Abstract
The mod(mdg4) locus of Drosophila melanogaster contains several transcription units encoded on both DNA strands. The mod(mdg4) pre-mRNAs are alternatively spliced, and a very significant fraction of the mature mod(mdg4) mRNAs are formed by trans-splicing. We have studied the transcripts derived from one of the anti-sense regions within the mod(mdg4) locus in order to shed light on the expression of this complex locus. We have characterized the expression of anti-sense mod(mdg4) transcripts in S2 cells, mapped their transcription start sites and cleavage sites, identified and quantified alternatively spliced transcripts, and obtained insight into the regulation of the mod(mdg4) trans-splicing. In a previous study, we had shown that the alternative splicing of some mod(mdg4) transcripts was regulated by Brahma (BRM), the ATPase subunit of the SWI/SNF chromatin-remodeling complex. Here we show, using RNA interference and overexpression of recombinant BRM proteins, that the levels of BRM affect specifically the abundance of a trans-spliced mod(mdg4) mRNA isoform in both S2 cells and larvae. This specific effect on trans-splicing is accompanied by a local increase in the density of RNA polymerase II and by a change in the phosphorylation state of the C-terminal domain of the large subunit of RNA polymerase II. Interestingly, the regulation of the mod(mdg4) splicing by BRM is independent of the ATPase activity of BRM, which suggests that the mechanism by which BRM modulates trans-splicing is independent of its chromatin-remodeling activity.
Keywords: splicing, SWI/SNF, chromatin remodeling, RNA polymerase II, ATPase activity
Introduction
Pre-mRNA splicing is a fundamental process in eukaryotic gene expression. Splicing is catalyzed by the spliceosome, a complex that is comprised of five small nuclear ribonucleoproteins (snRNPs) and a large number of non-snRNP proteins.1 The spliceosome assembles onto the pre-mRNA to catalyze the removal of introns and the ligation of selected exons. The splicing reactions are highly accurate and conserved nucleotide sequences in the pre-mRNA serve as recognition sites for the recruitment and assembly of the spliceosome.2 The most common type of splicing is known as “cis-splicing,” which involves the ligation of exons within one pre-mRNA. In some cases, however, exons from different pre-mRNA molecules are ligated together through a trans-splicing reaction, which is also catalyzed by the spliceosome. Trans-splicing is a relatively common phenomenon in organisms such as trypanosomes and nematodes, where it is often associated with the expression of polycistronic genes.3,4 Trans-splicing has also been reported in more complex metazoan, such as insects and mammals.5,6 However, the less abundant trans-spliced mRNAs reported in these complex organisms were suspected to be technical artifacts due to the fact that most reverse transcriptases can switch template and produce chimeric cDNAs.7 A genome-wide analysis of trans-splicing in Drosophila interspecies hybrids8 showed that many of the reported cases of trans-splicing were indeed artifactual but, more importantly, the same study identified 80 genes that undergo bona fide trans-splicing, including the previously identified genes mod(mdg4) and lola. Recent reports on the use of trans-splicing for therapeutic purposes in human cells and in mice demonstrate that also mammalian cells can catalyze trans-splicing reactions.9
Alternative splicing, which can involve both cis-splicing and trans-splicing phenomena, is regulated by mechanisms that either promote or inhibit the recognition of splice sites. Pre-mRNAs that undergo regulated alternative splicing usually contain sequences that act as either splicing enhancers or splicing silencers. These sequences recruit different splicing regulators, such as SR proteins and hnRNP proteins, which bind to the pre-mRNA and favor or inhibit the assembly of the spliceosome at selected splice sites.10,11,12 Moreover, the fact that many splicing reactions occur while the pre-mRNA is still tethered to the transcription machinery sets the basis for regulatory mechanisms that link splicing to transcription and to the local features of the chromatin.13,14 Indeed, studies in recent years have revealed that the structure of the chromatin not only regulates the activity of gene promoters but also influences the choice of alternative splice sites,15,16 and that some chromatin remodelers act as regulators of alternative splicing.17
The regulation of alternative splicing by the SWI/SNF chromatin-remodeling complex has been studied in both insect and mammalian cells.18,19,20 SWI/SNF is an ATP-dependent, multi-subunit complex that regulates the transcription of many genes by remodeling nucleosomes at promoter regions.21,22,23 The Brahma (BRM) protein is the only ATPase subunit of the SWI/SNF complex in D. melanogaster. The human ortholog of BRM, hBRM, favors the inclusion of alternative exons in the CD44 gene through a mechanism that reduces the rate of transcription elongation and involves the C-terminal domain (CTD) of RNA polymerase II (Pol-II).18 The BRM protein of D. melanogaster also affects pre-mRNA processing, and depletion of BRM changes the relative abundances of a subset of alternatively processed transcripts in D. melanogaster.19,20 A fraction of BRM is associated with nascent pre-mRNPs, which suggests that SWI/SNF affects pre-mRNA processing by acting at the RNA level.19
One of the genes whose expression is modulated by the levels of BRM in D. melanogaster is mod(mdg4).19,20 The mod(mdg4) gene codes for insulator proteins that participate in the establishment and maintenance of chromatin domains.6 The mod(mdg4) gene produces 31 splicing isoforms that share a common N-terminal sequence, but have different C-terminal regions.24 The C-terminal regions code for zinc fingers of the FLYWCH-type that determine the DNA-binding specificities of the different protein isoforms. The MOD(MDG4) isoforms originate by alternative splicing of pre-mRNAs encoded in both strands of the mod(mdg4) gene. One promoter drives the transcription of the major (sense) mod(mdg4) pre-mRNA that contains the common exons 1–4. Additional promoters drive the synthesis of several sense and anti-sense transcripts that code for alternative exons. Some MOD(MDG4) isoforms are translated from transcripts that contain exons encoded exclusively in the sense strand, and both cis-splicing and trans-splicing reactions contribute to the generation of such isoforms. Other mod(mdg4) transcripts contain exons of the anti-sense pre-mRNAs spliced together with the common exons from the sense strand, which can only occur by trans-splicing.8
The mod(mdg4) locus constitutes an interesting case of splicing regulation due to its complex organization. We have previously shown that the steady-state levels of some mod(mdg4) mRNAs are affected by the levels of BRM.20 Here we have further investigated this complex genetic locus and characterized the expression of one of the mod(mdg4) anti-sense regions within the locus with the aim of determining whether BRM is involved in the regulation of the mod(mdg4) trans-splicing. We have mapped the transcription start sites and polyadenylation sites, identified and quantified alternatively spliced transcripts, and obtained insight into the regulation of the mod(mdg4) trans-splicing. Using RNA interference and overexpression of recombinant BRM proteins, we show that BRM regulates a specific trans-splicing event at the mod(mdg4) locus through a mechanism that affects the phosphorylation state of the Pol-II. We also show that this process is independent of the ATPase activity of BRM.
Results
Trans-splicing at the mod(mdg4) locus
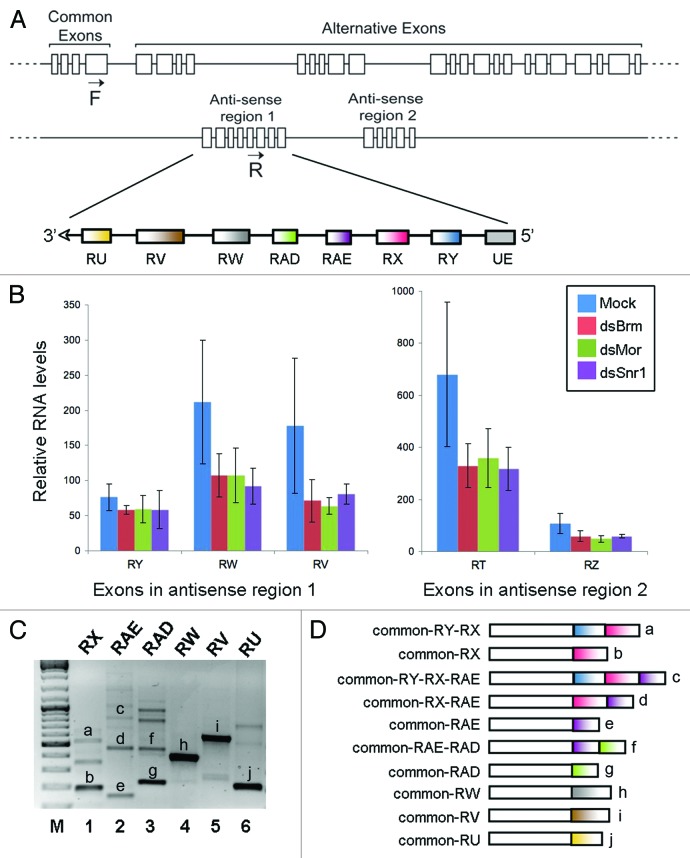
Figure 1A shows the structure of the mod(mdg4) locus. A major mod(mdg4) transcript that we refer to as the “sense transcript” includes four common exons followed by multiple alternative exons. Several additional promoters have been predicted to transcribe alternative exons from both the sense and anti-sense strands. The gene models available at FlyBase and modEncode show that the anti-sense strand codes for several anti-sense transcripts that are derived from two regions within the locus. These two regions will here be referred to as “anti-sense regions 1 and 2.” We have characterized the transcripts derived from anti-sense region 1. The sequence of the anti-sense region 1 and the positions of the anti-sense exons are provided in Figure S1.
Figure 1. RT-PCR analysis of trans-splicing involving exons from the mod(mdg4) anti-sense region 1. (A) Scheme showing the organization of the mod(mdg4) locus of D. melanogaster based on data from FlyBase and our experimental data in S2 cells. The exons included in the anti-sense region 1 are indicated. (B) The effect of depleting individual SWI/SNF subunits on the abundance of mod(mdg4) anti-sense exons measured by microarray hybridization (data from Moshkin et al.32). (C) RT-PCR analysis of trans-spliced transcripts that include exons from the anti-sense region 1. A forward primer complementary to the common exon 4 (F in Fig. 1A) was combined with different reverse primers complementary to different anti-sense exons, as indicated above each lane, and used to amplify specific transcripts using cDNA prepared from S2 cells. The RT-PCR products labeled with letters in the figure were purified and sequenced, and their exon composition is shown in (D). As expected, all the products contain the common exon plus at least one anti-sense exon.
Transcriptional profiling of Drosophila S2 cells in which individual subunits of the SWI/SNF complex were silenced by RNA interference show that the abundances of some mod(mdg4) anti-sense exons are reduced by approximately 50% in cells treated with dsRNA complementary to either BRM, Moira (MOR), or Snf5-related 1 (SNR1). As shown in Figure 1B, the levels of RW, RV, RT, and RZ exons were significantly reduced. Other sequences, for instance RY, were not affected. These results, obtained by microarray hybridization, measured the relative abundance of each individual exon but did not provide direct information about the abundance of the alternatively spliced mRNAs. We performed RT-PCR experiments to detect trans-spliced mod(mdg4) transcripts in S2 cells and to analyze their exon composition. In our experimental setup, in the absence of allelic polymorphisms, we cannot distinguish whether transcripts that include exons encoded in one same strand are generated by cis-splicing or trans-splicing. However, using appropriate PCR primers, we can unambiguously detect trans-spliced products that contain the common exons 1–4 from the sense strand spliced together with selected anti-sense exons, and therefore, we have focused on the study of such trans-spliced mRNAs. Six RT-PCR reactions were performed in parallel using the same forward primer complementary to a common exon in the sense transcript (primer F in Fig. 1A), and six different reverse primers complementary to alternative anti-sense exons. All the primer combinations were able to amplify one or more transcripts (Fig. 1C), which confirmed the existence of multiple trans-spliced mod(mdg4) transcripts. The major RT-PCR products were purified and sequenced to determine the exon composition of the trans-spliced transcripts (Fig. 1D). The sequenced products included trans-spliced exons, and some of them contained also multiple anti-sense exons spliced together.
In summary, these experiments confirmed that trans-spliced mod(mdg4) transcripts are present in S2 cells, and revealed that exons from the sense and anti-sense strands combine in highly complex ways.
Mod(mdg4) trans-splicing in vivo
We then performed a series of experiments to study the trans-splicing of mod(mgd4) transcripts in vivo. We isolated total RNA from flies and performed RT-PCR experiments using a forward primer complementary to one of the common exons, and a collection of reverse primers designed to amplify trans-spliced RNAs. We found evidence that all the exons of the anti-sense region 1 are part of trans-spliced transcripts in vivo, except for RAE (Fig. 2A). The amplification of trans-spliced transcripts from fly RNA could be reproduced with two different reverse transcriptases, Superscript III and M-MLV, which supports the conclusion that the detected transcripts exist in vivo and are not artifacts generated during the reverse transcription (Fig. S2A). This conclusion was also supported by the detection of trans-spliced RNAs by northern blotting. We designed and end-labeled three different probes for the analysis of trans-spliced RNAs. Probe 1 was 50 nt long and was complementary to the exon–exon junction generated by trans-splicing between the common exon 4 and the RX anti-sense exon. Probes 2 and 3 were shorter control probes that hybridized either to the constitutive exon or to the RX exon, respectively. A band of approximately 2 kb was observed, and this length fits well with the expected length of a trans-spliced mRNA that contains the four common mod(mdg4) exons spliced together with the RX anti-sense exon. Probe 1 gave a much stronger signal that Probe 2 or 3 under the same hybridization and exposure conditions (Fig. 2B). In summary, these results demonstrate that trans-splicing at the mod(mdg4) locus occurs not only in S2 cells but also in flies.
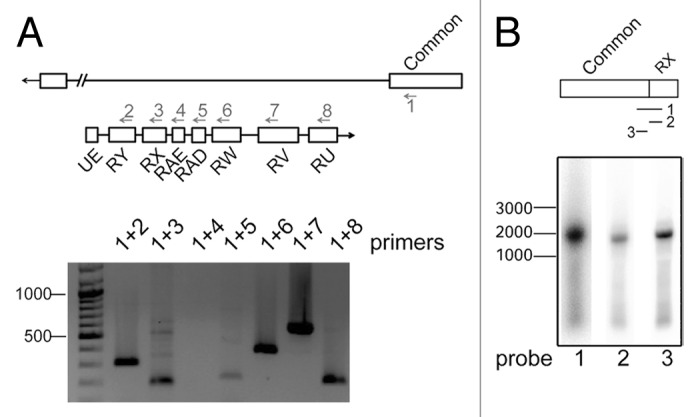
Figure 2. In vivo analysis of the mod(mdg4) trans-splicing. (A) The presence of trans-spliced mod(mdg4) transcripts in D. melanogaster flies was analyzed by RT-PCR, as in Figure 1, using a forward primer complementary to common exon 4 (primer 1) and different reverse primers in the anti-sense strand (primers 2 to 8). The different primer combinations were used to amplify trans-spliced products from D. melanogaster fly cDNA. The length of molecular mass standards, in bp, is shown to the left. (B) Northern blot with Drosophila fly mRNA. Probe 1 was complementary to the trans-spliced RX exon-exon junction. Probe 2 and probe 3 were complementary to RX and common exon 4, respectively. The probes were terminally labeled. The length of molecular mass standards, in bp, is shown to the left.
Characterization of transcripts from the anti-sense region 1
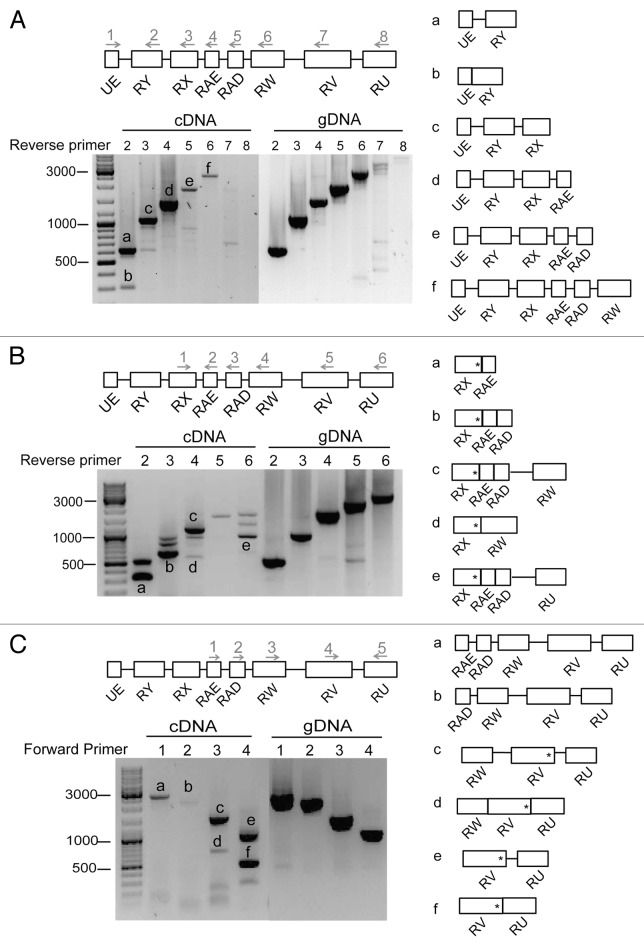
We performed rapid amplification of cDNA ends (RACE) reactions to map the transcription start sites (TSSs) and the cleavage and polyadenylation sites (CSs) of the mod(mdg4) anti-sense transcript in S2 cells. Two gene-specific primers complementary to either the RY or the RX exon were used for the 5′RACE. Table S1 provides a detailed description of the PCR primers that were used to amplify the RACE products.
Sequencing the PCR products obtained in the 5′RACE reactions revealed three TSSs in the anti-sense strand, two of them in close proximity to each other at the beginning of the UE exon, and the third one upstream of the RX exon (Fig. 3A; Fig. S1). The third TSS (tss3 in Fig. S1) was detected in two independent 5′RACE experiments, but the existence of a promoter in this position is not supported by the gene models available at FlyBase and modEncode and could not be confirmed by immunoprecipitation of capped mRNAs (data not shown). We cannot rule out the possibility that the RNAs starting at tss3 are downstream cleavage products of upstream transcripts cleaved at CS1–2, and therefore, this TSS is not included as a bona fide TSS in Figure 3.

Figure 3. Transcription start sites and 3′ end processing sites in the anti-sense region 1 of mod(mdg4). (A) 5′ and 3′ RACE reactions were used to map transcription start sites (TSS) and cleavage sites (CS) in total RNA purified from S2 cells. The figure compiles the TSSs and CSs identified by the RACE experiments. (B) The figure shows the nucleotide sequences of the TSSs and CSs. TSSs, polyadenylation signals and CSs are highlighted in orange.
The RACE experiments did not reveal the TSS of the trans-spliced transcripts, possibly because the common exons were too long to be amplified efficiently.
Annotations in FlyBase and Encode indicate the existence of an additional TSS upstream of the TSS1 detected in our 5′RACE experiments. We were able to verify the usage of this additional TSS in S2 cells by PCR reactions in which we used a forward primer upstream of TSS1 (see, for example, Fig. 4A). This upstream TSS is denoted “TSS0” in Figure 3A.
Figure 4. The mod(mdg4) anti-sense transcripts. (A) RT-PCR of the anti-sense transcripts in S2 cells. The image shows an example of an agarose-gel electrophoresis of the RT-PCR products. The lengths of molecular mass standards are shown to the left, in bp. A forward primer located at UE (primer 1) was combined with different reverse primers downstream (primers 2 to 8), as indicated in the figure. The primer pairs were used to amplify different regions of the transcript in cDNA prepared from S2 cells. Genomic DNA (gDNA) was used in parallel to monitor the efficiency of the PCR reactions. The major PCR products (bands a–f) were purified and sequenced. The right part of the figure shows the exon-intron composition of the sequenced bands. (B) RT-PCR analysis as in (A). In this case, the forward primer at RX (primer1) was combined with different reverse primers downstream (primers 2 to 6). In spite of their different length and exon composition, all these transcripts have a translation termination codon within the RX exon (asterisk). (C) RT-PCR analysis as in (A). Different forward primers (primers 1 to 4) and one reverse primer (primer 5) were used to amplify anti-sense transcripts. Translation termination codons are shown as asterisks.
The sequences of the products obtained in the 3′RACE reactions revealed the existence of at least 12 alternative cleavage sites. Each exon in the anti-sense transcript was followed by at least one CS. Some CSs, such as CS1 and CS2, shared a common polyadenylation signal (Fig. 3B).
Transcripts from the anti-sense region 1
We studied the splicing of the transcripts expressed from anti-sense region 1 in S2 cells in a series of RT-PCR experiments. In one of the experiments, we used a forward primer located close to the 5′ end of the anti-sense region 1, between TSS0 and TSS1, and different reverse primers downstream, as shown in Figure 4A. Genomic DNA was analyzed in parallel as a control for the efficacy of the amplification. Reverse transcription reactions without enzyme were also processed in parallel to assess for the presence of contaminating DNA (RT-controls, Fig. S2B and data not shown). The main PCR products were purified and sequenced, and the exon–intron composition of the sequenced bands is shown in the right part of Figure 4A. Table S2 provides the nucleotide sequences of all analyzed bands. Reverse primers complementary to sequences in exons RY, RX, and RAE, respectively, resulted in strong PCR products. Primers located in exons RAD and RW downstream gave poor signals, and primers in exons RV and RU failed to amplify any cDNA products, although the same primer combinations could efficiently amplify genomic DNA. These results suggest that most anti-sense transcripts initiated at TTS0–2 are cleaved at CS5, and only a few extend to CS8–9. Some primer combinations amplified both spliced and unspliced transcripts (see, for example, bands a and b in Fig. 4A).
In a second series of experiments, we used an upstream primer complementary to sequences in exon RX in combination with different reverse primers to give better coverage to the downstream sequences (Fig. 4B). The RX, RAE, and RAD exons were often spliced together, and we identified several alternatively spliced mRNAs that contained RX and additional downstream exons in different combinations (see Fig. 4B). However, these alternatively spliced transcripts do not encode alternative MOD(MDG4) protein isoforms, since a stop codon is present in exon RX (asterisks in Fig. 4B).
In a third series of experiments, we analyzed the RNAs derived from the downstream part of the anti-sense transcript 1 using forward primers located on different exons, combined with a reverse primer on exon RU (Fig. 4C). These experiments revealed the existence of transcripts containing the exons RV and RW, whereas RAE and RAD were less abundant. These results suggest that additional TSSs are located downstream of the RAD exon.
In summary, our results are consistent with the existence of a relatively abundant transcript that starts at TSS0–2 and is cleaved at CS5, and additional transcripts derived from the downstream parts of the region. Very few transcripts, if any, expand over the entire anti-sense region 1.
BRM depletion reduces RX trans-splicing in S2 cells and larvae
Previous studies have shown that depletion of BRM changes the relative abundances of some alternatively processed transcripts in Drosophila S2 cells, including some mod(mdg4) mRNAs.19,20 We designed experiments to knockdown BRM in S2 cells, and we analyzed the effects of the reduced BRM levels on the abundance of trans-spliced mod(mdg4) transcripts. We knocked down BRM by RNA interference (RNAi), and analyzed the abundances of selected mod(mdg4) transcripts by RT-qPCR 48 h after the dsRNA treatment. GFP dsRNA was used in parallel as a control, and the efficiency of the knockdown was assessed by western blotting (Fig. 5A). The relative abundances of trans-spliced mRNAs were quantified by RT-qPCR using exon–exon junction primers specific for the trans-spliced sequences. Five independent RNAi experiments revealed that BRM depletion reduced by 40% the abundance of the trans-spliced RX transcript (Figs. 5B and C). The levels of trans-spliced mRNAs containing each of the other anti-sense exons were analyzed in parallel with specific exon–exon junction primers, but none of them was significantly affected by BRM depletion (data not shown). Therefore, we concluded that the effect of BRM on trans-splicing was specific for the RX exon. The overall abundance of the individual anti-sense exons and the unspliced transcripts were not affected either (Fig. 5B), which argues against the possibility that the observed effects could be due to changes in the transcription of the TSS0–1 promoters.
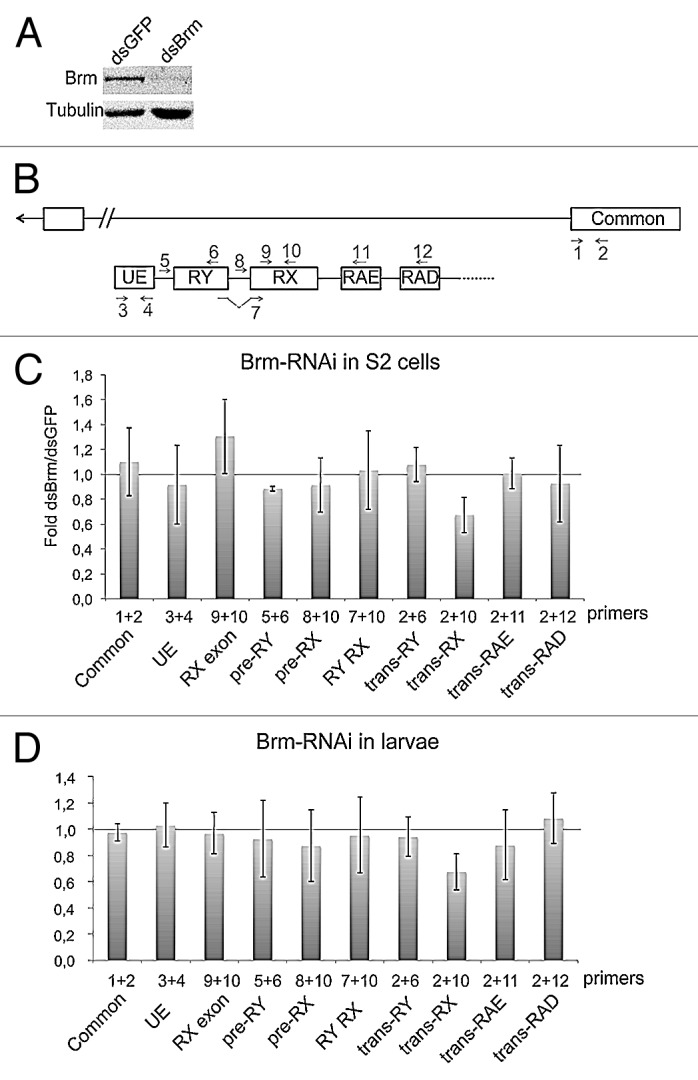
Figure 5. Depletion of Brm by RNAi reduces the abundance of trans-spliced RX transcripts. (A) RNAi was used to deplete BRM in S2 cells. Cells were treated in parallel with dsGFP as a control. The effect of BRM depletion was analyzed by western blotting after 48 h of dsRNA treatment. Tubulin was used as loading control. (B) Scheme showing the organization of the mod(mdg4) locus and the positions of the primer-pairs used in the PCR experiments that are presented in this figure. (C) The relative abundances of sense and anti-sense transcripts were quantified by RT-qPCR and calculated relative to Act5C. The ratios between dsBrm-treated cells and dsGFP-treated cells are presented in the histogram. The bars represent averages from five independent experiments. The error bars represent standard deviations. The trans-RX transcript was significantly reduced (P = 0.014, comparison of transcript abundances calculated relative to Act5C mRNA using a two-tailed, paired Student’s t test). The abundance of the other analyzed transcripts was not significantly changed (P > 0.05). (D) The effect of BRM depletion on the mod(mdg4) trans-splicing in larvae. hs-GAL4 virgin females were crossed with UAS-BrmRNAi males, and early third instar larvae from the cross were heat shocked for 2 h at 37 °C. Control experiments were performed in parallel by crossing hs-GAL4 virgin females with wild type males W1118. Total RNA was purified and the abundances of selected transcripts were analyzed by RT-qPCR, as in (C). The bars represent averages from three independent experiments. The trans-RX transcript was significantly reduced (P = 0.017, comparison of transcript abundances calculated relative to the common exon 4 using a two-tailed, Student’s t test).
We next analyzed the effect of BRM depletion in vivo by knocking down the expression of BRM using a systemic heat shock. Total RNA was purified from third instar larvae, and the transcript levels were quantified in the same way as the specimens shown in Figures 5B and C. The abundances of trans-spliced RNAs that contained the RX exon were specifically reduced, as in S2 cells. The levels of the common and UE exons were not changed, which suggests that the levels of BRM do not affect the activity of the mod(mdg4) promoters. These results suggest that BRM does not affect the transcription of the mod(mdg4) promoters, but influences the efficiency of the RX trans-splicing in both S2 cells and larvae.
The amount of anti-sense transcripts does not limit trans-splicing
To further strengthen the conclusion that the observed effect on trans-splicing was not due to transcriptional changes that affect the abundance of the anti-sense pre-mRNAs, we asked whether the abundance of the anti-sense transcript was a limiting factor for trans-splicing. We constructed an S2 cell line that expresses a mod(mdg4) anti-sense transcript under the control of an inducible promoter. We induced the expression of the anti-sense transcript and we analyzed the effects on the levels of mod(mdg4) RNAs by RT-qPCR. The relative abundance between induced cells and uninduced cells was calculated using PCR primers specific for different RNA sequences. The abundance of the anti-sense pre-mRNA transcript was highly increased, as expected (Fig. 6A), and the abundances of transcripts containing the RX and RY exons spliced together were also significantly increased (RY RX in Fig. 6B). However, there was no significant change in the abundance of any of the analyzed trans-spliced RNAs (Fig. 6B). We conclude that the abundance of the trans-spliced mRNAs is determined by factors other than the availability of the anti-sense pre-mRNAs.
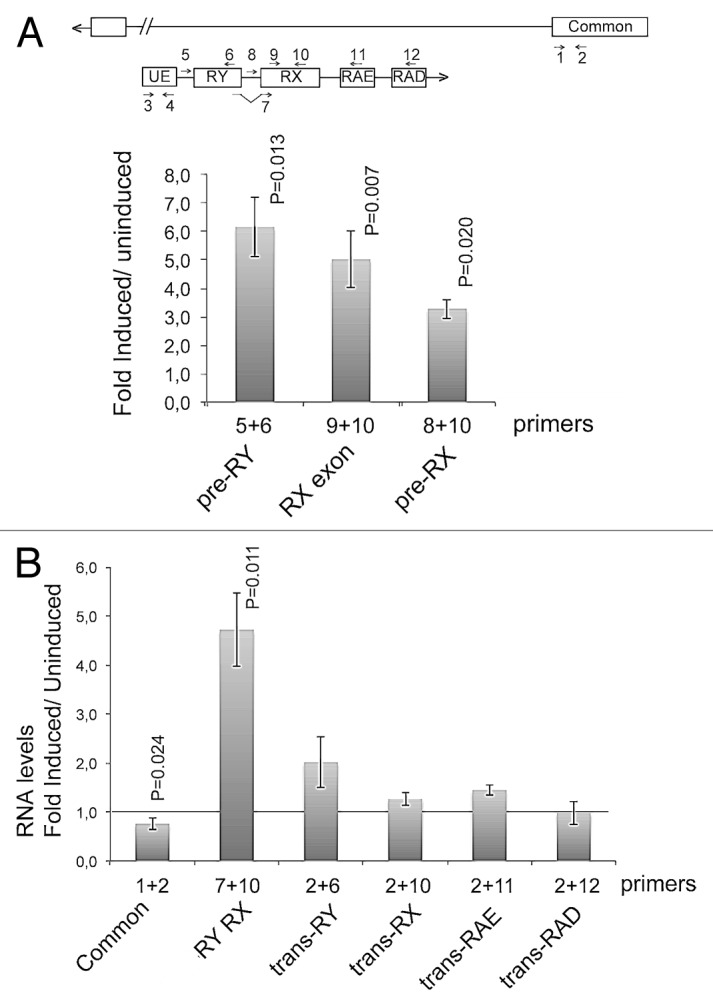
Figure 6. Overexpression of an anti-sense mod(mdg4) transcript. A genomic sequence expanding from TSS1 to CS6 was cloned into an expression vector under the control of an inducible promoter and transfected into S2 cells. The effect of the overexpression of the anti-sense RNA on the levels of several mod(mdg4) RNAs was measured by RT-qPCR. (A) The expression of the anti-sense transcript was induced by treating the stably transfected S2 cells with 200 µM CuSO4. The abundance of the anti-sense transcript was measured by RT-qPCR using different primer combinations to quantify different parts of the transcript, as indicated in the figure. The transcript abundances were calculated relative to Act5C mRNA abundance in the same samples. Uninduced cells were analyzed in parallel and used as a reference. The histogram shows the changes in transcript abundance expressed as fold change relative to the levels in uninduced cells. The bars represent averages and the error bars represent standard deviations from three independent experiments. The statistical significance of the changes observed between induced and uninduced cells was assessed using a two-tailed, paired Student’s t test. Significant P values (< 0.05) are given in the figure. (B) The effect of the overexpression on the abundances of cis-spliced and trans-spliced transcripts was quantified as in (A). The abundance of the common exons was quantified in parallel and was slightly reduced. The abundance of an anti-sense trancript containing the RY and RX exons (RY RX) was increased significantly. The abundance of the trans-spliced transcripts was not significantly changed (P > 0.05).
BRM regulates the RX trans-splicing through a mechanism that does not require the ATPase activity of BRM
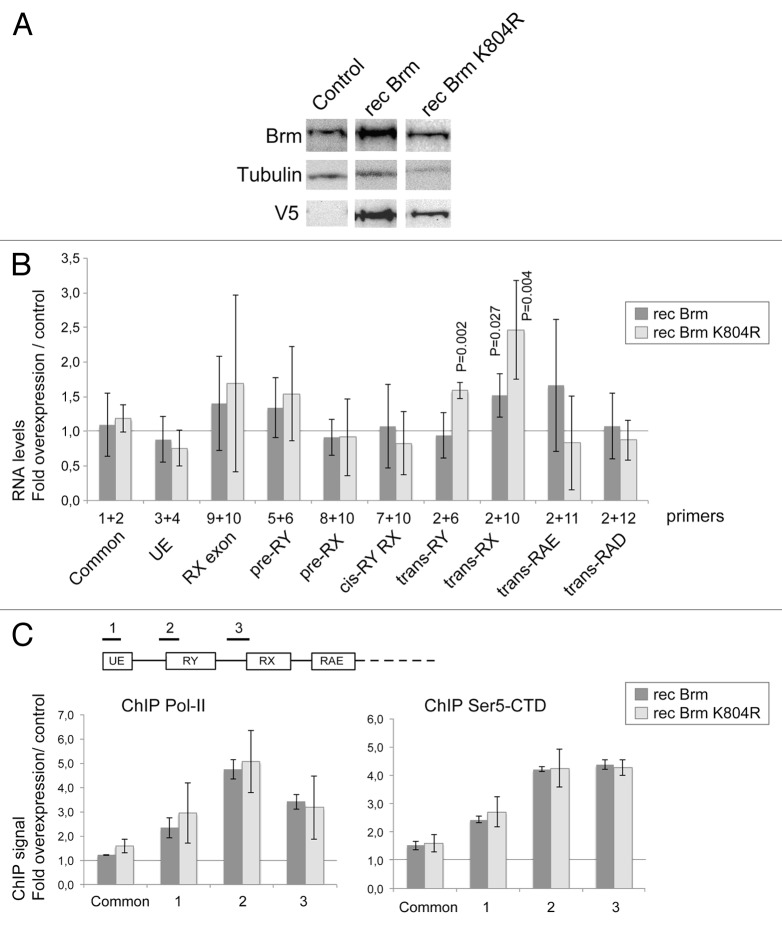
In the next series of experiments, we asked whether overexpression of BRM facilitates the RX trans-splicing. A stable cell line was constructed to overexpress a V5-tagged BRM protein (recBRM) under the control of an inducible promoter. The levels of BRM were monitored by western blotting (Fig. 7A) and the effects of the overexpression on the mod(mdg4) splicing were measured by RT-qPCR, as in Figure 5. The levels of the UE and common exon were not changed by the overexpression, which suggests that the overexpression of BRM does not change the transcriptional activity of the mod(mdg4) promoters. However, the steady-state level of the trans-spliced RX transcript, measured with specific exon–exon junction primers, increased significantly. This effect was specific, as shown by the fact that the levels of other trans-spliced RNAs were not significantly changed (Fig. 7B), and was in accordance with the RNAi results reported above.
Figure 7. The overexpression of BRM promotes RX trans-splicing. S2 cells were stably transfected with plasmids expressing either wild-type BRM (recBRM) or the ATPase mutant recBRM-K804R. (A) The expression of the recombinant BRM proteins was induced with 200 µM CuSO4 for 24 h and assessed by western blotting using anti-BRM and anti-V5 antibodies. Control S2 cells were analyzed in parallel. Tubulin was used as a loading control. (B) The relative abundances of selected mod(mdg4) transcripts were quantified by RT-qPCR in cells that overexpressed BRM and in control cells. The PCR primers were the same as in Figures 5 and 6. In all cases, the RNA levels were calculated relative to Act5C. The ratios between cells that overexpressed recBRM or recBRM-K804R and control cells are presented in the histogram. The bars represent averages from four independent experiments. The error bars represent standard deviations. Significant P values (< 0.05, two-tailed, paired Student’s t test) are given in the figure. The other changes are not significant. (C) ChIP analysis of the effect of BRM overexpression on the density of Pol-II. The expression of recBRM or recBRM-K804R was induced as above, and chromatin was prepared 24 h after induction. ChIP experiments using an antibody against the CTD of Pol-II were used to quantify the density of Pol-II (left panel). The levels of Pol-II-CTD phosphorylated at Ser5 (Ser5-CTD) were quantified in ChIP experiments using a Ser-5P specific antibody (right panel). Three different genomic regions within the anti-sense region 1 were analyzed, as indicated in the figure. The density of Pol-II at the upstream sense promoter (Common) was also analyzed. The ChIP signals were quantified by qPCR using a standard curve for each primer pair. The values obtained in negative control reactions without primary antibody were substracted, and the resulting signals were referred to input levels and expressed as fold change in cells overexpressing BRM relative to control cells. The histograms show average folds and the error bars represent standard deviations calculated from three independent experiments.
BRM is an ATP-dependent helicase, and we investigated whether the ATPase activity of BRM is required for the effect of this protein on the mod(mdg4) RX trans-splicing. We constructed S2 cells that express a mutated form of BRM, recBRM-K804R, which lacks ATPase activity and has a dominant-negative effect on transcription.25,26 Overexpression of recBRM-K804R increased the levels of the mod(mdg4) trans-RX transcript, thus reproducing the effect of the expression of the wild-type recBRM (Fig. 7B). Moreover, expression of recBRM-K804R also resulted in a modest but significant increase in the levels of the trans-RY transcript. These results lead us to conclude that BRM affects the trans-splicing of some mod(mdg4) transcripts through a mechanism that does not require the ATPase activity of BRM.
We argued that if BRM regulates the splicing of the mod(mdg4) transcripts directly, it should be associated with the mod(mdg4) locus. Chromatin-immunoprecipitation (ChIP) experiments showed that BRM is indeed associated with the mod(mdg4) anti-sense region 1 (Fig. S3). The ATPase mutant BRM-K804R was more stably associated with the locus, which agrees with reports concerning other chromatin-remodeling ATPases.27
BRM influences the phosphorylation state of the Pol-II CTD at the mod(mdg4) locus
Previous studies in human cells have suggested that BRM regulates alternative splicing through a mechanism that affects the phosphorylation of the Pol-II CTD and the transcription elongation rate.18 This proposal led us to ask whether the levels of BRM have any effect on the phosphorylation state of Pol-II CTD at the mod(mdg4) locus. We performed ChIP experiments to measure the density of Pol-II at different points along the mod(mdg4) locus, and we analyzed whether the levels of BRM affected Pol-II density. The results of three independent overexpression experiments show that expression of recBRM causes an increase in the density of Pol-II along the anti-sense region 1 (Fig. 7C). The levels of Pol-II were increased over the TSS1–2 region and the intron between the UE and RY exons increased most strongly. Interestingly, the same variation in Pol-II density occurred in cells expressing the recBRM-K804R mutant, which further supports the conclusion that the effect of BRM on the regulation of mod(mdg4) is independent of its ATPase activity.
An increase in Pol-II density may be the consequence of more active transcription. Alternatively, an increase in Pol-II density may reflect pausing of Pol-II, which would not necessarily result in increased expression levels. Increased BRM levels did not cause a general increase in the steady-state levels of the mod(mdg4) anti-sense transcripts (Fig. 7B), which argues against an activation of the anti-sense transcription. Moreover, ChIP experiments aimed at measuring the density of CTD phosphorylated at Ser5 (Ser5-CTD) showed an increase in Ser5-CTD that paralleled the increase observed for total Pol-II (Fig. 7C). High Ser 5 phosphorylation levels are a property of paused polymerases (for a review, see ref. 28), which suggests that the Pol-II that accumulates in the mod(mdg4) anti-sense region 1 during overexpression of recBRM is not engaged in efficient elongation (see Discussion). Interestingly, the increase in Ser5-CTD relative to the overall increase in CTD density was more pronounced in the region involved in the RX trans-splicing (compare ChIP Pol-II and ChIP ser5-CTD in region 3 of Fig. 7C).
Discussion
We have studied the transcripts derived from one of the anti-sense regions within the mod(mdg4) locus in order to shed light on the mechanisms that control the expression of this complex locus. We have identified BRM as one of the regulators of mod(mdg4) trans-splicing. We have shown that BRM is associated with the mod(mdg4) locus, and that the levels of BRM regulate a specific trans-splicing reaction through a mechanism that does not require its ATPase activity.
The expression of the mod(mdg4) anti-sense transcripts
The mod(mdg4) locus encodes multiple MOD(MDG4) protein isoforms that share a BTB/POZ protein–protein interaction domain, but contain different zinc-finger domains of the FLYWCH type. The different MOD(MDG4) isoforms are expressed in a manner that depends on the tissue and the stage of development, as judged from data in FlyAtlas (www.flyatlas.org). The mod(mdg4) pre-mRNAs start at multiple promoters that transcribe sequences from both DNA strands within the locus. We have studied the pre-mRNAs derived from the mod(mdg4) anti-sense region 1, and we have demonstrated the existence of multiple promoters that drive the synthesis of the anti-sense mod(mdg4) pre-mRNAs. These anti-sense pre-mRNAs show a remarkable diversity in their patterns of alternative splicing. Among the many transcripts detected, we have found trans-spliced mRNAs that contain anti-sense exons spliced together with the common mod(mdg4) exons. We have also found spliced transcripts that only contain anti-sense exons. However, these anti-sense transcripts lack the BTB/POZ domain and do not seem to contain any open reading frame, which suggests that the function of the anti-sense pre-mRNAs is to serve as a source of exons for trans-splicing reactions.
The regulation of mod(mdg4) splicing in its physiological context
The MOD(MDG4) proteins are components of the gypsy insulator complex, and as such, participate in the developmental regulation of genetic programs through the establishment and/or maintenance of chromatin domains.29,30 Little is known about the specific functions of the different MOD(MDG4) isoforms in vivo, but it seems reasonable to propose that the expression of different subsets of MOD(MDG4) insulators with slightly different DNA-binding specificities will result in different patterns of chromatin organization. Our results show that BRM is involved in the regulation of the mod(mdg4) isoforms (Waldholm et al.20 and our present results). This involvement allows BRM to modify the organization of chromatin domains, which in turn, can lead to changes in the expression of target genes. The role of BRM in the fine-tuning of mod(mdg4) expression is perfectly in line with BRM being a master regulator of tissue-specific gene expression, involved in regulating organ development after hormonal stimulation.31
Mechanistic considerations on the function of BRM in splicing regulation
We have shown that BRM is one of the regulators of mod(mdg4) splicing and that the levels of BRM affect specifically the trans-splicing of the RX exon in S2 cells and in vivo. The magnitude of the changes observed in the RX trans-splicing upon BRM knockdown was not striking, in spite of the fact that the dsRNA treatment was efficient, as shown by western blot. The mild effect could be due to the presence of small amounts of active BRM in the cells, or to the fact that SWI/SNF participates in the fine-tuning of the alternative mod(mdg4) isoforms, which may nevertheless be of functional importance in vivo. Another factor that contributes to the limited extent of the observed changes is that our experiments underestimate the amounts of trans-spliced transcripts because we can only detect trans-spliced RNAs that contain exons encoded in opposite strands. In spite of its reduced magnitude, the reported effect of BRM on RX splicing is reproducible and significant both in S2 cells and in vivo, and multiple observations support the conclusion that this effect is direct. First, depletion of BRM does not lead to any significant change in the levels of expression of pre-mRNA processing factors.19,20,32 Second, BRM is associated with the mod(mdg4) locus, as shown by ChIP. Third, overexpression and depletion of BRM lead to opposite effects. And, most importantly, overexpression of a mutant form of BRM that cannot remodel nucleosomes reproduces the same effect, which rules out the possibility that BRM could remodel chromatin and, indirectly, affect the RX splicing. Based on these observations, we conclude that the effect of BRM on the mod(mdg4) trans-splicing is a direct, local effect of BRM on a specific splicing event.
What is the mechanism by which BRM affects the mod(mdg4) trans-splicing? Depletion of SWI/SNF core subunits other than BRM in S2 cells also leads to changes in the splicing of several pre-mRNAs,20 and the regulation of the Eig71Eh pre-mRNA splicing by BRM involves at least one other SWI/SNF subunit, SNR1.33 These observations suggest that a SWI/SNF chromatin-remodeling complex, not BRM alone, is responsible for the observed effects on trans-splicing. Although the idea of a chromatin-remodeling complex playing a role in splicing regulation is compatible with recent studies showing that the local properties of the chromatin can influence alternative splicing decisions,34,35 chromatin remodeling per se cannot explain the effect of BRM on the regulation of mod(mdg4) trans-splicing. We have shown that a mutant form of BRM that lacks ATPase activity and is unable to remodel chromatin can nevertheless reproduce the effects of BRM on the RX trans-splicing, which confirms that the functions of BRM and SWI/SNF in splicing and chromatin remodeling are independent of each other.
The effect of BRM on trans-splicing at the mod(mdg4) locus is accompanied by an increase in the density of Pol-II in the anti-sense region, and by an increased CTD-Ser5 phosphorylation, which agrees with recent results showing that the BRM protein of D. melanogaster stabilizes the initiating form of Pol-II on chromatin.33 The changes in CTD-Ser5 phosphorylation that we have observed at the mod(mdg4) locus are also similar to those observed at the human CD44 gene, where it has been proposed that BRM regulates alternative splicing by reducing the rate of elongation of Pol-II at regions that encode variable exons.18 It can be readily understood that changes in the elongation rate affect splice-site selection in the context of cis-splicing reactions, but it is more difficult to envision how changes in the Pol-II elongation rate can affect trans-splicing reactions that involve two polymerases that transcribe different strands. The levels of BRM do, indeed, have a clear effect on the CTD density and on Ser5 phosphorylation. It is unclear, however, whether these changes in the transcription machinery affect the elongation rate of Pol-II in the anti-sense region 1, and whether the Pol-II elongation rate has any effect on trans-splicing. Zraly and Dingwall33 showed that the regulation of the Eig71Eh pre-mRNA splicing by BRM is associated with an increased CTD-Ser5 phosphorylation, but that reducing the elongation rate of the Pol-II does not have any effect on the splicing outcome. In any case, splicing regulation by BRM seems to be accompanied by changes in the CTD phosphorylation and in the Pol-II density, in both human and insect cells, and our results show that these changes do not require the ATPase activity of BRM and are therefore independent of the SWI/SNF chromatin-remodeling activity. Based on the reported association of BRM with nascent pre-mRNPs,19 we favor the hypothesis that BRM regulates pre-mRNA processing through mechanisms that involve interactions with the pre-mRNA, and the identification of specific RNA-binding proteins involved in such a mechanisms will be of outmost importance. Most of the bona fide targets of BRM are developmentally regulated genes that are poorly expressed in S2 cells (see, for example, ref. 31) and the deeper understanding of the mechanisms by which BRM regulates splicing will require splicing studies in specific tissues and at specific developmental stages.
Materials and Methods
Cell culture and fly strains
Drosophila melanogaster S2 cells were cultured in Schneider’s Drosophila medium (Invitrogen) containing 10% heat-inactivated FBS, 50 U/ml penicillin and 50 μg/ml streptomycin at 28 °C. Drosophila melanogaster wild-type flies (W1118) from Bloomington stock center were kept at 25 °C under standard conditions.
RNA isolation, reverse transcription, and qPCR
Total RNA was isolated from S2 cells using the RNAqueous kit (Ambion). Total RNA from flies was isolated using TRIzol as described by Simms et al.36 The total RNA was treated by Turbo DNA-free kit (Applied Biosystems) and reverse transcribed by Superscript III (Invitrogen) using random primers. RT(-) controls were performed to rule out DNA contamination (Fig. S2). The resulting cDNA was amplified with KAPA SYBR Fast qPCR Master Mix (KAPA Biosystems) in a QIAGEN Rotor-Gene Q using primers specific for each transcript. The relative abundance of each transcript was normalized to the abundance of actin 5C mRNA in the same sample. The qPCR assays followed the MIIQE guidelines for primer design.37 All primer pairs fulfilled quality criteria according to amplification and melting curves. Standard curves for each primer pair were used for quantitative purposes in order to obtain accurate measurements. The results presented are based on compiled data from at least three independent biological replicates, each analyzed in duplicate, as indicated in the figures. The primer sequences are provided in Table S3.
Northern blot
Drosophila fly mRNA was purified from total RNA using Micro Poly(A)Purist Kit (Ambion). The total mRNA was separated by agarose gel electrophoresis and transferred to Zeta-Probe membranes (Bio-Rad) by northern blotting. The membrane was subsequently UV-cross linked in a Stratalinker (Stratagene). The membrane was pre-hybridized with ULTRAhyb-Oligo buffer (Invitrogen) at 42 °C for 30 min, hybridized with DNA probes overnight and washed twice with a washing buffer that consisted of 2 × SSC, 0.5% SDS for 30 min at 42 °C. The hybridization was detected in a phosphorimager (FujiFilm Fluorescent Image Analyzer FLA-3000). The probes were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase.
3′RACE and 5′RACE analysis
Total RNA was isolated from cultured S2 cells by RNAqueous kit (Ambion) and treated with DNase to remove contaminating DNA. The purified RNA was used to synthesize cDNA using M-MLV reverse transcriptase and the 3'RACE adaptor of the FirstChoice RLM-RACE Kit (Ambion). The 3'RACE adaptor sequence can be found in Table S3. For 5'RACE, RNA was first treated with calf intestine alkaline phosphatase (CIP) to remove free 5' phosphates from degraded transcripts and subsequently digested with tobacco acid pyrophosphatase (TAP) to remove the cap structure of intact mRNAs. A 5′RACE adaptor was ligated to the decapped mRNA using T4 RNA ligase. The cDNA was synthesized by M-MLV reverse transcriptase and random decamers. 3′RACE and 5′RACE cDNAs were amplified by PCR. The resulting PCR products were purified and ligated into a plasmid using the TOPO TA Cloning Kit (Invitrogen). The recombinant plasmids were transformed into E. coli XL-10 Gold. Individual colonies were analyzed by PCR, and their inserts were amplified and sequenced. The sequences obtained were analyzed using CLC Sequence Viewer 6, and are provided in Table S4.
SDS-page and western blot
Total protein lysates from S2 cells were separated by SDS-PAGE and transferred to a polyvinylidenefluoride membrane (Millipore). The membrane was blocked, incubated with an anti-BRM antibody,19 and with a horseradish peroxidase-conjugated secondary antibody (DAKO) following standard procedures. The labeling was detected using the ECL system (GE Healthcare). An antibody against tubulin (clone B-5-1-2, Sigma Aldrich) was used as a loading reference.
RNA interference
Double-stranded RNAs targeting either Brm or GFP were synthesized in vitro using the MegaScript RNAi kit (Ambion) according to the manufacturer’s instructions. Primer sequences used for dsRNA synthesis can be found in Table S3. S2 cells were cultured in standard conditions and treated with 30 µg dsRNA as described by Hase et al.38 After 48 h, the cells were harvested and the RNA was purified as described above.
BRM depletion in D. melanogaster larvae was performed by crossing hs-GAL4 virgin females with UAS-BrmRNAi males (strains #37721 and #37720). Early third instar larvae from the cross were heat shocked for 2 h at 37 °C and analyzed 24 h after the heat shock, as described by Waldholm et al.20
Cloning and expression of mod(mdg4) anti-sense transcript 1 in S2 cells
A mod(mdg4) anti-sense genomic DNA was amplified by PCR using forward and reverse primers containing cloning sites for SpeI and EcoRV, respectively. The amplified DNA fragment contained the genomic region covering from the UE to the RW exon. The PCR product was cloned into the pMT/V5-HisB vector (Invitrogen). The recombinant plasmid was purified using the Nucleobond Xtra Midi Plus Kit (Macherey-Nagel) and transfected into Drosophila S2 cells using the calcium phosphate transfection kit (Invitrogen). Stably transfected cells were selected and maintained in the presence of 300 µM hygromycin B.
Cloning and expression of recombinant BRM in S2 cells
Recombinant Brm plasmid RE61274 (Drosophila Genomic Resource Center) was amplified by PCR using Pfu polymerase (Agilent) with forward and reverse primers containing cloning sites for Spe I and Sac II, respectively. The BRM-K804R mutant was generated from the RE61274 plasmid by PCR using primers designed by the QuikChange Primer Design Program (Agilent technologies). The mutated PCR product was cloned into the pMT-puromycin vector (Addgene) and the insert was sequenced to confirm the mutations. The recombinant plasmid encoding BRM-K804R was purified using the QIAprep Spin Miniprep Kit (QIAGEN) and transfected into Drosophila S2 cells using the calcium phosphate transfection kit (Invitrogen). Stably transfected cells were selected in 5 µg/ml puromycin and maintained in the presence of 2 µg/ml puromycin. The occurrence of the K804R mutation was again confirmed by sequencing cDNA produced from the stably transfected cells.
Chromatin immunoprecipitation
Drosophila S2 cells were fixed at room temperature with 2% formaldehyde, and the chromatin was extracted as described by Hessle et al.39 The chromatin was sheared to 250–900 bp by sonication. Immunoprecipitation was performed at 4 °C using the following antibodies: anti-CTD (Abcam, ab5408), anti-cBrm,19 and α-Ser5 (Bethyl Laboratories, A300-655A). Blocked protein A-Sepharose and protein G-Sepharose beads mixture was added to the sample and the incubation was continued for 1 h. The beads were washed with RIPA buffer, resuspended in TE buffer containing 0.5% SDS, 0.1 mg/ml RNase A, and 0.2 mg/ml protease K, and incubated at 55 °C for 3 h and at 65 °C overnight. The immunoprecipitated DNA was finally extracted with a mixture of phenol, chloroform, and isoamylalcohol (Sigma), precipitated with ethanol and analyzed by qPCR using KAPA SYBR Fast qPCR Master Mix (KAPA Biosystems) in a QIAGEN Rotor-Gene Q. The sequences of the primers used are provided in Table S3. Data from three independent experiments were compiled and analyzed using a paired Student’s t test.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank AK. Östlund Farrants for fruitful discussions and critical reading of the manuscript, U. Theopold, and Z. Wang for help with the in vivo RNAi experiments, and G. Farrants for English editing. Our research is financed by grants from The Swedish Research Council (VR-NT).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27866
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–40. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–40. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 5.Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt FG. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci U S A. 1998;95:12185–90. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorn R, Krauss V. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica. 2003;117:165–77. doi: 10.1023/A:1022983810016. [DOI] [PubMed] [Google Scholar]
- 7.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–31. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 8.McManus CJ, Duff MO, Eipper-Mains J, Graveley BR. Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci U S A. 2010;107:12975–9. doi: 10.1073/pnas.1007586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avale ME, Rodríguez-Martín T, Gallo JM. Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum Mol Genet. 2013;22:2603–11. doi: 10.1093/hmg/ddt108. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–8. doi: 10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 11.Lopez AJ. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 12.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–9. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 13.Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–71. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 14.Zorio DA, Bentley DL. The link between mRNA processing and transcription: communication works both ways. Exp Cell Res. 2004;296:91–7. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 15.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 16.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allemand E, Batsché E, Muchardt C. Splicing, transcription, and chromatin: a ménage à trois. Curr Opin Genet Dev. 2008;18:145–51. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–9. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi A, Ryme J, Brodin D, Östlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5:e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldholm J, Wang Z, Brodin D, Tyagi A, Yu S, Theopold U, Farrants AK, Visa N. SWI/SNF regulates the alternative processing of a specific subset of pre-mRNAs in Drosophila melanogaster. BMC Mol Biol. 2011;12:46. doi: 10.1186/1471-2199-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Dingwall AK, Beek SJ, McCallum CM, Tamkun JW, Kalpana GV, Goff SP, Scott MP. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–91. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouazoune K, Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006;14:433–49. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- 24.Krauss V, Dorn R. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene. 2004;331:165–76. doi: 10.1016/j.gene.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–4. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 26.Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR, Daubresse G, DePace A, et al. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–65. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–54. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–6. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Bortle K, Corces VG. The role of chromatin insulators in nuclear architecture and genome function. Curr Opin Genet Dev. 2013;23:212–8. doi: 10.1016/j.gde.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zraly CB, Middleton FA, Dingwall AK. Hormone-response genes are direct in vivo regulatory targets of Brahma (SWI/SNF) complex function. J Biol Chem. 2006;281:35305–15. doi: 10.1074/jbc.M607806200. [DOI] [PubMed] [Google Scholar]
- 32.Moshkin YM, Mohrmann L, van Ijcken WFJ, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–61. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zraly CB, Dingwall AK. The chromatin remodeling and mRNA splicing functions of the Brahma (SWI/SNF) complex are mediated by the SNR1/SNF5 regulatory subunit. Nucleic Acids Res. 2012;40:5975–87. doi: 10.1093/nar/gks288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–72. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Almeida SF, Carmo-Fonseca M. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends Biochem Sci. 2012;37:248–53. doi: 10.1016/j.tibs.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Simms D, Cizdziel PE, Chomczynski P. TRIZOL: a new reagent for optimal single-step isolation of RNA. Focus. 1993;15:99–102. [Google Scholar]
- 37.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 38.Hase ME, Yalamanchili P, Visa N. The Drosophila heterogeneous nuclear ribonucleoprotein M protein, HRP59, regulates alternative splicing and controls the production of its own mRNA. J Biol Chem. 2006;281:39135–41. doi: 10.1074/jbc.M604235200. [DOI] [PubMed] [Google Scholar]
- 39.Hessle V, Björk P, Sokolowski M, González de Valdivia E, Silverstein R, Artemenko K, Tyagi A, Maddalo G, Ilag L, Helbig R, et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol Biol Cell. 2009;20:3459–70. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.