Abstract
Heterogeneous nuclear ribonucleoprotein L (hnRNP L) is a multifunctional RNA-binding protein that is involved in many different processes, such as regulation of transcription, translation, and RNA stability. We have previously characterized hnRNP L as a global regulator of alternative splicing, binding to CA-repeat, and CA-rich RNA elements. Interestingly, hnRNP L can both activate and repress splicing of alternative exons, but the precise mechanism of hnRNP L-mediated splicing regulation remained unclear. To analyze activities of hnRNP L on a genome-wide level, we performed individual-nucleotide resolution crosslinking-immunoprecipitation in combination with deep-sequencing (iCLIP-Seq). Sequence analysis of the iCLIP crosslink sites showed significant enrichment of C/A motifs, which perfectly agrees with the in vitro binding consensus obtained earlier by a SELEX approach, indicating that in vivo hnRNP L binding targets are mainly determined by the RNA-binding activity of the protein. Genome-wide mapping of hnRNP L binding revealed that the protein preferably binds to introns and 3′ UTR. Additionally, position-dependent splicing regulation by hnRNP L was demonstrated: The protein represses splicing when bound to intronic regions upstream of alternative exons, and in contrast, activates splicing when bound to the downstream intron. These findings shed light on the longstanding question of differential hnRNP L-mediated splicing regulation. Finally, regarding 3′ UTR binding, hnRNP L binding preferentially overlaps with predicted microRNA target sites, indicating global competition between hnRNP L and microRNA binding. Translational regulation by hnRNP L was validated for a subset of predicted target 3′UTRs.
Keywords: hnRNP L, CLIP, splicing regulation, microRNA
Introduction
The heterogeneous nuclear ribonucleoprotein L (hnRNP L) is a multifunctional RNA-binding protein containing four RNA-recognition motifs (RRMs), which specifically recognizes CA-repeat and CA-rich RNA elements.1 It participates in diverse functions of mRNA metabolism, such as export of intronless mRNA,2,3translational regulation,4-7regulation of mRNA stability,1,4,8,9poly(A) site selection,10and alternative splicing.10Recent work by Jafarifar et al.11 demonstrated yet another role: competing with microRNAs (miRNAs) for binding to a CA-rich RNA element within the VEGFA 3′ UTR. A similar mechanism was also very recently described for DGK-α.12
HnRNP L is localized uniformly in the nucleoplasm and enriched in perinucleolar structures,13,14which were later identified as SLM/Sam68 Nuclear Bodies, where hnRNP L interacts with Sam68.15 Splicing targets of hnRNP L affect diverse processes, including tumorgenesis,16,17T-cell activation,18-21vasculogenesis,22 and signal transduction.23 HnRNP L also regulates its own expression by a negative feedback loop, in which it activates splicing of a poison exon that leads to nonsense-mediated mRNA decay (NMD) of the hnRNP L mRNA.24 HnRNP L can regulate alternative splicing either as an activator or a repressor, and it is still unclear what determines its activity in each specific case. As recently shown, alternative splicing regulation of hnRNP L can be based on interference with splice site recognition.25
To address how the multiple functions of hnRNP L correlate with RNA binding, we made use of a powerful new approach: individual nucleotide resolution crosslinking and immunoprecipitation (iCLIP) combined with RNA-Seq.26 This recent development of the basic CLIP method allows genome-wide mapping of all in vivo RNA-interaction sites at single-nucleotide resolution for any RNA-binding protein.27 CLIP approaches have been applied to several factors, including Nova,28 SRSF1,29 and—combined with next-generation sequencing—to Nova,30 RbFox2,31 PTB,32 HuR,33 SRSF3/4,34 and others (reviewed in refs. 35 and 36). By integrating the CLIP binding data with information on expression of splice isoforms, functional RNA maps can be derived that reveal position-dependent splicing effects.26,37
Based on iCLIP-Seq and genomic mapping, we obtained a comprehensive genome-wide map of hnRNP L RNA interaction sites in HeLa cells. Analysis of the crosslink sites revealed several novel regulatory roles of hnRNP L: First, hnRNP L RNA-binding follows distinct patterns for exons activated or repressed by hnRNP L, indicating a positional code and a predictive RNA map. Second, hnRNP L binding is enriched at both terminal as well as internal poly(A) sites. Third, hnRNP L binding preferentially overlaps with predicted miRNA target sites in the 3′ UTR, consistent with an hnRNP L-vs.-miRNA competition model. For a subset of these targets, we demonstrated 3′ UTR-depending translational regulation by hnRNP L. A very recent study has also investigated hnRNP L by CLIP-Seq in T-cells.38 Although focusing on hnRNP L-regulated splicing changes specific to T-cell biology, remarkably similar results were obtained on target distribution and specificity (see Discussion).
Results and Discussion
HnRNP L in vivo RNA-crosslink sites determined by iCLIP match in vitro consensus
To monitor genome-wide RNA binding sites of hnRNP L in HeLa cells, we performed three iCLIP experiments, combined with Solexa high-throughput sequencing (for a representative experiment, see Fig. 1). Sequence reads from the three experiments were combined and analyzed for hnRNP L crosslink sites. Genomic mapping and sequence analysis was performed,26 resulting in ~1.1 million sites mapped on chromosomes 1–22 and X (Fig. S1A; for details on data processing, see Supplementary Materials and Methods; for the high reproducibility between the three experiments, see Fig. S1B).
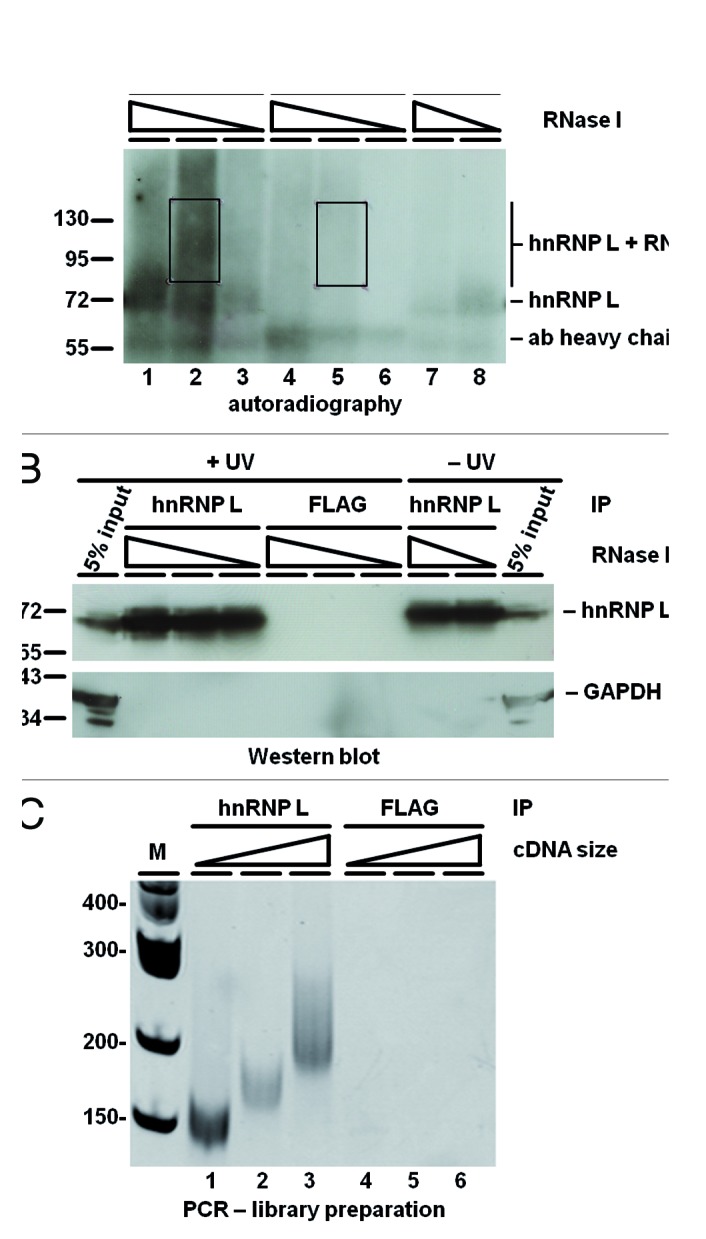
Figure 1. iCLIP-Seq analysis for transcriptome-wide mapping of hnRNP L RNA-binding sites. (A) A representative experiment is shown: Briefly, protein–RNA interactions were crosslinked by UV-irradiation of HeLa cells, followed by lysate preparation, limited RNase digestion, immunoprecipitation of hnRNP L-RNA adducts, 3′-RNA-linker addition to the RNA tags, 5′-terminal 32P-labeling, and gel separation of the covalent RNA–protein complexes under denaturing conditions. Autoradiography of iCLIP membrane with bound 32P-labeled RNA–protein complexes, comparing after UV-irradiation (+UV) hnRNP L- (lanes 1–3) with control FLAG immunoprecipitated material (IP, lanes 4–6). As additional control, UV irradiation was left out (lanes 7–8). In each case, different amounts of RNase I were applied (as indicated). Boxed regions were cut out and subjected to RNA isolation and library preparation. Marker positions and the mobilities of hnRNP L-RNA adducts, limit-digest hnRNP L, and of the antibody heavy chain are marked. (B) Western blot analysis of the same membrane as in panel (A), using anti-hnRNP L and GAPDH antibodies and including 5% input material. (C) Library preparation from iCLIP-processed hnRNP L- (IP; lanes 1–3) or control FLAG-immunoprecipitated material (FLAG-IP; lanes 4–6). PCR products for high-throughput sequencing were visualized by ethidium bromide staining on a 6% polyacrylamide-TBE gel. The three different size fractions resulted from prior size selection of the cDNA, following reverse transcription. PCR products derived from all three immunoprecipitations were pooled for sequencing.
Sequence motif analysis (in pentamers, as described in ref. 39) revealed significant enrichment of CA-repeat and CA-rich motifs (Fig. 2A): CA-repeats (ACACA, CACAC) were enriched by about 110–140% (100% corresponding to a 2-fold enrichment) and certain CA-rich motifs by 80–95% (ACAT, TACA). This specific enrichment pattern perfectly agrees with hnRNP L-binding properties derived earlier by an in vitro SELEX study.1 In contrast, the control experiment yielded no significant enrichment of a specific pentamer. Shankarling et al. (2014) obtained a similar binding consensus in their CLIP-Seq analysis.38
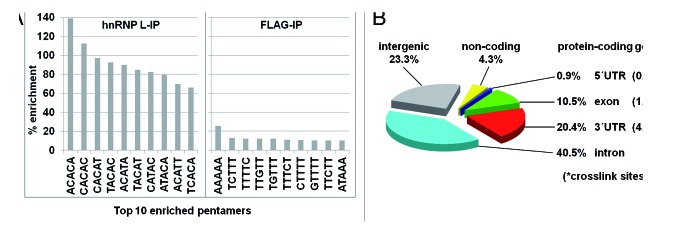
Figure 2. Genome-wide hnRNP L RNA-binding analysis by iCLIP. (A) Sequence motif analysis. HnRNP L crosslink sites were analyzed for motif enrichment (5-mers within a -30 to -10 and +10 to +30 window compared with 100 randomized positions from the same regions of the gene. The top 10 pentamers are listed for the hnRNP L- vs. the control FLAG immunoprecipitation (IP). All P values were < 0.01. (B) Distribution of hnRNP L crosslink sites, including their respective crosslink densities.
Activator vs. repressor function of hnRNP L in alternative splicing depends on its binding position
The ~1.1 million sites were clustered and filtered, and a total of 622 789 crosslink sites were selected for functional analysis (see Supplementary Materials and Methods). Figure 2B presents the distribution of the crosslink sites: A large fraction (72.3%) of hnRNP L binding sites maps within protein-coding genes, particularly in their introns (40.5%) compared with the mRNA (31.8%). The relatively high number of crosslink sites in introns is striking, considering the transient nature of intronic sequences, suggesting a global regulatory role of hnRNP L through intron binding. Within mRNA transcripts, 3′ UTRs exhibit a much higher density of binding sites than exons (4.46 vs. 1.47 crosslink sites per kb), consistent with known functions of hnRNP L in the 3′ UTR (see Introduction). This binding distribution of hnRNP L on the genome-wide level as well as on pre-mRNA regions is similar to that observed in both primary and immortalized T-cells, no matter if activated or repressed.38
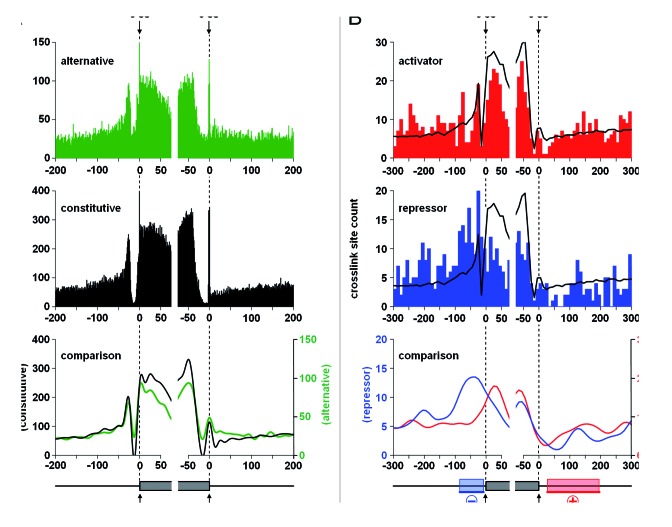
To investigate the RNA-binding patterns of hnRNP L near splice sites, we used all ENCODE-annotated exons of protein-coding genes and grouped their splice sites into two classes: alternative and constitutive. The frequency of crosslink sites around these two groups of splice sites is plotted in Figure 3A. The apparent overrepresentation of hnRNP L binding sites in constitutive exons most likely simply reflects their higher abundance. The discrete peaks at the 5′ splice site and at the -25 to -40 region relative to the 3′ splice site are probably not based on hnRNP L crosslink sites, but may reflect primer-extension stops at the branched lariat after RNase treatment, with hnRNP L bound to the respective intron. More interestingly, we detected a higher density of hnRNP L binding sites around alternative 5′ splice sites, compared with constitutive 5′ splice sites, both in the exonic (positions -30 to 0) and intronic regions (0 to +70); in addition, in the alternative 3′ splice site regions there are strikingly more hnRNP L binding sites around position -20. This pattern suggests that hnRNP L regulates splice site usage predominantly through binding around the 5′ splice site region, involving either exonic or intronic elements, and through binding within the pyrimidine tract region.
Figure 3. HnRNP L activates or represses alternative splicing in a position-dependent manner. (A) Number of hnRNP L crosslink sites around 3′ (-200 to +75) and 5′ splice sites (-75 to +200) of protein-coding genes is diagrammed, separately for alternative and constitutive splice sites (upper and middle panels, respectively). Below, alternative (green) and constitutive (black) splice sites are compared, plotted in smooth lines (lower panel). (B) Number of hnRNP L crosslink sites around 3′ (-300 to +75) and 5′ splice sites (-75 to +300) of hnRNP L-regulated exons, separately for activated (upper panel) and repressed exons (middle panel). Non-target exons were used as background (black smooth lines). In the lower panel, hnRNP L-activated (red) and -repressed exons (blue) exons are compared, plotted in smooth lines. Below, the positional information of hnRNP L binding and its correlation with activator/repressor function is schematically summarized.
Next we correlated RNA-binding and alternative splicing targets: HnRNP L was downregulated in HeLa cells by RNAi, followed by an additional cycloheximide treatment to suppress nonsense-mediated RNA decay (NMD) and to detect premature termination codon-containing splice variants. Splice-sensitive exonarray and data analysis predicted 890 hnRNP L-activated and 574-repressed target cassette exons (see Supplementary Materials and Methods).
Based on these two data sets, we analyzed the density of hnRNP L crosslink sites, comparing activated vs. repressed exons (Fig. 3B). This revealed clear differences in hnRNP L binding around the splice sites: The repressor activity of hnRNP L concentrates immediately upstream of the 3′ splice site (peaking around position -40), whereas hnRNP L preferentially activates when bound at intronic positions further downstream of the 5′ splice site (positions +25 to +200). The apparent higher density in activated vs. repressed exons likely reflects their different abundances.
In sum, our data suggest that hnRNP L preferentially represses 3′ splice sites through interfering with the recognition of the pyrimidine tract/3′ splice site region, and around the 5′ splice site, and activates 5′ splice sites from downstream intronic positions. The binding position relative to the regulated exon therefore appears to provide an important determinant in specifying hnRNP L’s activator and repressor function (schematically represented below Fig. 3B). In contrast, in a recent study focusing on alternative splicing of genes important in T-cell biology, no positional code was observed; most likely due to the much lower number of investigated hnRNP L-regulated splicing events.38
In fact, several examples of hnRNP L-mediated splicing regulation support our conclusions from this global analysis: First, hnRNP L as a splicing activator was initially discovered for the human eNOS gene, where an intronic CA-repeat region activates the 5′ splice site of the upstream exon 13.40 Second, alternative exon 6 of the human MAPK10 gene is strongly regulated by a CA-repeat enhancer (positions +61 to +118 of intron 6; ref. 1). Third, our initial exonarray analysis10 provided several examples of hnRNP L repressing alternative exons by binding close to their 3′ splice site, which can be based on U2AF interference, as demonstrated for TJP1.25 Additionally, hnRNP L binding in very close proximity to the 5′ splice site can interfere with recognition by the U1 snRNP, as shown for SLC2A2,25 which is consistent with a minor effect in the RNA map at a region 0 to +25 relative to the 5′ splice site. Other splicing factors function by related mechanisms: CLIP analyses of Nova28,30and RBFOX231 revealed similar functional binding patterns as observed for hnRNP L, whereas PTB instead represses alternative exons also from downstream intronic positions, and activates by binding to the intron in close proximity to the flanking constitutive exons.32 A recent study investigated six hnRNP proteins (A1, A2/B1, F, H1, M, and U) by CLIP-Seq.41 Interestingly, in RNA maps derived from these data, hnRNP F and U activated exon inclusion from positions overlapping with hnRNP L repressor sites. In contrast, hnRNP A2/B1 and M repress alternative exons from hnRNP L’s activator position, suggesting potential antagonistic mechanisms of hnRNP L and other hnRNPs in alternative splicing regulation.
There are other cases of complex alternative splicing mechanisms regulated by hnRNP L, involving co-regulators such as the paralog hnRNP L-like, for example, the case of the CD45 gene.18-21,42 In this mechanism, an interplay of both splicing factors regulates three differentially spliced exons of CD45, dependent on the activation state of different immune cells. Such mechanisms probably represent a special minority of events and cannot be accounted for in the presented RNA map. Also, since CD45 is not expressed in HeLa cells, our iCLIP approach did not include it.
HnRNP L binding overlaps with polyadenylation signals
In addition to positional binding of hnRNP L around the splice sites, we investigated the crosslink-site density in proximity to polyadenylation [poly(A)] sites. All ENCODE-annotated polyadenylation events from protein-coding genes were grouped into terminal and internal poly(A) sites (Fig. 4), and the hnRNP L crosslink-site density was plotted in windows of -300 to +300 nucleotides relative to internal (top diagram, in red) and terminal poly(A) sites (middle diagram, in blue). The high crosslink-site density downstream of the poly(A) site in case of the internal poly(A) sites reflects hnRNP L binding in this region of the (pre-)mRNA. In contrast, hnRNP L binding disappeared downstream of terminal poly(A) sites, since most of the mRNA is cleaved. In both groups, the crosslink site frequency increased toward the poly(A) site, up to a position around -30. The strongly reduced frequency of crosslink sites between position -30 and the poly(A) site is due to the alignment problem with short sequence reads containing poly(A) tails. Interestingly, this region contains important control elements for the poly(A) site recognition, such as the AAUA3 signal, 15–30 nucleotides upstream of the poly(A) site, which is bound by CPSF; as well as a U-rich upstream sequence element (USE), which was shown to enhance polyadenylation (reviewed in ref. 43). This hnRNP L crosslink site distribution suggests that hnRNP L may commonly regulate poly(A) site selection through competing with binding of components of the polyadenylation machinery. Consistent with this we had previously described an example of hnRNP L-dependent alternative poly(A) site selection, based on an exonarray analysis after hnRNP L knockdown. In the ASAH1 mRNA, an internal poly(A) site is more frequently used if hnRNP L is downregulated, indicating suppression of internal polyadenylation by hnRNP L.10

Figure 4. HnRNP L binding overlaps with polyadenylation signals. Number of hnRNP L crosslink sites around polyadenylation [poly(A)] sites (positions -300 to +300) of protein-coding genes is diagrammed, separately for internal and terminal poly(A) sites (upper and lower panels, respectively). Below, internal (red) and terminal (blue) poly(A) sites are compared, plotted in smooth lines after normalization (lower panel).
HnRNP L binds preferentially in close proximity to predicted miRNA binding sites in 3′ UTR and can thereby regulate translation efficiency
It has been recently reported that hnRNP L competes with miRNAs for binding to the VEGFA 3′ UTR in hypoxia, thereby modulating miRNA function.11 Analyzing our iCLIP data, we in fact observed hnRNP L crosslink sites at a high density within and around these specific miRNA target sites. Interestingly, a second set of predicted miRNA-target sites overlapping with dense hnRNP L binding was observed in the downstream half of the VEGFA 3′ UTR (Fig. 5A and B). Since ~20% of hnRNP L crosslink sites were mapped to 3′ UTRs (Fig. 2B), competition of hnRNP L with miRNAs for binding to their respective target sites may represent a general mechanism.
Figure 5. HnRNP L binds preferentially to miRNA target sites in 3′ UTRs. (A) UCSC Genome Browser view of VEGFA exon/intron structure with crosslink-site distribution determined by iCLIP (hnRNP L-IP vs. control FLAG-IP). The middle panel (miRNA sites; in green) shows conserved mammalian miRNA regulatory target sites for conserved miRNA families in the 3′ UTR regions of Refseq genes, as predicted by ‘TargetScanHuman 5.1’ (http://www.targetscan.org). Target sites of miR-297 and miR-299, where hnRNP L modulates miRNA repression are highlighted by a green arrowhead below.11 (B) UCSC Genome Browser view of the VEGFA 3′ UTR, represented as in panel (A). (C) Frequency of crosslink sites around predicted miRNA target sites (-80 to +80 of target site). (D) Comparison of the crosslink site density between miRNA target site regions (from position -20 to +20 relative to the miRNA target site, including the miRNA target site itself) on the X-axis, and the corresponding non-target regions on the Y-axis, based on 3′ UTRs of 5062 protein-coding genes that contain both miRNA target sites and crosslink sites.
To investigate this on a genome-wide level, we extracted the conserved mammalian miRNA target sites in the 3′ UTRs of RefSeq genes, as predicted by TargetScanHuman 5.1 (http://www.targetscan.org). We first determined the frequency of hnRNP L crosslink sites within a region of 80 nt up- and downstream of the target sites (Fig. 5C), confirming an increased frequency of hnRNP L crosslinks on and around miRNA target sites.
Second, we compared the density of hnRNP L crosslinks at the miRNA target sites (positions -20 to +20 of target sites) vs. outside of the miRNA target sites (Fig. 5D), based on 5062 3′ UTRs that contain both crosslink sites and miRNA target sites (see Supplementary Materials and Methods). We conclude that the density at the miRNA target sites is significantly higher than outside, suggesting that competition of hnRNP L with binding of the Ago/miRNA complexes provides a global mechanism of hnRNP L function.
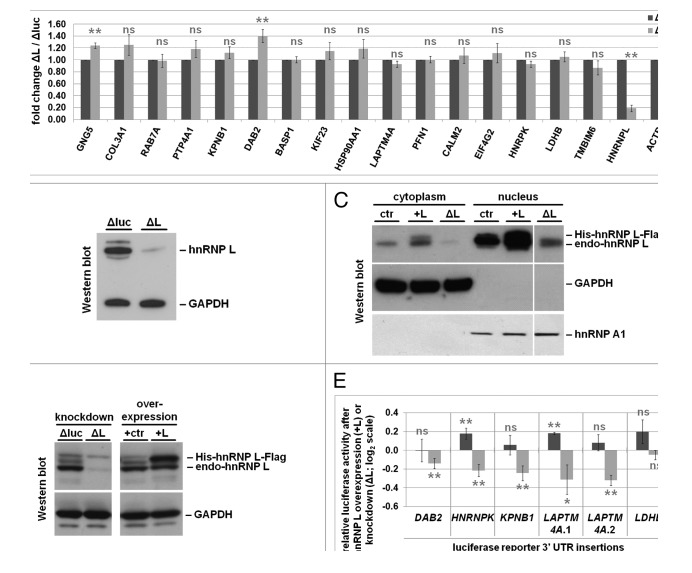
To predict potential target mRNAs for such a regulatory mechanism, two strategies were applied. First, dense hnRNP L binding clusters (based on our iCLIP data) were screened for overlap with TargetScan-predicted miRNA binding sites of miRNAs expressed in HeLa cells.44 Second, the top Ago2 binding clusters (based on PAR-CLIP45) were screened for overlap with dense hnRNP L binding clusters and TargetScan-predicted miRNA binding sites. As a result, 16 potential target genes were derived (Fig. S2). Since hnRNP L was reported to be involved in mRNA stability, either by destabilizing (SLC2A1 mRNA4) or by stabilizing (BCL2 mRNA9), in each case by binding to a CA-rich region in the 3′ UTR, we assayed by quantitative RT-PCR the steady-state mRNA levels of the 16 mRNAs after hnRNP L knockdown (Fig. 6A and B). Only in two cases (GNG5 and DAB2), steady-state mRNA levels were significantly increased after hnRNP L knockdown, suggesting a destabilizing role of hnRNP L (as in SLC2A1 mRNA4). However, hnRNP L binding in the 3′ UTR does not appear to be a widespread determinant of mRNA stability in this set of predicted target mRNAs.
Figure 6. HnRNP L knockdown and overexpression affects translational efficiency of luciferase reporters bearing 3‘ UTRs with predicted hnRNP L/miRNA binding sites. (A) Steady-state mRNA levels of predicted hnRNP L/miRNA targets after hnRNP L knockdown. Total RNA was isolated from hnRNP L knockdown (ΔL) and luciferase control knockdown (Δluc) cells (see panel B), and subjected to quantitative RT-PCR analysis with gene-specific PCR primers against the 16 potential target mRNAs (as indicated). HNRNPL- and ACTB-specific primers served as controls; samples were normalized to U1 snRNA. Student's t test was performed (**: P value < 0.01; *: P value = 0.01–0.05; ns: not significant, P value > 0.05). (B) Representative western blot of siRNA-mediated hnRNP L knockdown (ΔL) and luciferase control knockdown (Δluc), using antibodies against hnRNP L and GAPDH (loading control). (C) HnRNP L protein levels upon knockdown or overexpression change correspondingly both in the nucleus and in the cytoplasm. HeLa cells were transfected with an siRNA against hnRNP L (ΔL) or a vector expressing His-/Flag-tagged hnRNP L (+L) and compared with control cells (ctr). Nuclear and cytoplasmic extracts were prepared, followed by western blot analysis with antibodies against hnRNP L, detecting both endogenous (endo-hnRNP L) and the overexpressed tagged protein (His-hnRNP L-Flag); GAPDH served as a cytoplasmic, hnRNPA1 as a nuclear marker. Note that although hnRNP A1 is a shuttling protein, its steady-state levels are much higher in the nucleus, with the cytoplasmic levels below the detection limit under these conditions. (D and E) Effects of hnRNP L knockdown or overexpression on luciferase reporter expression. To test for effects of hnRNP L knockdown, cells were transfected with either an unrelated luciferase-control siRNA (Δluc) or an hnRNP L-specific siRNA (ΔL); to test for effects on hnRNP L overexpression, an empty vector control (+ctr) or a vector expressing His/Flag-tagged hnRNP L protein (+L) were transfected, always in addition to a dual luciferase reporter construct expressing both firefly luciferase with or without selected 3′ UTR insertions, using Renilla luciferase for normalization. Lysates were prepared, followed by western blot analysis with antibodies against hnRNP L, detecting both endogenous (endo-hnRNP L) and the overexpressed tagged protein (His-hnRNP L-Flag), as well as GAPDH as a loading control (panel D). Firefly luciferase activity of reporters with 3′ UTR insertions (as indicated) was monitored (panel E), normalized to Renilla luciferase activity in the same lysate, comparing the effects of hnRNP L knockdown (ΔL) and overexpression (+L). The 3′ UTR regions selected are marked as red boxes in Figure S2. The fold changes between hnRNP L knockdown/overexpression and the respective control were calculated, normalized to a firefly luciferase reporter without 3′ UTR insertion, and displayed as log2 values. All values and standard deviations were derived from three biological replicates. Student's t test was performed, comparing relative luciferase activity from the cells expressing the reporter without 3′ UTR insertion (reference) to the respective 3′ UTR insert-carrying reporters (**, P value < 0.01; *, P value = 0.01–0.05; ns, not significant, P value > 0.05).
To analyze potential effects of altered hnRNP L levels on translation efficiency, six selected 3′ UTR regions containing both hnRNP L crosslink and predicted miRNA binding sites (DAB2, HNRNPK, KPNB1, LDHB, and two different regions from LAPTM4A; Fig. S2E, F, J, and O) were cloned in the 3′ UTR of a firefly luciferase reporter. Note that hnRNP L levels change both in the nucleus as well as, more importantly, in the cytoplasm (Fig. 6C). The luciferase constructs and a control without 3′ UTR insertion were transfected in HeLa cells, where hnRNP L levels had been increased by overexpression or decreased by siRNA-mediated knockdown (Fig. 6D). In five of the six constructs tested (DAB2, HNRNPK, KPNB1, LAPTM4A region 1 and 2) translation efficiency significantly decreased after hnRNP L knockdown; in two cases (HNRNPK and one of the LAPTM4A 3′ UTRs) translation efficiency increased after hnRNP L overexpression (Fig. 6E). The reciprocal effects in these two cases are consistent with a model, whereby hnRNP L competes with miRNA/Ago binding to target 3′ UTRs, as previously shown for the VEGFA mRNA.11 Why do we see only two cases of reciprocal, hnRNP L-dependent effects on translation, and in the other three constructs (DAB2, KPNB1, and LAPTM4A.2) only a decrease in translation efficiency upon hnRNP L knockdown? We know that hnRNP L levels in the cytoplasm are lower than in the nucleus (ref. 10; Fig. 6C), yet most likely these effects depend on the relative steady-state levels of hnRNP L as well as of the respective miRNA and target 3′ UTR. Of these targets, the HNRNPK mRNA is particularly interesting, since hnRNP proteins are often tightly linked in their expression by cross-regulatory networks.
A recent study pointed to the cross-regulatory relationships between hnRNP proteins.41 Interestingly, hnRNP L binding clusters are also found in the 3′ UTRs of hnRNP A1, A2/B1, F, H1, M, and U, in each case overlapping with predicted miRNA target sites (data not shown). We conclude that an hnRNP L/miRNA-competition mechanism may add another layer of complexity to the cross-regulatory networks between hnRNP proteins.
Materials and Methods
iCLIP, sequencing, and genomic mapping
Three individual iCLIP experiments were performed with HeLa cells, using hnRNP L-specific antibody 4D11 (Sigma), and FLAG antibody (Sigma M2) as a negative control, followed by sequencing on an Illumina GAIIx sequencer (50 bp single-end reads). For experimental procedures and genomic mapping, see reference 26.
Statistics and bioinformatics
Data analysis details are described in the Supplementary Materials and Methods, sections 1–7. Solexa sequencing reads (.fastq files) of iCLIP experiments and filtered crosslink sites combining all three experiments of hnRNPL (hnRNPL_crosslink_site.bed) were deposited to the NCBI GEO database (GSE 37562), as well as exon array raw data (.CEL files) of hnRNP L-knockdown experiments and lists of regulated exons predicted to be activated (activated_exon.txt) or repressed (repressed_exon.txt) by hnRNP L.
hnRNP L knockdown and RT-PCR analysis
Knockdown of hnRNP L in HeLa cells was performed for four days as described earlier,10 with two different siRNAs against hnRNP L or Luciferase as a control. To monitor hnRNP L knockdown on the protein level, western blotting was performed as described below. Total RNA was isolated using TRIzol reagent (Invitrogen) and RNeasy columns (QIAGEN). RNA was reverse-transcribed (iScript cDNA Synthesis Kit, Bio-Rad) and specific mRNAs were monitored by semiquantitative PCR. For primer and siRNA sequences, see Supplementary Materials and Methods (section 8). For quantitative PCR, cDNAs were measured with the KAPA SYBR FAST QPCR MasterMix (PeqLab) in a Mastercycler ep realplex2 S (Eppendorf). Relative RNA levels from three biological and three technical replicates each were calculated according to Pfaffl (2001).46
HnRNP L overexpression and subcellular fractionation
To control whether cytoplasmic hnRNP L levels change after knockdown or overexpression, siRNA-mediated knockdown was performed as described above, or His/Flag-tagged hnRNP L was overexpressed,24 using pcDNA3-His-hnRNP L-Flag, followed by subcellular fractionation (ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit, Fermentas). Cytoplasmic and nuclear extracts were subjected to western blot analysis with hnRNP L 4D11 (Sigma), GAPDH 6C5 (Ambion), and hnRNP A1 4B10 (Santa Cruz) antibodies, using the Lumi-Light Western Blotting Substrate (Roche).
Firefly luciferase/3′ UTR constructs and luciferase reporter assays
3′ UTR regions from selected genes (indicated by red boxes in Fig. S2) were PCR-amplified, and products were cloned into the multiple cloning site of pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega), within the 3′ UTR of the firefly luciferase reporter gene and between SalI and SacI restriction sites.
For the luciferase reporter assay in combination with hnRNP L knockdown, 3.8 × 104 HeLa cells per well were reverse-transfected in a 12-well plate, using siRNA at a final concentration of 12.5 nM in culture medium (hnRNP L 3′-UTR or luciferase GL2) and 2.1 μl Lipofectamine RNAiMAX Reagent in 210 μl OptiMEM medium (both Life Technologies). Note that no binding sites for luciferase GL2 siRNA are contained in firefly or Renilla luciferase reporters within pmirGLO. After hnRNP L knockdown for three days, the cells were transfected with 1 μg of the pmirGLO reporter plasmids (with or without 3′ UTR insertion), diluted in 41 μl OptiMEM medium and 2.9 μl FuGENE HD Transfection Reagent (Promega). Cells were further incubated for 24 h.
For the luciferase reporter assay in combination with hnRNP L overexpression, 1.14 × 105 HeLa cells per well were seeded the day before transfection in a 12-well plate. 230 ng of empty pcDNA3 vector or pcDNA3-His-hnRNP L-Flag vector were transfected in combination with 910 ng of the pmirGLO reporter plasmids (with or without 3′ UTR insertion), diluted in 114 μl OptiMEM medium, using 2.3 μl TurboFect Transfection Reagent (Thermo Scientific), and incubated for 24 h. Cells were lysed by adding 200 μl Lysis-Juice 2 (PJK), following the manufacturer’s instructions. An aliquot of these lysates was analyzed by western blotting as described above to verify hnRNP L knockdown/overexpression. Bioluminescence was monitored for firefly luciferase, using Beetle-Juice Kit, and for Renilla luciferase, using Renilla-Juice Kit (both PJK) in an Orion L Microplate Luminometer (Titertek-Berthold).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Julia Renz for transfection, cell fractionation, and western blot assays, Lorraine Fischer, Sina Fuchs, and Melanie Goebel for construction of the luciferase reporter constructs, and Marcel Oschwald for western blots. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Bi 316/12, IRTG1384, and a German-Israeli Project Cooperation Grant, to AB), the Federal Ministry for Education and Research (BMBF RUS 10/030; to Bindereif A and Gelfand MS), the European-Commission-funded Network of Excellence EURASNET (to Bindereif A), Russian State contract 14.740.11.0003 (to Gelfand MS), the Slovenian Research Agency (P2-0209, Z7-3665; to Curk T and Zupan B), and the Russian Academy of Sciences (programs Molecular and Cellular Biology and Basic Medical Science; to Gelfand MS). The results in this work are part of a dissertation (Oliver Rossbach) submitted at the Justus Liebig University in Giessen.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27991
References
- 1.Hui J, Hung LH, Heiner M, Schreiner S, Neumüller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–98. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, Mertz JE. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–80. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 3.Guang S, Felthauser AM, Mertz JE. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol Cell Biol. 2005;25:6303–13. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton BJ, Nichols RC, Tsukamoto H, Boado RJ, Pardridge WM, Rigby WF. hnRNP A2 and hnRNP L bind the 3’UTR of glucose transporter 1 mRNA and exist as a complex in vivo. Biochem Biophys Res Commun. 1999;261:646–51. doi: 10.1006/bbrc.1999.1040. [DOI] [PubMed] [Google Scholar]
- 5.Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782–8. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29:2899–912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang B, Lim JH, Hahm B, Jang SK, Lee SW. hnRNP L is required for the translation mediated by HCV IRES. Biochem Biophys Res Commun. 2009;378:584–8. doi: 10.1016/j.bbrc.2008.11.091. [DOI] [PubMed] [Google Scholar]
- 8.Shih SC, Claffey KP. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem. 1999;274:1359–65. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 9.Lee DH, Lim MH, Youn DY, Jung SE, Ahn YS, Tsujimoto Y, Lee JH. hnRNP L binds to CA repeats in the 3’UTR of bcl-2 mRNA. Biochem Biophys Res Commun. 2009;382:583–7. doi: 10.1016/j.bbrc.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 10.Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–96. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–34. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kefas B, Floyd DH, Comeau L, Frisbee A, Dominguez C, Dipierro CG, Guessous F, Abounader R, Purow B. A miR-297/hypoxia/DGK-α axis regulating glioblastoma survival. Neuro Oncol. 2013;15:1652–63. doi: 10.1093/neuonc/not118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piñol-Roma S, Swanson MS, Gall JG, Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989;109:2575–87. doi: 10.1083/jcb.109.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S. Review: perinucleolar structures. J Struct Biol. 2000;129:233–40. doi: 10.1006/jsbi.2000.4247. [DOI] [PubMed] [Google Scholar]
- 15.Rajan P, Dalgliesh C, Bourgeois CF, Heiner M, Emami K, Clark EL, Bindereif A, Stevenin J, Robson CN, Leung HY, et al. Proteomic identification of heterogeneous nuclear ribonucleoprotein L as a novel component of SLM/Sam68 Nuclear Bodies. BMC Cell Biol. 2009;10:82. doi: 10.1186/1471-2121-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dery KJ, Gaur S, Gencheva M, Yen Y, Shively JE, Gaur RK. Mechanistic control of carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) splice isoforms by the heterogeneous nuclear ribonuclear proteins hnRNP L, hnRNP A1, and hnRNP M. J Biol Chem. 2011;286:16039–51. doi: 10.1074/jbc.M110.204057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goehe RW, Shultz JC, Murudkar C, Usanovic S, Lamour NF, Massey DH, Zhang L, Camidge DR, Shay JW, Minna JD, et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest. 2010;120:3923–39. doi: 10.1172/JCI43552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motta-Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell. 2010;37:223–34. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–84. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem. 2005;280:38297–304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]
- 21.Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–9. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Hai Y, Liu G, Fang T, Kung SKP, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J Biol Chem. 2009;284:1505–13. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29:1442–51. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heiner M, Hui J, Schreiner S, Hung LH, Bindereif A. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010;7:56–64. doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 26.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–86. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–6. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 29.Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–7. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23:206–12. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 36.König J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr., Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shankarling G, Cole BS, Mallory MJ, Lynch KW. Transcriptome-wide RNA interaction profiling reveals physical and functional targets of hnRNP L in human T cells. Mol Cell Biol. 2014;34:71–83. doi: 10.1128/MCB.00740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003;10:33–7. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 41.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–78. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preussner M, Schreiner S, Hung LH, Porstner M, Jäck HM, Benes V, Rätsch G, Bindereif A. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res. 2012;40:5666–78. doi: 10.1093/nar/gks221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–82. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.