosomal subunit,24 while M. tuberculosis toxin MazF-mt6 also cleaves 23S rRNA in the 50S subunit.1 Cleavage of mRNAs or rRNAs disrupts protein synthesis,1,12,24 which can induce a state of reversible dormancy.41 Expression of MazF triggers this “quasi-dormant” state, during which cells stop dividing but are able to transcribe mRNA and synthesize proteins.42 Due to its recognition of a relatively short RNA sequence (ACA), E. coli MazF cleaves a majority of cellular mRNAs. In addition, not only can MazF render certain mRNAs leaderless by cleaving an ACA sequence upstream of the start codon, but it also removes 43 nt from the 3′ end of 16S rRNA to create “stress ribosomes” that selectively translate either naturally present or MazF-generated leaderless mRNAs.24E. coli MazF has also been shown to initiate cell death if its toxic effects are not neutralized by MazE in sufficient time.6,8,9,43 In contrast, M. tuberculosis toxin MazF-mt6 is expected to cleave only a subpopulation of mRNAs due to its recognition of the five-base RNA sequence UUCCU. However, MazF-mt6-mediated cleavage of 23S rRNA in free 50S ribosomal subunits is sufficient to inhibit protein synthesis,1 so combined cleavage of mRNA and rRNA may elicit rapid growth arrest. The effect of MazF-mt6 on M. tuberculosis cells in vivo has not been rigorously investigated, but two possible endpoints are shown.
E. coli MazF mediates bacterial cell growth control through its dynamic association and dissociation with its cognate antitoxin MazE.10 Earlier biochemical studies on E. coli MazF demonstrated that this toxin is a single-stranded, sequence-specific endoribonuclease that targets ACA sequences and that its cleavage at ACAs appeared to be specific for mRNA.11,12 Consequently, Inouye and colleagues coined the term “mRNA interferase” for E. coli MazF, and this term was often extended to MazF orthologs from other bacteria.11-20 Despite high sequence similarity between MazF toxins in various prokaryotes, nearly every MazF ortholog recognizes a unique 3- to 7-base RNA sequence.1,11-23 Therefore, it was thought that if one could determine the precise cleavage recognition sequence of a given MazF ortholog, one could predict how the toxin specifically alters the transcriptome in vivo.
Implicit in this prediction, the length and base content of the recognition sequence required for MazF toxin-mediated RNA cleavage should dictate the degree of post-transcriptional editing. Expression of MazF toxins with short RNA recognition sequences, such as the ACA-cleaving E. coli MazF, should result in wholesale destruction of mRNAs. In contrast, MazF toxins with larger recognition sequences are predicted to selectively degrade a subpopulation of transcripts that possess one or more copies of the recognition sequence and spare those lacking this sequence. Since it has been demonstrated that the rate of transcript degradation can be correlated with the number of cleavage sites,15,18,22 statistical analyses can be performed to predict the degree of vulnerability of mRNA transcripts.18,19,22 However, this kind of statistical analysis is only applicable when mRNAs are the sole targets of MazF toxins. In 2011, Moll, Engelberg-Kulka, and colleagues demonstrated that mRNA is not the only target of E. coli MazF.24 This toxin also specifically cleaves 16S rRNA at a single ACA sequence, resulting in the production of a population of truncated 16S rRNAs lacking the last 43 nucleotides at the 3′ end. Furthermore, their studies also showed that these shortened 16S rRNAs assemble into specialized “stress ribosomes” that selectively translate leaderless transcripts. Therefore, mRNAs are clearly not the only MazF target in E. coli.
It was also thought that the function of the representative E. coli MazF toxin mirrors that of the family as a whole. Our paper1 demonstrates that this is somewhat true, but the devil is in the details. We focused on one of the nine MazF family members in the M. tuberculosis genome. Although M. tuberculosis harbors an extremely high number (> 80) of predicted TA modules relative to most other prokaryotes,2,25 the physiological role of this large repertoire of TA loci in M. tuberculosis is unclear because their fundamental properties, enzymatic activities, and intracellular targets are only now beginning to be studied in molecular detail.
23S rRNA as a New Target for MazF Toxins
In Schifano et al.,1 we first demonstrate that the consensus recognition sequence for MazF-mt6 cleavage of RNA is UU↓CCU, where “↓” indicates the cleavage site. Coincidently, the 23S rRNA band diminishes in intensity when MazF-mt6 is ectopically expressed in E. coli or the mycobacterial model organism Mycobacterium smegmatis. The loss of 23S rRNA appears to result from cleavage at a single site, since the loss of full-length 23S rRNA coincides with the increase of two stable degradation products whose estimated sizes total that of the intact rRNA. Even though E. coli and M. tuberculosis 23S rRNAs each contain three UUCCU sequences, only the lone site located in a single-stranded region is targeted in each case. Therefore, the MazF-mt6 toxin not only cleaves mRNA, it also directly targets 23S rRNA for cleavage at a single UUCCU site evolutionarily conserved in E. coli, M. smegmatis, and M. tuberculosis. Because of this conservation, we employed an in vitro coupled transcription/translation system with purified E. coli components and a separate ribosome fraction to determine how addition of MazF-mt6 before or after ribosome assembly influences the translation step. This also enabled us to distinguish the net effect of mRNA plus 23S rRNA cleavage by MazF-mt6 vs. 23S rRNA cleavage alone on translation. We observed complete translation inhibition in both cases, indicating that MazF-mt6-mediated cleavage of 23S rRNA alone is capable of disabling protein synthesis. This result is in agreement with the critical function of the target region, helix/loop 70 of domain IV, which facilitates tRNA binding in the ribosomal A-site (Fig. 2).
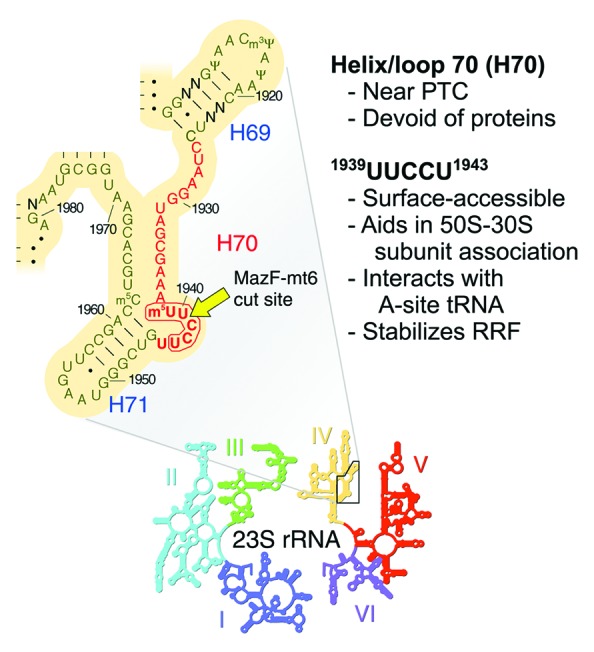
Figure 2. MazF-mt6 cleaves M. tuberculosis 23S rRNA at a single UUCCU in the ribosomal A site. To pinpoint this cleavage, the sequence and secondary structure of a conserved region of domain IV (above) in 23S rRNA (inset below) are shown. Helix/loop 70 (H70), whose nucleotides are highlighted in red, is 100% identical between E. coli, M. smegmatis, and M. tuberculosis. Non-conserved residues are labeled N, E. coli position numbers are adjacent to tick marks, the MazF-mt6 recognition sequence is encircled by a bold red line, and the lone MazF-mt6 cleavage site is indicated by a yellow arrow. This structure is adapted from E. coli 23S rRNA gene rrlB, courtesy of Harry Noller and the University of California, Santa Cruz (http://rna.ucsc.edu/rnacenter/ribosome_images.html) and published in Yusupov et al.29 PTC, peptidyl transferase center.
The vital role of this highly conserved UUCCU sequence in 23S rRNA of M. tuberculosis is illuminated by a wealth of data for its counterpart in E. coli 23S rRNA, 1939UUCCU1943 (Fig. 2). First, a U1940A mutation results in 50S ribosomal subunits defective in assembly.26 Second, the X-ray crystal structure of a 50S subunit revealed that helix/loop 70 forms part of the front rim of the peptidyl transferase active site cleft.27 Third, crystallographic studies revealed that helix/loop 70 contacts and positions the CCA tail of the tRNA acceptor stem and that residues C1941, C1942, and U1943 within the MazF-mt6 cleavage sequence also contact the acceptor stem.28,29 Fourth, this region also directly interacts with ribosome recycling factor (RRF), forming hydrogen bonds to distinct amino acids.30 Higher resolution X-ray crystal structures of the 50S subunit bound to a domain of RRF also uncovered an interaction between U1941 and a highly conserved amino acid in RRF that is important for 50S binding and function.31 Finally, a slowdown in cell growth occurs upon cleavage of 23S rRNA by an unknown E. coli RNase at helix/loop 70 within the same UUCCU sequence cleaved by MazFmt6.32
In addition, there is ample precedent for potent translation inhibition through disruption of rRNA in the large ribosomal subunit. Interestingly, 23S rRNA and its eukaryotic counterpart are targeted by several deadly plant or bacterial toxins—ricin, saporin, Shiga toxin, α-sarcin, and pokeweed antiviral toxin—that either remove a specific adenine from the rRNA backbone or cleave between the two adjacent nucleotides in the universally conserved helix 95, also known as the sarcin-ricin loop.33 However, these toxins cleave intact 70S ribosomes, while MazF-mt6 only cleaves rRNA in free 50S subunits. There are also several antibiotics—sparsomycin, clindamycin, chloramphenicol, linezolid, the pleuromutilins, and the macrolides—that perturb translation through binding to 23S rRNA in E. coli 50S subunits.34 Recently, an M. tuberculosis toxin in the VapC family has also been shown to cleave 23S rRNA, but unlike MazF-mt6, this toxin cleaves at the sarcin-ricin loop.35
Rethinking the Role of MazF
The physiological activities of MazF-mt6 and other family members in M. tuberculosis were originally thought to exclusively stem from their cleavage of mRNA.19,20 Consequently, the characterized M. tuberculosis MazF toxins with five-base recognition sequences were proposed to selectively edit the transcriptome to alter protein expression.19 Our data revealed that this is not the complete extent of MazF-mt6 activity, because cleavage at a functionally essential and evolutionarily conserved region of 23S rRNA also blocks translation.1 Therefore, this toxin appears to have evolved to selectively target both UUCCU-containing mRNAs and ribosomes. Lesson learned? Proceed with caution when inclined to designate MazF family members as mRNA interferases since we and others1,24 have now demonstrated that MazF toxins do not possess inherent specificity for exclusive mRNA cleavage. Instead, there are at least three requirements dictating MazF cleavage of RNA: (1) the cleavage recognition sequence must be present, (2) the RNA must be single-stranded, and (3) the RNA must be accessible to the toxin.
Because cleavage of 23S rRNA alone can arrest translation, the significance of dual cleavage of mRNA and ribosomes and the consequences of their interplay are unclear. In E. coli, 99% of mRNAs (4192 of 4243) contain the ACA MazF motif, and are therefore susceptible to MazF cleavage,36 so widespread mRNA degradation likely occurs in conjunction with the production of “stress ribosomes.” In contrast, only 38% of M. tuberculosis mRNAs (1530 of 4022) contain one or more UUCCU MazF-mt6 cleavage sequences, suggesting mRNA cleavage is not as prevalent with this toxin. Since MazF-mt6 appears to preferentially target 23S rRNA in free 50S ribosomal subunits, the simultaneous cleavage of rRNA with a subset of mRNAs may facilitate the rapid release of ribosomal subunits that are then inactivated (Fig. 1). This “one-two punch” by the MazF-mt6 enzyme should enable a faster physiological response—by reducing the level of ribosomes below the threshold required for active growth—than by mRNA cleavage alone.
Stay tuned, as the scope of new RNA targets and associated molecular detail for MazF toxins expands, our understanding of their role in M. tuberculosis, other pathogens, and yes—even E. coli—will surely surprise us again.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to authors whose work could not be cited due to space constraints. This work was supported in part by National Institutes of Health (NIH) grant R21AI072399 and R01GM095693 to Woychik NA, and NIH training grant T32AI007403, Virus-Host Interactions in Eukaryotic Cells to Schifano JM, awarded to G. Brewer.
Glossary
Abbreviations:
- TA
toxin-antitoxin
- RRF
ribosome recycling factor
- H70
helix/loop 70 in 23S rRNA
- PTC
peptidyl transferase center
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27949
References
- 1.Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, Woychik NA. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci U S A. 2013;110:8501–6. doi: 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–76. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao MC, Rubin EJ. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 4.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012;36:514–32. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–23. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 6.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A. 1996;93:6059–63. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–19. doi: 10.1016/S0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 8.Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol. 2004;186:3663–9. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol. 2001;183:2041–5. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J Biol Chem. 2001;276:5975–84. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280:3143–50. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–23. doi: 10.1016/S1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 13.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 14.Park JH, Yamaguchi Y, Inouye M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011;585:2526–32. doi: 10.1016/j.febslet.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi Y, Nariya H, Park JH, Inouye M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat Commun. 2012;3:607. doi: 10.1038/ncomms1621. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Zhang Y, Zhu L, Suzuki M, Inouye M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem. 2004;279:20678–84. doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhu L, Zhang J, Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem. 2005;280:26080–8. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, Inoue K, Yoshizumi S, Kobayashi H, Zhang Y, Ouyang M, Kato F, Sugai M, Inouye M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J Bacteriol. 2009;191:3248–55. doi: 10.1128/JB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol. 2008;69:559–69. doi: 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem. 2006;281:18638–43. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 21.Pimentel B, Madine MA, de la Cueva-Méndez G. Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J. 2005;24:3459–69. doi: 10.1038/sj.emboj.7600815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothenbacher FP, Suzuki M, Hurley JM, Montville TJ, Kirn TJ, Ouyang M, Woychik NA. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J Bacteriol. 2012;194:3464–74. doi: 10.1128/JB.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuster CF, Park JH, Prax M, Herbig A, Nieselt K, Rosenstein R, Inouye M, Bertram R. Characterization of a mazEF toxin-antitoxin homologue from Staphylococcus equorum. J Bacteriol. 2013;195:115–25. doi: 10.1128/JB.00400-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–57. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leviev I, Levieva S, Garrett RA. Role for the highly conserved region of domain IV of 23S-like rRNA in subunit-subunit interactions at the peptidyl transferase centre. Nucleic Acids Res. 1995;23:1512–7. doi: 10.1093/nar/23.9.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 28.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/S1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 29.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 30.Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc Natl Acad Sci U S A. 2004;101:8900–5. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–60. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17:338–45. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stirpe F, Battelli MG. Ribosome-inactivating proteins: progress and problems. Cell Mol Life Sci. 2006;63:1850–66. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson DN. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci. 2011;1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 35.Winther KS, Brodersen DE, Brown AK, Gerdes K. VapC20 of Mycobacterium tuberculosis cleaves the Sarcin-Ricin loop of 23S rRNA. Nat Commun. 2013;4:2796. doi: 10.1038/ncomms3796. [DOI] [PubMed] [Google Scholar]
- 36.Baik S, Inoue K, Ouyang M, Inouye M. Significant bias against the ACA triplet in the tmRNA sequence of Escherichia coli K-12. J Bacteriol. 2009;191:6157–66. doi: 10.1128/JB.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchimoto S, Nishimura Y, Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992;174:4205–11. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell. 2003;11:875–84. doi: 10.1016/S1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchimoto S, Ohtsubo E. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pem operon. Mol Gen Genet. 1993;237:81–8. doi: 10.1007/BF00282787. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–10. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Mol Cell. 2005;18:253–61. doi: 10.1016/j.molcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol. 2004;186:8295–300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


