Abstract
CRISPR adaptive immune systems were analyzed for all available completed genomes of archaea, which included representatives of each of the main archaeal phyla. Initially, all proteins encoded within, and proximal to, CRISPR-cas loci were clustered and analyzed using a profile–profile approach. Then cas genes were assigned to gene cassettes and to functional modules for adaptation and interference. CRISPR systems were then classified primarily on the basis of their concatenated Cas protein sequences and gene synteny of the interference modules. With few exceptions, they could be assigned to the universal Type I or Type III systems. For Type I, subtypes I-A, I-B, and I-D dominate but the data support the division of subtype I-B into two subtypes, designated I-B and I-G. About 70% of the Type III systems fall into the universal subtypes III-A and III-B but the remainder, some of which are phyla-specific, diverge significantly in Cas protein sequences, and/or gene synteny, and they are classified separately. Furthermore, a few CRISPR systems that could not be assigned to Type I or Type III are categorized as variant systems. Criteria are presented for assigning newly sequenced archaeal CRISPR systems to the different subtypes. Several accessory proteins were identified that show a specific gene linkage, especially to Type III interference modules, and these may be cofunctional with the CRISPR systems. Evidence is presented for extensive exchange having occurred between adaptation and interference modules of different archaeal CRISPR systems, indicating the wide compatibility of the functionally diverse interference complexes with the relatively conserved adaptation modules.
Keywords: CRISPR, Cas, Type I, Type III, archaea
Introduction
CRISPR adaptive immune systems are present in most archaea and in many bacteria where they primarily target and degrade invading genetic elements. The immune reaction involves three primary stages. First, adaptation involving selection of 30–45 bp DNA fragments (protospacers) from an invading genetic element and their insertion between repeats of genomic CRISPR arrays as de novo spacers. Second, transcripts of CRISPR arrays are processed generally within the repeat sequences to yield small CRISPR RNAs (crRNAs). Third, crRNAs are assembled into protein interference complexes and guide the complex to matching sequences on nucleic acid(s) of the invading genetic element which are then cleaved.1
Early classifications of CRISPR adaptive immune systems were based primarily on sequence analyses of CRISPR repeats or CRISPR-associated (Cas) proteins and yielded several distinct groupings.2-4 There is now a consensus that CRISPR systems can be classified structurally into major classes denoted Types I, II, and III for bacteria and Types I and III for archaea.5 The initial adaptation step is relatively conserved mechanistically among the three main CRISPR types and requires proteins Cas1, Cas2 and, in many systems, Cas4. Primary processing of CRISPR transcripts is accomplished with Cas6 in Type I and Type III systems and by a combination of RNase III and a tracrRNA in the bacterial Type II system.1 The interference modules are much more varied with respect to both the number and sequences of the protein components involved, and the nucleic acid targets. Moreover, the high-sequence diversity of interference components, especially for the proteins labeled Cas7 and Cas8, provided a major obstacle to reaching a consensus about their classification. Despite this diversity, however, the three dimensional structures of the different interference complexes (denoted Cascade) appear to be partly conserved.6-8 Nevertheless, the mechanisms of interference are likely to be diverse, with Type I and Type II systems appearing to target primarily DNA while Type III interference systems can target DNA or RNA.9-13
Type I and Type III systems have been further classified into subtypes based primarily on sequences of signature Cas proteins Cas1, Cas3, and Cas8 for Type I and Cas10, and the small protein component S (protein S) for Type III systems, and on their gene synteny.14 However, there remain some limitations in the methods employed for identifying CRISPR subtypes. For example, categorizing systems according to the sequence of the larger conserved Cas1 protein is not always unambiguous and, moreover, Cas8, Cas10, and protein S are unsatisfactory signature proteins because of their variable size and high-sequence diversity.
Type III systems are found in most archaeal phyla, and especially among extreme thermophiles, and their interference complexes are commonly encoded in gene cassettes that are not linked genomically to either CRISPR loci or adaptation cas gene cassettes.15 Earlier, we analyzed archaeal CRISPR-Cmr/Csm systems of archaea, based on Cas10 (Cmr2/Csm1) sequences, and concluded that there were five main families A, B, C, D, and E, of which the smallest and least well defined was family C.15,16 Subsequently, these were defined as Type III systems and were separated into the major subtypes III-A and III-B for bacteria and archaea,5 where subtypes III-A and III-B corresponded to the archaeal families E and B, respectively. Although the potential existence of families A and D was acknowledged by others earlier,4 they were omited from the subsequent general classification.14
Previously, there have been reports of other proteins being encoded within or adjacent to archaeal CRISPR-Cas cassettes, including mobile elements and toxin-antitoxin systems,15,16 and in a study of Pyrobaculum species, evidence was presented for herA and nurA genes being specifically linked to cas genes.17 Here, we systematically analyzed the numbers and degree of specificity of all genes that are associated physically with archaeal cas gene cassettes, and identified 12 additional accessory protein families.
Results
Analysis strategy
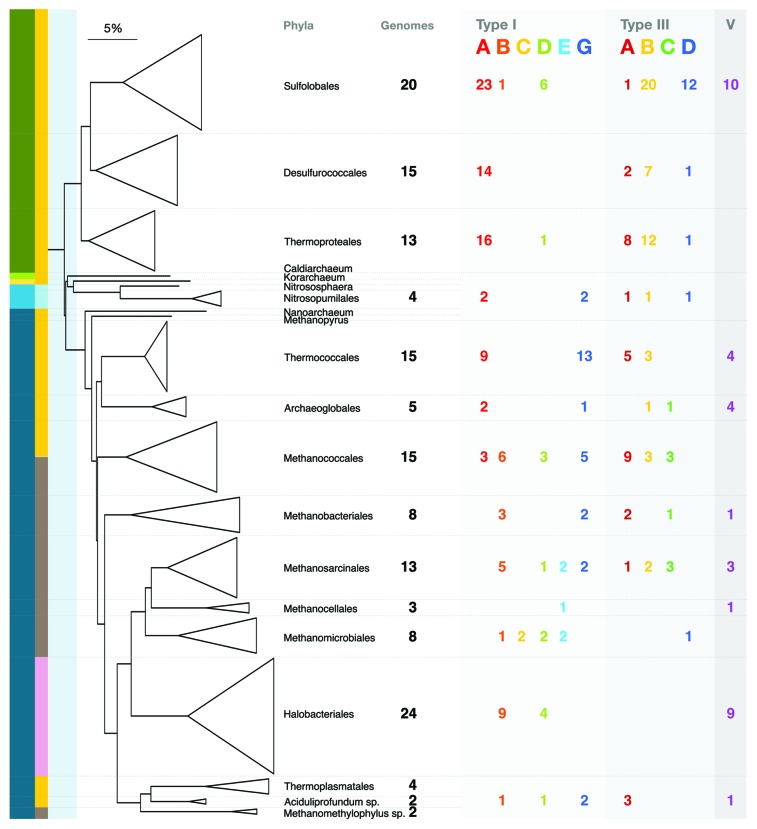
First, we downloaded the 159 archaeal genomes available in May 2013 (www.ebi.ac.uk/genomes/archaea.html). The phylogenetic diversity of the archaea and the number of sequenced genomes that were analyzed within each order are illustrated in the 16S rRNA-based phylogenetic tree (Fig. 1). All genomic loci-containing cas genes were first extracted automatically and the proteins were then clustered using Markov clustering. Subsequently, the loci were annotated manually aided by profile–profile searches against Conserved Domains (CDD) and TIGRFAMs (TF) databases.3,5 We then followed the proposal of Makarova et al.14 pertaining to the unification of Cas protein families involved in interference complexes of Type I and Type III systems throughout the analysis. Gene cassettes encoding adaptation and interference modules were extracted together with the cas6 gene of the CRISPR RNA processing enzyme (Table S1). A significant fraction of the genetic modules were deficient or otherwise defective and they are also identified and tabulated (Table S1). Whereas 93% of gene cassettes encoding Type I adaptation and interference complexes were found to be linked to one another and to CRISPR loci, for Type III systems, only 55% of the cas gene cassettes and CRISPR loci were contiguous, and many interference complexes were encoded as separate genetic units (Table 1), consistent with them sharing adaptation modules and CRISPR loci with Type I systems.13,15
Figure 1. 16S rRNA phylogenetic tree for all archaeal genomes included in the study. The tree shows the primary archaeal phyla with the total number of genomes analyzed in each phylum indicated (single genomes for a given phylum are not numbered). The putative kingdoms are color-coded on the bar on the far left. Crenarchaeota, green; Aigarchaeota, light green; Korarchaeota, yellow; Thaumarchaeota, light blue; Euryarchaeota, dark blue. Environmental habitats are color-coded on vertical lines to the right of the kingdom bar: hydrothermal, orange; marine, blue; wetland sediment, brown, and hypersaline lake, pink. For each phylum, the Type I and Type III CRISPR subtypes are color-coded on the right and the total numbers of each subtype are given. The branch length ruler corresponds to a 5% difference in 16S RNA sequence.
Table 1. Archaeal CRISPR systems. Total numbers of integral CRISPR immune systems and of independent interference modules.
| CRISPR system | Total adaptation + interference | Interference alone |
|---|---|---|
| Type I | 106 | 11 |
| Type III | 16 | 59 |
| Type I+III | 57 | 4 |
| Variants | 11 | 20 |
Given that many of the CRISPR systems were non integral, and that there is strong evidence for occurrence of exchange between adaptation and interference modules,15 we analyzed the two functional modules separately. First, we determined the operon structures and gene syntenies within each genetic unit to define the modules. Next, we prepared separate dendrograms for the adaptation and interference modules by comparison of sequences of all their protein components and adding up scores for each module, which resulted in module-to-module distance matrices, which were then converted to dendrograms using the neighbor joining method (Figs. S1 and S2). Subsequently, a detailed analysis of the different Type I and Type III subtypes was performed employing a combination of properties of the gene cassettes, and the protein components, of the interference modules.
Our criteria for defining subtypes were as follows. A vertical line is drawn near the base of the dendrogram in order to separate optimally the already established CRISPR subtypes,5 and this line is defined as the subtype threshold. Branches occurring before the line represent separate subtypes whereas branches initiating after the line belong to the same subtype. Exceptions were branches starting within the line. They were defined as separate subtypes if they showed consistent differences in gene synteny (as seen for subtypes I-G and VI-2, see below), whereas if they showed similar gene syntenies they are inferred to represent divergent variants of the same subtype. These criteria can be readily applied to classifying newly identified CRISPR systems if a dendrogram is first prepared using the approach outlined above. On applying these criteria to the archaeal genomes, we generated a comprehensive, manually curated catalog of all the archaeal CRISPR systems. The catalog is presented in Table S1 and Figure S2 in a readily accessible form. It can easily be searched for individual organisms and genes, and researchers can both utilize and further analyze the data online. The results obtained were generally consistent with the current classification for Type I and Type III subtypes for bacteria and archaea,14 but a number of archaea-specific properties emerged, which are summarized below.
Adaptation modules
Initially, we generated a dendrogram of all the archaeal adaptation modules employing a combination of concatenated protein sequences and gene synteny of the adaptation Cas proteins of Type I and Type III systems (Table 2; Fig. S1). The threshold for defining adaptation subtypes was then determined to maximize the matches to the universal CRISPR subtypes,5 which were primarily Type I subtypes because most archaeal adaptation modules are linked genomically to Type I interference complexes (Table 1). Representative gene syntenies were then determined for each group of adaptation modules, and they are shown in Figure 2 superimposed on a simplified dendrogram. The complete adaptation dendrogram is presented in Figure S1. Many of the adaptation gene clusters are linked to single Type I subtypes but about 42% are linked to multiple subtypes (Fig. 2). Although the gene syntenies show significant variation for the different adaptation clusters, the protein sequences were highly conserved, and therefore, we did not use adaptation modules for further classification of CRISPR subtypes. However, they were useful for investigating the phenomenon of adaptation–interference modular exchange (see below).
Table 2. Cas proteins associated with archaeal Type I and Type III CRISPR functional modules.
| Function | Type-I | Type III-A | Type III-B |
|---|---|---|---|
| Adaptation | |||
| Cas1 | Cas1 | Cas1 | |
| Cas2 | Cas2 | Cas2 | |
| Cas4 | - | Cas4 | |
| Cas4' | - | - | |
| Processing | |||
|---|---|---|---|
| Cas6 | Cas6 | Cas6 | |
| Interference | |||
|---|---|---|---|
| Cas7 | Csm3+5 | Cmr4+6+1 | |
| Cas8 | Csm1 | Cmr2 | |
| Cas5 | Csm4 | Cmr3 | |
| Cse2/Csa5 | Csm2 | Cmr5 | |
| Cas3 | - | - | |
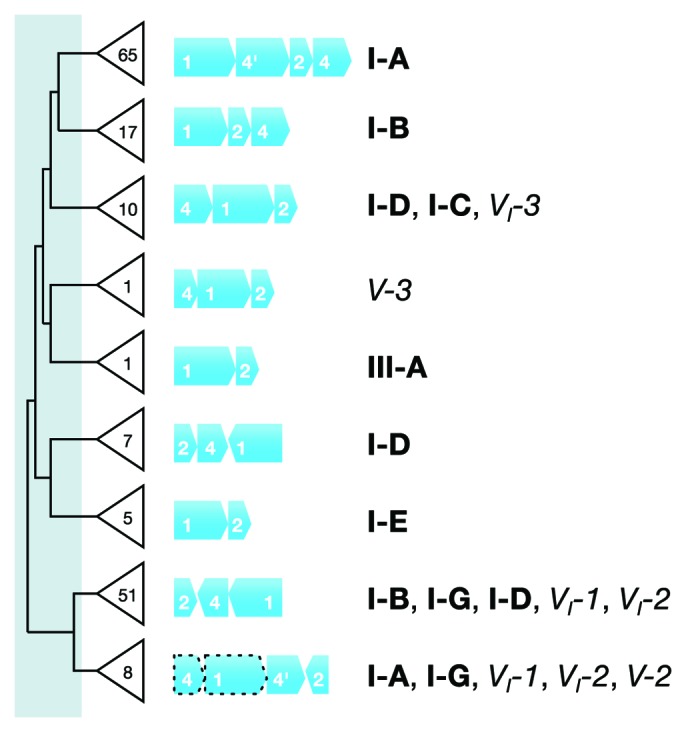
Figure 2. Dendrogram and gene syntenies of archaeal adaptation modules of different CRISPR subtypes and the total numbers of identified subtypes are given on the tree. The dendrogram represents a simplified version of the full dendrogram in Figure S1 showing the different classes of adaptation modules. Gene contents (indicated by bold white numbers), gene sizes, and gene syntenies are shown for representative adaptation modules. Further, the subtypes of interference modules associated with each class of adaptation module are shown. The threshold used to classify the variants is indicated by the light blue area toward the base of the tree, and it was selected so that the resulting classes would match optimally with the associated interference subtypes, which are mainly Type I. Nevertheless, some adaptation module classes are associated with interference modules from different subtypes.
Type I subtypes
Currently, all Type I systems of bacteria and archaea are classified into six subtypes I-A to I-F based on Cas1, Cas3, and Cas8 phylogeny and on the gene syntenies.5 Although this classification scheme has been largely successful, identification of newly sequenced CRISPR systems is not always straightforward, and in particular, employing Cas8 as a signature protein can be problematic. Often, newly sequenced CRISPR systems are not covered by existing Cas8 models. Furthermore, sequence matches to published Cas8 models are often ambiguous or misleading. For example, the Cas8a1_I-A model gives positive matches to Type I-B systems.
Nevertheless, we could assign most archaeal Type I systems to current subtypes using our criteria. The results show a strong archaeal bias to subtypes I-A, I-B, and I-D, with very few examples of subtypes I-C and I-E, and no I-F subtypes were found (Table 3). Our sequence analyses also support the division of the archaeal subtype I-B into distinct subtypes, designated here as subtype I-B and I-G, which correspond closely to the earlier proposed groupings Hmar and Tneap, respectively.3 Four variant Type I subtypes were identified as VI-I, VI-2, VI-3, and VI-4 that each occurred in low numbers and tended to be phyla-specific (Table 4). For each subtype, variations in gene order were sometimes observed but the same ORFs with similar sequences were still present. Typical subtype gene syntenies are illustrated on a simplified interference module dendrogram in Figure 3 and the complete archaeal interference dendrogram is presented in Figure S2.
Table 3. Number and kingdom distribution of the major subtypes of archaeal Type I and Type III systems.
| Type I | Type III | ||||||
|---|---|---|---|---|---|---|---|
| subtype | number | cren | eury | subtype | number | cren | eury |
| A | 69 | 53 | 14 | A | 32 | 11 | 21 |
| B | 26 | 1 | 25 | B | 49 | 39 | 9 |
| C | 2 | 0 | 2 | C | 8 | 0 | 8 |
| D | 18 | 7 | 11 | D | 16 | 14 | 1 |
| E | 5 | 0 | 5 | ||||
| G | 27 | 0 | 25 | ||||
cren, crenarchaea; eury, euryarchaea.
Table 4. Number and kingdom distribution of variant archaeal interference subtypes.
| Type-1 | |||
|---|---|---|---|
| subtype | number | cren | eury |
| VI-1 | 5 | 0 | 5 |
| VI-2 | 2 | 0 | 2 |
| VI-3 | 2 | 0 | 2 |
| VI-4 | 1 | 0 | 1 |
| Type III | |||
|---|---|---|---|
| subtype | number | cren | eury |
| VIII-1 | 10 | 10 | 0 |
| VIII-2 | 1 | 0 | 1 |
| Unclassified | |||
|---|---|---|---|
| V-1 | 9 | 0 | 9 |
| V-2 | 1 | 0 | 1 |
| V-3 | 1 | 0 | 1 |
cren, crenarchaea; eury, euryarchaea.
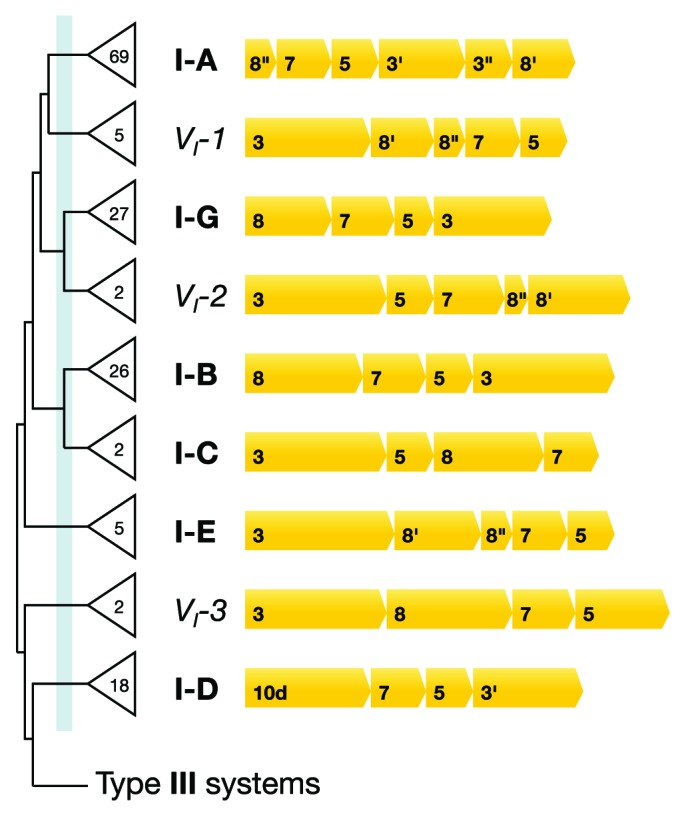
Figure 3. Dendrogram and gene syntenies of archaeal interference modules of different Type I subtypes where the total numbers of identified subtypes are given on the tree. Gene contents (indicated by bold numbers), gene sizes, and gene syntenies are shown for representative interference modules of each subtype. The dendrogram is a simplified version of the top half of the full dendrogram in Figure S2. The subtype threshhold is indicated by the light blue vertical line near the base of the tree. Most of the identified modules fall within already established subtypes. The new subtype I-G was separated from the earlier subtype I-B, and variant subtypes VI-I, VI-2, and VI-3 constitute exceptional subtypes with few members.
The interference module of subtype I-D is quite distantly related to the other Type I subtypes. In the dendrogram, it lies at the junction of the Type I and Type III systems (Fig. 3; Fig. S2) and it carries a Cas10d protein that shows low but significant similarity to both Cas8 and Cas10 of Type I and Type III systems, respectively (data not shown). Therefore, we infer that subtype I-D may represent an intermediate between Type I and Type III interference modules.
The total numbers of each Type I subtype found associated with different archaeal phyla are indicated on the phylogenetic tree (Fig. 1) and are summarized for the main archaeal kingdoms in Table 3. The results show significant differences between the kingdoms. There is a strong bias to subtype I-A systems among crenarchaea, which lack most other subtypes except I-B (one example) and I-D (7 examples). In contrast, euryarchaea exhibit a more varied composition with multiple examples of subtypes I-A, I-B, I-G, and I-D, but with few instances of subtypes I-C and I-E.
Newly classified Type I subtypes
It is clear from the interference module dendrogram (Fig. S2) that the original subtype I-B contains independent groups, which branch off well before the subtype separation threshold, hence, it does not constitute a homogeneous cluster. Therefore, we propose dividing subtype I-B into subtypes I-B and I-G (Fig. 3). Subtype I-G modules are also widespread among bacteria where, for example, they constitute the dominant Type I subtype in thermophilic Clostridium species. Cas8 sequences of the new I-B subtype match the CDD Cas8b_I-B model as predicted,5 while Cas8 sequences from I-G modules match Cas8a1_I-A,5 further supporting the division into subtypes I-B and I-G. Furthermore, the Cas8a1_I-A CDD model does not match any Type I-A Cas8 sequences, demonstrating that the model is unsuitable for correct identification of Type I subtypes.
The VI-1 and VI-2 variants branch off from subtypes I-A and I-G (Fig. 3; Fig. S2). Both variants carry interference modules with five genes, whereas I-A and I-G modules exhibit six and four genes, respectively. When compared with subtype I-G, the additional genes in variants VI-1 and VI-2 appear to have resulted from partitioning of cas8 into cas8' and cas8”. Despite the similarity in gene synteny, variants VI-1 and VI-2 are distinct in their amino acid sequences, diverging before the subtype definition threshold (Fig. 3; Fig. S2). Moreover, their Cas8 sequences are not covered by any CDD5 or TF3 models. However, given that there are so few members and that they appear to be archaea-specific, VI-1 and VI-2 are classified as variants rather than subtypes (Table 4).
VI-3 was also reported earlier as a GSU0053-type module,3 and more recently, as a variant of subtype I-C.14 We were unable to confirm any link to subtype I-C and believe it represents an independent subtype, which should retain variant status until more examples are found in archaea and/or bacteria (Fig. 3).
Methanocella conradii contains a Type I interference module designated VI-4, which despite yielding close protein sequence similarity to a group of I-D systems from cyanobacteria, carries a Cas8 protein which does not have an HD domain, a key structural signature of subtype I-D systems. However, the cofunctional Cas3 protein does contain an HD domain and, therefore, VI-4 may represent an intermediate between subtype I-D and other Type I subtypes (Figs. 3 and 4).
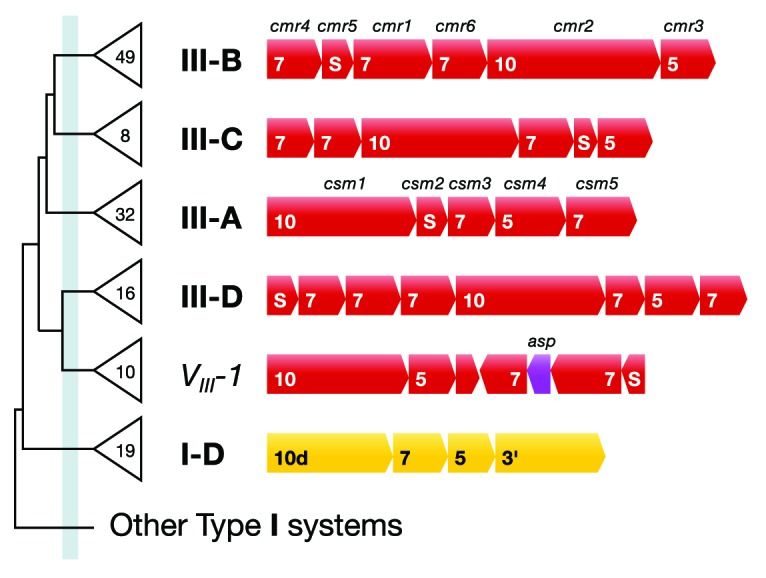
Figure 4. Dendrogram of the classified Type III subtypes. Total numbers of identified subtypes are given on the tree. Gene contents, gene sizes, and gene syntenies are shown for representative interference modules of the different subtypes. The dendrogram represents a simplified and inverted bottom half of the full dendrogram in Figure S2. Gene contents are indicated by bold numbers within genes, and for subtypes III-A and III-B, csm/cmr gene names are also given above the genes. The subtypes have distinct gene syntenies and branch before the defined threshold indicated by the light blue vertical line. Subtypes III-C and III-D are newly defined, and VIII-1 that was found only in some members of the Sulfolobales, and is therefore classified as a variant subtype. All subtypes have cas10, protein S, cas5, and multiple paralogs of cas7. asp denotes the gene of a putative aspartate protease. The subtype I-D gene cassette (in yellow) that branches at the junction of the Type III and Type I subtypes in Figure S2 is included as an outgroup.
Type III subtypes
Earlier, five CRISPR families A to E were proposed for archaea based primarily on the divergent sequences of the Cas10 (Cmr2/Csm1) proteins.15,16 Later, Makarova et al.5 defined the universal subtype III-A corresponding to family E and subtype III-B equivalent to family B. Here, all archaeal genomic copies of Type III systems were identified many of which were limited to interference gene cassettes (Tables 1 and 2). Inspection of their positions within the interference dendrogram (Fig. S2) demonstrated that about 70% of the interference modules fell unambiguously into the current universal subtypes III-A or III-B, which are common to both crenarchaea and euryarchaea (Fig. 1).
Here we reclassify the remaining 30% of the subtypes into subtypes III-C (formerly family A) and III-D (formerly family D); the earlier family C was integrated into subtype III-B.16 Ten examples of an additional subtype were observed that were specific to the order Sulfolobales and the subtype is classified as variant subtype VIII-1 (Table 4). Typical gene cassettes for each subtype are illustrated in the simplified dendrogram (Fig. 4), and the complete Type III interference dendrogram is included in Figure S2. As for Type I subtypes, gene orders occasionally differ for a given subtype but the same ORFs with closely related sequences are retained. The distribution of Type III subtypes within the different archaeal phyla is illustrated in Figure 1 and their total numbers and kingdom distributions are summarized in Table 3 where subtypes III-C and III-D show strong kingdom biases.
Newly classified Type III subtypes
Subtype III-C corresponds to the Type III systems defined earlier as MTH-326-like Type III14 and archaeal family A.15,16 Eight examples of this subtype were found among archaea, and a further 29 were identified in bacteria (data not shown). Although the overall gene synteny of subtype III-C is similar to that of subtype III-B (Fig. 4), the encoded Cas10 analog (Csx11, TIGR02682) is divergent showing no sequence similarity detectable by conventional sequence alignment and search methods. However, when employing sensitive profile–profile alignments, using HHsearch, we obtained a match with a 98% probability score, between most of the N-terminal half of the protein and Csm1, the Cas10 analog of subtype III-A. In contrast to the earlier report,14 we found that the Cas5 analog of subtype III-C gives a full-length sequence match to Csm4, the Cas5 analog of subtype III-A and that it is not fused to Cas7. Finally, we were unable to detect any sequence similarity between protein S of subtype III-C and Csm2 or Cmr5 of subtypes III-A and III-B, respectively.
Earlier, the SSO1438-type Type III module was documented as being distinct from subtypes III-A and III-B.4,16 With 16 examples in diverse archaea and at least one in bacteria (Rhodothermus), we propose naming this subtype III-D. The Cas10 analog is quite similar to those of subtypes III-A and III-B but subtype III-D is exceptional in carrying a higher number of RAMP genes, where Cas7 has diverged into four or five paralogs, in addition to the presence of the Cas5 analog (Fig. 4). Protein S of subtype III-D yields an almost full-length sequence match to the subtype III-A analog Csm2 using a profile–profile alignment.
An additional group of distinct Type III modules was confined to Sulfolobus sp. genomes and was classified as a variant VIII-1 subtype (Fig. 4). The Cas10 analog has diverged yielding no detectable sequence similarity to other Cas10 proteins using conventional methods, although profile–profile searches reveal that the N-terminal half aligns significantly with Cas10 components of subtypes III-A and III-B (with a probability of 95%). Another small ORF (~100 aa) encoding a putative aspartate protease is present in most, but not all, VIII-1 modules. VIII-1 modules carry two Cas7 paralogs and a single Cas5 analog (Fig. 4). Similarity was detectable along the entire length of protein S and its subtype III-D counterpart using a profile–profile search. Another small ORF (~125 aa) is found in all VIII-1 modules. It is predicted to be mainly α-helical and yields no significant sequence matches in public databases.
A further variant subtype VIII-2 was detected in Ferroglobus placidus that carries a Cas5 and a Cas7 analog together with a small protein (89 aa) and a large protein (659 aa). A similar module also occurs in Thermotoga lettingae, and a profile–profile search with the larger protein gave a full-length match to Csm1 (99% probability). They constitute an interesting example of a minimal Type III system with only a single cas7 gene.
Unclassified CRISPR systems
Several interference modules could not be classified as either Type I or Type III and they were categorised separately as variant CRISPR systems (Table 4). Nine examples of V-1 were found among the Halobacteriales (Hlac_2813–2815 and homologs) with some haloarchaeal strains carrying multiple copies. V-1 exhibits a small three-gene module encoding Cas5 and Cas7 analogs and a third ORF (~300 aa), which may be a Cas8/10 analog.
Another variant V-2 was found in Thermococcus onnurineus (TON_0322–0325). It constitutes a Csf-type interference module, also known as Type U.14 The V-2 interference module contains Cas5, Cas7, and Cas8/10 analogs and an additional small protein and it is encoded adjacent to an adaptation module.
A third variant V-3 exhibits a single protein interference system identified as Cas_Cpf1 on TIGRFAMs.3 It lacks Cas3, Cas5, Cas7, and Cas8 and the interference function appears to be directed by the single protein, reminiscent of Cas9 in bacterial Type II systems except that Cpf1 is only half the size of Cas9 and the two proteins do not appear to share any structural domains. At least 25 examples of V-3 were also detected in bacteria (data not shown).
Non-core proteins specific for CRISPR systems
Genome context analyses revealed that non-core cas genes are often linked to cas gene cassettes of CRISPR-Cas systems and all identified examples of non-core cas-associated genes are marked in Table S1 together with their cognate core cas gene modules. These accessory genes encode a variety of proteins, which include Csx1, Csx3, HerA, and NurA, as reported earlier.3,5,17 Several additional genes are listed in Table 5, some of which are specific to CRISPR-Cas systems, which include the genes encoding Csx1 and sRRM. Other linked proteins, including Cas_RecF, contain structural domains which are also encoded elsewhere but we were able to demonstrate a functional link to CRISPR-Cas systems because sequence analyses revealed that the proteins had coevolved significantly with their partner CRISPR-Cas system. Thus, Cas_recF proteins are more closely related to each other, sequence-wise, than they are to other proteins with recF domains.
Table 5. Accessory proteins encoded by non-core cas genes associated primarily with Type III systems.
| Name | Number | Example | Description |
|---|---|---|---|
| Type III-specific | |||
| Csx1 proper | 65 | TTX_1228 | protein (~450 aa) matching TF Cas_DxTHG and CDD Csx1_III_U models |
| Csx1 superfamily (Csx1s) | 18 | TON_0318 | diverse group of proteins matching Csx1_III_U, including TF Cas02710. Does not include CasR and Csm6 |
| RecF-associated protein (RFas) | 16 | P186_0873 | diverse group of small proteins (~170 aa), always associated with Cas_RecF. |
| Cas_RecF | 12 | P186_0874 | protein with RecF domain found near crenarchaeal Type III systems. Always associated with RFas |
| Cas ABC ATPase (ABC_ATPase) | 11 | YN1551_2137 | protein with ABC ATPase domain found near crenarchaeal Type III systems. Always associated with sRRMs |
| small RRM protein (sRRM) | 12 | YN1551_2138 | diverse group of proteins (~150 aa) containing RRM domains and always accociated with Cas ABC ATPase |
| Aspartic protease (Asp_prot) | 11 | Saci_1895 | small protein (~100 aa) with retropepsin domain, associated with Type III-D and VIII-1 subtypes in Sulfolobales |
| Cmr7 | 7 | SSO1986 | protein (~200 aa) with metalloprotease domain, associated with certain variants of Type III-B subtypes |
| Csx3 | 5 | Mhar_1706 | small (~100 aa) protein matching to the TF Cas_Csx3 model. Unrelated to Csx1 |
| Membrane dipeptidase (Zn2+_pep) | 4 | MJ_1673 | small (~130 aa) protein with Zn2+ dipeptidase domain, associated with some Type III-A systems in Methanococcales |
| Mvol_05XX-fam | 5 | Mvol_0536 | five proteins spanning three families associated with Type III-B systems in Methanococcales. Also found in Clostridium sp. |
| Types I + III-specific | |||
| Cas HerA helicase (HerA) | 14 | Tagg_0815 | always associated with NurA |
| Cas NurA nuclease (NurA) | 14 | Tagg_0814 | always associated with HerA |
| Csx1 minimal (Csx1m) | 13 | Pyrfu_0517 | small (~180 aa) protein containing a minimal Csx1 domain matching TF Cas_NE0113 and CDD Csx1_III_U |
| Csx1 with PIN toxin (Csx1p) | 8 | Metig_1253 | ~400 aa protein consisting of a minimal Csx1 domain fused to a PIN toxin domain |
| Tneu_1160-fam | 3 | Tneu_1160 | large protein of unknown function, associated with CRISPR systems of Thermoproteales |
| SbcC repair ATPase (SbcC) | 3 | CSUB_C0986 | large protein with SbcC domain found in cas gene cassettes of diverse archaea |
| SbcD nuclease (SbcD) | 3 | CSUB_C0987 | protein with SbcD domain, associated with SbcC |
| Type I-specific | |||
| Cas ABC ATPase (ABC_ATPase) | 5 | TTX_1256 | specific for Type I systems but related to corresponding protein for Type III systems |
| small RRM protein (sRRM) | 4 | TTX_1257 | specific for Type I systems but related to corresponding protein for Type III systems |
| Thermococcales specific (see Table S2) |
53 | PF1132–1135 | 11 protein families found in Thermococcales adjacent to Type I systems regardless of subtype |
Most of the accessory proteins are specific to CRISPR systems and are not normally found encoded outside of cas gene cassettes. In addition to the most widespread Csx1-type accessory proteins, numerous accessory proteins have helicase and nuclease domains, which tend to be found associated with DNA replication, recombination, and repair. Protease domains, toxin domains, and RRM domains are also found in many proteins, while some larger proteins have non-identifiable domains.
There was minimal evidence for the presence of non-core cas genes associated specifically with Type I systems (Table S1). Only a few examples of two different proteins were found, one a predicted ABC ATPase and the other containing an RRM domain (Table 5). Exceptionally, several proteins were found associated with Type I systems of the order Thermococcales and they are listed separately in Table S2. The genes encoding these proteins flank CRISPR systems in Thermococcales regardless of their subtype. This may reflect that the CRISPR systems are borne on integrated elements or specialized genomic loci where the flanking genes, although conserved, have no direct functional link with CRISPR activity.
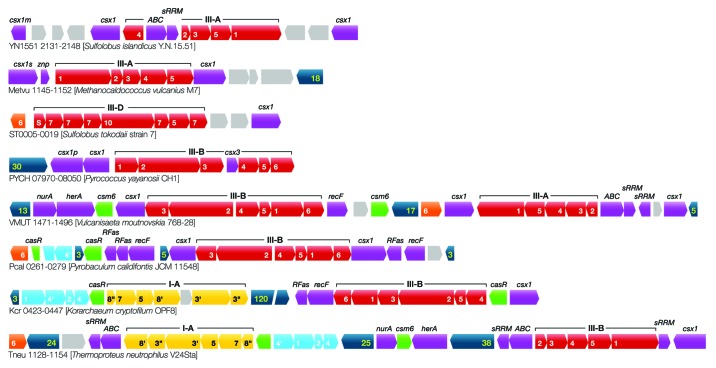
In contrast, analysis of Type III systems yielded several different proteins encoded with interference modules, with some of the genes located in operons of two or three cas genes, consistent with their being co-functional (Table S1). Examples of archaeal CRISPR-Cas gene cassettes carrying interspaced accessory protein genes, sometimes present in multiple copies, are color-coded violet in Figure 5. In total, 18 different accessory proteins were identified, 11 of which were specific for Type III systems, and seven were encoded together with combined Type I + Type III systems (Table 5). Many of the proteins are designated Csx1 and they comprise a large group of disparate proteins exhibiting a wide range of sizes and very diverse sequences but share similar N-terminal domains. Significantly, a Csx1 protein of S. islandicus was recently implicated in influencing the nucleic acid targeting specificity of a subtype III-B interference complex.13
Figure 5. Examples of Type III-A, III-B, and III-D subtypes in archaea from diverse phyla that are associated with multiple and diverse accessory protein genes. Type III gene cassettes are color-coded red and cmr and csm gene labels are given. Type I-A gene cassettes are color-coded for adaptation (light blue) and interference (yellow). Some of the subtype III-B systems are directly linked to subtype I-A systems. All core cas genes are identified by bold numbers. Non-core cas-associated genes are color-coded violet and are labeled by names above the genes (see also Table 5). Genes for putative regulatory proteins (green) and Cas6 (orange) are also indicated together with CRISPR loci (dark blue) where the number indicates the number of repeat-spacer units. In some Type III gene cassettes, the number of accessory protein genes exceeds the number of core cas genes.
The present work did not include an analysis of toxin–antitoxin gene pairs and IS elements that are commonly associated with archaeal CRISPR systems.15 Nor does it cover proteins implicated in modulating CRISPR RNA transcription and processing including Cbp1 of the Sulfolobales18 and Cbp2 of the Thermoproteales,19 which, like RNase III that mediates RNA processing in bacterial Type II CRISPR systems,20 are not linked genomically to CRISPR systems, and are therefore likely to perform additional cellular functions.
CRISPR RNA processing
Cas6 is the primary processing endoribonuclease for all archaeal CRISPR transcripts, which are generally transcribed as single transcripts from within the leader.21,22 The cas6 gene is found either separately, or associated with gene cassettes for adaptation, Type I interference, Type III interference, or combinations thereof (Table S1). Experimental data suggest that the crRNA processing capability of a single Cas6 protein can be shared by different CRISPR systems in the same host.13 Consistent with this finding, we observed that cas6 genes associated with Type III systems are not phylogenetically distinct from those associated with Type I systems. This is visualized on the full archaeal Cas6 dendrogram where Cas6 proteins associated specifically with Type III systems are intermixed with those associated with Type I systems (Fig. S3).
Modular exchange
The genomic organization of archaeal adaptation and interference genes as separate cas gene cassettes is consistent with their constituting separate functional modules.15 Moreover, exchange of adaptation and interference modules between different CRISPR systems appears to have been widespread,15,23 and this is exemplified for two methanoarchaea in Figure 6, where almost identical adaptation modules are associated with interference modules of three different Type I subtypes. To quantify the extent of modular exchange, we analyzed the adaptation and interference module dendrograms (Figs. S1 and S2). Groups of closely related adaptation modules were examined to establish whether their cognate Type I interference modules were also clustered. When locations on the dendrogram were inconsistent, the groups are color-coded in red, indicating that the adaptation modules can function with different interference modules and are, therefore, susceptible to modular exchange. The corresponding interference modules were also color-coded red. In summary, we estimate that about 50% of archaeal Type I systems are subceptible to exchange and that the phenomenon is particularly widespread for subtypes I-B, I-G, I-D, VI-1, and VI-2.
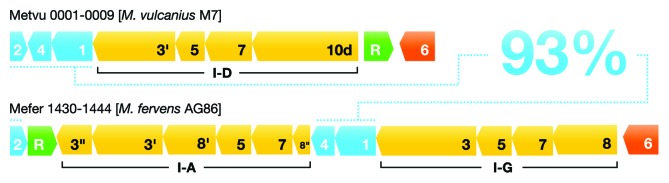
Figure 6. A putative example of modular exchange of Type I adaptation and interference modules. Gene cassettes for adaptation (coded blue) and interference (coded yellow) for Methanocaldococcus vulcanius and Methanocaldococcus fervens genomes are depicted showing the adaptation modules with highly similar sequences (93% concatenated amino acid sequence identity) that are associated with three different subtypes (I-A, I-D, and I-G) of Type I interference modules. Gene contents are indicated by bold numbers within genes. R (green) denotes genes of putative transcriptional regulators and 6 (orange) indicates genes of the Cas6 RNA processing enzyme.
Discussion
In this study we have examined all the currently detectable CRISPR adaptive immune systems in sequenced archaeal genomes and have generated a catalog of different Type I and Type III subtypes found within each archaeon (Table S1). The analyses have focused primarily on the properties of interference modules because of the widespread occurrence of modular exchange and the finding that adaptation modules are relatively highly conserved in protein composition and sequence. While most archaeal Type I CRISPR systems constitute integral genetic units, many interference modules of Type III systems are encoded separately. The results obtained underline the high structural conservation of archaeal adaptation modules and the relatively broad structural diversity of interference modules.
Most archaeal Type I systems fall into a few of the universal subtypes defined earlier,5 albeit using different criteria. In addition, we have assigned some Type III systems to other subtypes III-C, III-D, and VIII-1, the former two of which (families A and D) were described earlier for archaea.16 In their more general classification, Makarova et al.14 employed the signature protein Cas10/Cmr2/Csm1 for classifying Type III systems, but this protein is extremely divergent in subtypes III-C, VIII-1, and VIII-2, and we suspect, therefore, that some Type III systems remain undetected. We successfully used the Pfam RAMP model (PF03787) as a signature for Type III systems because this model is quite specific for Type III Cas7 proteins. Therefore, we predict that employing PF03787 as a universal signature for Type III systems will also lead to the discovery of new Type III subtypes in bacteria.
In late 2011, Makarova et al.14 proposed unifying protein families of the different CRISPR types, Type I Cas and Type III Cmr and Csm, basically inferring that all Type I interference systems were made up of Cas3, Cas5, Cas7, and Cas8, and that Type III interference complexes contained Cas5, Cas7, Cas10, and the small protein component S. This proposal has received strong independent support from a number of recent structural studies. For example, the subtype I-E and subtype III-B interference complexes exhibit some common structural features7,24 that are illustrated schematically in Figure 7. Type I interference modules generally encode a Cas3 helicase, Cas5 RAMP, Cas7 RAMP, and Cas8, and in the subtype I-E complex, Cas5 is embedded in Cas8 to form the base of the complex from which a backbone of multiple Cas7 subunits extends (Fig. 7A). For some subtypes, Cas8 is split into Cas8’ and Cas8,” and Cas3 can also be partitioned. The interference complex of subtype III-B does not carry a Cas3 helicase but it exhibits analogous structural features, where Cas8’ is replaced by Cas10 and Cas8” is exchanged with protein S (Fig. 7B). Moreover, Cas5 is also embedded in Cas10 and the shared Cas7 backbone interfaces with other parts of the complex through divergent Cas7 paralogs, the numbers of which differ between different Type III subtypes (Fig. 7B). Although protein sequence similarities were very difficult to detect by conventional sequence comparison methods, for many predicted homologs,14 using the profile–profile approach employed here we were able to detect and confirm the predicted homologies. The resulting reduction in the number of core Cas protein families considerably facilitated Cas protein identification and annotation of cas genes. It is likely that some additional Cas proteins described in a recent metagenomic study of bacterial CRISPR systems25 could have been assigned within this unified Cas protein classification if more sensitive sequence comparative methods had been employed.
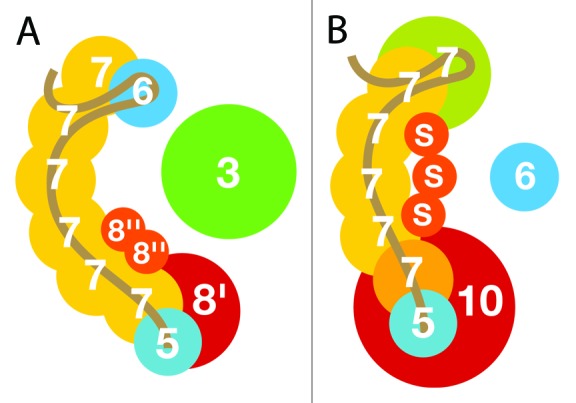
Figure 7. Schematic comparison of (A) a subtype I-E interference complex from E. coli24 with (B) a subtype III-B interference complex from Pyrococcus furiosus.7 The two structures share homologous protein components consistent with the related compositions of their gene cassettes. In (A), the Cas6 protein is considered to be part of the interference complex and Cas3 is essential for the interference mechanism. In (B), Cas 6 has not been shown to be part of the interference complex. Moreover, the two Cas7 proteins in green (Cmr1) and orange Cmr6 (see Fig. 4) are Cas7 paralogs. The estimated binding sites of the crRNAs are color-coded brown.
Extensive evidence supporting the periodic exchange of adaptation and interference modules between different archaeal Type I systems is summarized in Figures S1 and S2 and the data are consistent with earlier evidence presented for archaeal CRISPR modular exchange.15,23 Presumably this widespread phenomenon reflects that the structure and function of the adaptation modules are highly conserved. This property also undermines using the phylogeny of the conserved adaptation protein Cas1 for determining CRISPR subtypes. However, the sole employment of interference modules for classifying archaeal CRISPR systems also carries the reservation that they may function with different types of adaptation modules.
We still have limited insights into the detailed mechanistic and functional diversity of the interference modules of the different Type I and Type III subtypes.9,10,12 Closely related S. islandicus strains were shown earlier to carry multiple and different Type III subtypes,26 which increases the likelihood that their interference mechanisms are functionally diverse. Furthermore, the recent broad unification of core Cas protein families has provided a basis for discerning non-core proteins that are co-functional with CRISPR systems. Several accessory protein families were identified in this study that are exclusively or primarily encoded within, or adjacent to, a limited number of archaeal Type III interference gene cassettes. Two of these, HerA and NurA, were identified earlier for CRISPR systems of different Pyrobaculum species.17 It is likely that some of the proteins, which include putative proteases and ATPases, can modify or extend functions of the associated CRISPR systems. This supposition recently received experimental support from one of two Type III-B systems in S. islandicus REY15A that was demonstrated to share a CRISPR spacer, and a Cas6 RNA processing enzyme, with a Type I-A system, but was functionally dependent on an accessory Csx1 protein.13
Proteins annotated as Csx1 do not comprise a single protein family, but rather a number of diverse protein families exhibiting different sizes and variable domain architectures. What they share is an N-terminal domain of approximately 150 amino acids in length containing a DxTHG motif or variants thereof. This domain, which defines what can be called the Csx1 superfamily of proteins, is also found in the cas transcriptional regulators CasR and Csm6 where it is fused to a C-terminal HTH domain. While it is not clear why such a conserved domain should be fused to a wide range of different proteins, the conserved domain could provide a site for interfacing with Type III and Type I interference modules to modify their activities.
CRISPR-based immune systems probably originated in early primitive cellular structures where exchange of genetic material was likely to have been common.27,28 Moreover, their widespread presence in most archaea and many bacteria suggests that they predated the inferred branching of the bacterial domain, although the bacterial Type II system is likely to have evolved into its present form later. We have discussed earlier the evidence supporting inter-genomic exchange of CRISPR systems between archaea.23 This was considered to be facilitated by the often integral nature of the CRISPR-cas gene cassettes, which are often located in variable genomic regions and are sometimes bordered by transposable elements.16 Nevertheless, the relative similarity of protein components of a few archaeal and bacterial subtypes, combined with their biased distributions, suggests that some inter-domain exchange of CRISPR systems has occurred, despite the presence of formidable genetic barriers.23 For example, the relatively common bacterial subtypes I-C and I-E occur rarely among archaea, and no examples of bacterial subtype I-F, were found (Fig. 1), suggesting that rare transfers from bacteria to archaea may have occurred, and primarily among the euryarchaea.
Crenarchaea, which tend to occupy archaea-rich thermophilic or acidothermophilic environments, are relatively homogeneous in their CRISPR systems. They exhibit predominantly subtypes I-A (90%) and III-B (53%), which suggests that they have undergone very few, if any, transfers of CRISPR systems from other archaeal kingdoms or from bacteria. In contrast, the euryarchaea carry several examples of subtypes I-A, I-B, I-D, and I-G, and subtype III-A (52%) which constitutes the most common Type III system. Euryarchaea frequent a wide range of natural environments and are more diverse phylogenetically than crenarchaea. Moreover, many of their natural habitats are relatively rich in bacteria, possibly rendering their CRISPR systems more susceptible to exchange between archaea and bacteria.
Our aim in this work was to complete a comprehensive analysis of the archaeal CRISPR systems. Here we provide an interactive system, in Table S1, and in Figure S2, which can be used for future analysis and characterization of these systems, including further examination of the different variant systems as well as the possible functional roles of the non-core Cas proteins.
Materials and Methods
Bioinformatical analyses
Genomic loci encoding archaeal adaptation, Type I interference, and Type III interference modules were first identified in the set of 159 complete archaeal genomes by searching for cas1 and cas7 genes using custom HMMs29 and the Pfam30 RAMPs model. The identified cas1 and cas7 genes and the 10 genes flanking them on either side were pooled, and the whole pool was subject to an all-against-all pairwise sequence alignment comparison,31 which was used as an input for Markov clustering32 using custom similarity measures. Each protein family resulting from the Markov clustering was searched against protein family databases CDD,5 COG,4 TIGFAMs,3 and Pfam30 using profile–profile alignments with HHsearch.33 Each genomic locus containing the genes was inspected manually, and using the information from the profile–profile comparison, as well as conventional sequence searches against public databases, cas modules, and cas gene families, were defined. cas cassettes were also defined as consisting of collections of modules (Table S1). Adaptation and interference module dendrograms (Figs. S1 and S2) were created by comparing all protein components of each type of module against corresponding modules from other cassettes. By keeping track of the modules corresponding to each protein component, module-to-module similarity scores were calculated by adding up scores for all constituent proteins. Scores between all modules were inverted and normalized to create a module-to-module distance matrix. The distance matrix was used as input for constructing a neighbor-joining tree,34 which was subsequently mid-point rooted. Variant systems that could not be classified as either Type I or Type III were not included in the interference dendrogram.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Qunxin She is thanked for stimulating discussions and suggestions, and meetings with the German FOR 1680 CRISPR Consortium were both productive and helpful. Michael Terns and Kira Makarova provided insightful and encouraging advice and Johannes Söding's help with the HHsearch profile–profile program was much appreciated. The research was supported by the Danish Natural Science Research Council.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27990
References
- 1.Barrangou R, van der Oost J, eds. CRISPR-Cas systems. Springer press, Heidelberg, 2012; pp. 1-299. [Google Scholar]
- 2.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52:124–34. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52:146–52. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–97. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Rouillon C, Kerou M, Reeks J, Brügger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–99. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 14.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett RA, Vestergaard G, Shah SA. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011;19:549–56. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, Erdmann S, She Q. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans. 2011;39:51–7. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- 17.Bernick DL, Cox CL, Dennis PP, Lowe TM. Comparative genomic and transcriptional analyses of CRISPR systems across the genus Pyrobaculum. Front Microbiol. 2012;3:251. doi: 10.3389/fmicb.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–80. doi: 10.1093/nar/gkr1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenchappa CS, Heidarsson PO, Kragelund BB, Garrett RA, Poulsen FM. Solution properties of the archaeal CRISPR DNA repeat-binding homeodomain protein Cbp2. Nucleic Acids Res. 2013;41:3424–35. doi: 10.1093/nar/gks1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–72. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 23.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Doak TG, Ye Y. Extending the catalog of cas genes with metagenomes. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1262. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo L, Brügger K, Liu C, Shah SA, Zheng H, Zhu Y, Wang S, Lillestøl RK, Chen L, Frank J, et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J Bacteriol. 2011;193:1672–80. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woese CR. Archaebacteria and cellular origins: an overview. Archaebacteria. 1982:1–17. [Google Scholar]
- 28.Woese C. The universal ancestor. . ed. Kandler O.) Gustav Fischer press, Stuttgart Proc Natl Acad Sci U S A. 1998;95:6854–9. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson W. Finding protein and nucleotide similarities with FASTA. Curr Protoc Bioinformatics 2004; Chapter 3:Unit3.9. [DOI] [PubMed] [Google Scholar]
- 32.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonsen M, Mailund T, Pedersen CNS. Inference of large phylogenies using neighbour-joining. CCIS. 2011;127:334–44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.