Abstract
Telomeres are protective nucleoprotein structures at the ends of eukaryotic chromosomes. Despite the heterochromatic state of telomeres they are transcribed, generating non-coding telomeric repeat-containing RNA (TERRA). Strongly induced TERRA transcription has been shown to cause telomere shortening and accelerated senescence in the absence of both telomerase and homology-directed repair (HDR). Moreover, it has recently been demonstrated that TERRA forms RNA–DNA hybrids at chromosome ends. The accumulation of RNA–DNA hybrids at telomeres also leads to rapid senescence and telomere loss in the absence of telomerase and HDR. Conversely, in the presence of HDR, telomeric RNA–DNA hybrid accumulation and increased telomere transcription promote telomere recombination, and hence, delayed senescence. Here, we demonstrate that despite these similar phenotypic outcomes, telomeres that are highly transcribed are not processed in the same manner as those that accumulate RNA–DNA hybrids.
Keywords: TERRA, telomere, senescence, Exo1, RNA-DNA hybrid, R-loop, RNase H
Telomeres are nucleoprotein structures that protect the ends of eukaryotic chromosomes from nucleolytic degradation as well as unscheduled DNA repair activities. The latter may lead to chromosomal aberrations such as chromatid-type end-to-end fusions.1,2 Additionally, telomeres are permissive to telomere lengthening mechanisms, which promote the complete replication of chromosome ends, and thus overcome the end-replication problem.3-5 Whether telomeres can fulfill these functions depends on both their length and structural status. The structure is generally conserved in eukaryotes and in budding yeast consisting of double-stranded, non-nucleosomal TG1–3 repeats that end in a single-stranded 3′ overhang,6,7 which can fold back into the subtelomeric region to form a loop.8,9 These structural features promote the association of protective telomeric proteins and allow telomerase-mediated elongation, respectively.10 In the absence of a specific telomere elongation mechanism, the chromosome ends progressively shorten with every round of replication, eventually leading to telomere dysfunction, checkpoint activation, and cellular senescence. The length of the shortest telomere in the cell serves as a major determinant of the onset of cellular senescence.11,12
There are two mechanistically distinct processes that can prevent telomere attrition. One is telomere elongation by the ribonucleoprotein enzyme telomerase, which is constitutively expressed in yeast. Telomerase adds telomeric repeats to chromosome ends and has a strong preference for short telomeres.13 The majority of human somatic cells, however, are telomerase-negative and experience telomere shortening during the S phase of each cell division cycle.3 To model this scenario in yeast, telomerase-negative cells are used for understanding the effects of telomere shortening on cellular senescence. The second telomere maintenance mechanism (TMM) depends on homology-directed repair (HDR) to elongate telomeres and is referred to as alternative lengthening of telomeres (ALT), which is active in about 10–15% of human cancer cells.14-16 Likewise, in S. cerevisiae there is a telomerase-independent mechanism of telomere elongation that is mediated by HDR. Yeast cells that exclusively employ this mechanism are referred to as survivors.17-19 Their formation in a culture of senescing cells is a rare event and almost always depends on the central recombination gene, RAD52.
Despite the heterochromatic state of telomeres and subtelomeres, they are transcribed and telomeric repeat-containing RNA (TERRA) is generated. TERRA is a long non-coding RNA that is conserved throughout eukaryotes.20,21 It is largely transcribed by RNA polymerase II (RNAPII) and consists of subtelomeric and telomeric sequences.21,22 TERRA expression is regulated in a cell cycle-dependent manner with the lowest levels occurring in S phase, when telomeres get replicated.23 In both humans and yeast, TERRA transcription is responsive to changes in the local chromatin status.24-26 In yeast, there is a further level of regulation through Rat1-mediated degradation.22,26 In human cells, TERRA has been shown to localize to telomeres21 where it may form an integral part of the heterochromatin27,28 and play a role in telomere function.
It was suggested that TERRA might be involved in the inhibition of telomerase activity; a theory supported strongly by in vitro data.29 However, using modified transcriptionally inducible telomeres (tiTELs) in various in vivo model systems it has been demonstrated that TERRA is likely not acting as an inhibitor of telomerase.30-32 In contrast, recent evidence suggests that TERRA may be promoting telomere elongation via both telomerase and HDR depending on whether or not the cells express telomerase.33,34 Here, we focus on recently published data about the role of TERRA in telomere maintenance in senescing cells. We suggest that there may be fundamental differences regarding how telomeres get processed following increased rates of TERRA transcription as opposed to decreased rates of TERRA RNA–DNA hybrid removal from chromosome ends.
Increased Rates of TERRA Transcription Lead to Telomere Processing in an Exo1-Dependent Manner
Using a previously described galactose-inducible upstream-activating sequence (UAS) positioned directly in front of the telomeric repeats on the left arm of chromosome 7 (7L-Gal tiTEL),35 it was shown that induced telomeric transcription causes rapid telomere shortening in cis even in the absence of the catalytically active subunit of telomerase, Est2.30 This additive effect suggested that tiTEL induction is causing telomere shortening for reasons other than telomerase inhibition. Consistently, in the absence of both TMMs (telomerase and HDR), forced transcription of a 7L-Gal tiTEL is sufficient to significantly accelerate rates of cellular senescence (Fig. 1A).30 The tiTEL-induced shortening effect only occurred upon passage through S phase, which indicated a replication-dependent loss of telomeric sequence.
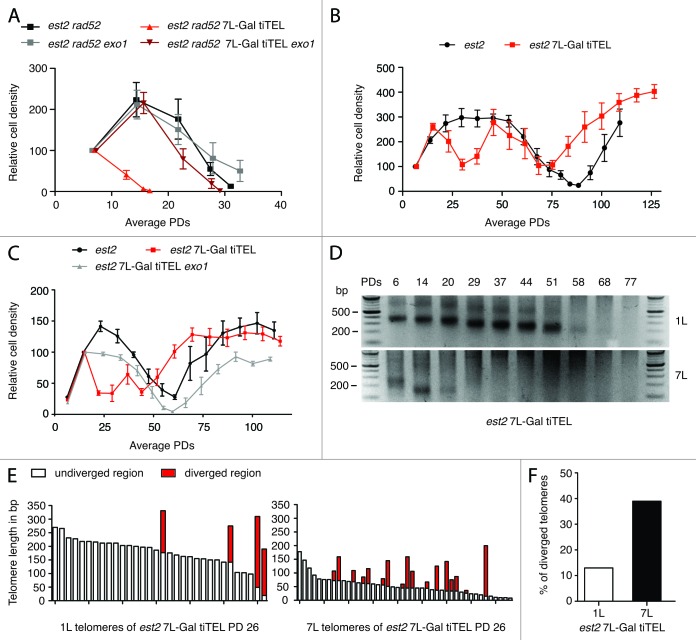
Figure 1. Increased rates of TERRA transcription lead to telomere processing in an Exo1-dependent manner and trigger recombination in HDR-proficient cells. (A–C) Growth curves, following a 24 h dilution protocol.33 Average relative cell density is shown on the y-axis (set arbitrarily to 100) and average PDs on the x-axis. Mean and s.e.m. of six biological replicates are depicted for the indicated mutants. (D) 1L and 7L telomere PCR products of est2 7L-Gal tiTEL cells from (C). (E) Sequencing of 1L and 7L telomere PCR products from est2 7L-Gal tiTEL cells in (B) at PD 26. Open bars represent undiverged telomeres; red bars indicate divergence (indicative of HR). The point of divergence corresponds to where the red bar joins the open bar. (F) Comparison of the telomeric recombination frequency is shown as percent of telomeres with a recombination event (total number of telomeres analyzed for 1L: n = 39 and for 7L: n = 64).
In a parallel study,32 a similar tiTEL-based approach was employed at telomere 1L with the key differences being that (1) the promoter used was doxycycline responsive and (2) full-length TERRA (subtelomeric and telomeric) was induced instead of just the telomeric repeat portion of TERRA as in the case with the 7L-Gal tiTEL. Similar to 7L-Gal tiTEL, it was shown that TERRA transcription from the TetO7-1L tiTEL also resulted in telomere shortening in cis in the absence of telomerase.32 Importantly, this study was able to demonstrate that the observed shortening was due to increased exonuclease 1 (Exo1)-mediated resection at induced TetO7-1L tiTEL. The telomere effects of increased TERRA expression are likely not tiTEL-specific as the deletion of SIR2, which results in increased TERRA transcription, also leads to telomere shortening and premature senescence in the absence of HDR and telomerase.30 It is important to point out that tiTEL induction in human cells did not lead to significant changes in telomere length. This might be due to the fact that transcription induction is less efficient in this system or possibly because small changes in telomere length are more difficult to detect at long human telomeres.31
We set out to determine whether or not the premature senescence phenotype observed in 7L-Gal tiTEL yeast cells was also due to Exo1 activity. In the absence of all TMMs (est2 rad52 cells), we found that the increased rate of cellular senescence caused by 7L-Gal tiTEL induction was completely abolished upon further deletion of EXO1 (Fig. 1A). In order to determine the effects of forced TERRA transcription on telomere recombination, we repeated the above experiments in RAD52+ cells (Fig. 1B). In these conditions, induction of 7L-Gal tiTEL caused cells to lose viability at early population doublings (PDs) but they quickly regained viability before eventually succumbing to senescence as the est2 control cells (Fig. 1B). This rapid drop in viability followed by recovery might be explained by shortening of the 7L-Gal tiTEL via Exo1-dependent resection and subsequent HDR to promote its re-elongation. Indeed, the 3′ overhang created by Exo1 would facilitate the recombination reaction. This idea is also supported by the finding that Rad52 accumulates particularly at the shortest telomere.36 To examine whether the early drop in viability was due to Exo1-mediated resection, the senescence curve was repeated either in the presence or absence of EXO1 (Fig. 1C). Strikingly, the early drop in viability induced by 7L-Gal tiTEL was completely abolished upon deletion of EXO1. To demonstrate that the transient recovery of viability was due to recombination at 7L-Gal tiTEL, we performed telomere PCR in est2 7L-Gal tiTEL cells at the transcribing 7L telomere as well as at natural telomere 1L (Fig. 1D). As expected, 7L-Gal tiTEL shortened much faster than telomere 1L. The 7L-Gal tiTEL PCR signal was lost at a time corresponding to the transient recovery of 7L-Gal tiTEL cells, presumably due to recombination-mediated lengthening of 7L-Gal tiTEL. This early recombination was restricted to the transcribing 7L telomere as the 1L telomere was much longer and the signal was only lost at about 58 PDs when survivors form. Southern blot analysis reinforced that type II survivors formed at PD 58 (data not shown). To confirm that 7L-Gal tiTEL telomeres recombine more frequently, we cloned and sequenced telomeres as previously described.13,33 Indeed, we found that recombination rates were more than three times greater at 7L-Gal tiTEL compared with natural telomere 1L (Fig. 1E and F).
Together, these data demonstrate that increased rates of telomere transcription promote Exo1-mediated resection, which can have differential outcomes depending on the TMM status of the cell. In the absence of both TMMs, transcription-induced end resection leads to rapid telomere loss, and subsequently, premature cellular senescence (Fig. 1A). However, if only telomerase activity is impaired and the cells are recombination competent, the increased resection promotes telomere recombination (Fig. 1C, E, and F). Although the recombination increase is largely restricted to the transcribing telomere, there may also be a trans effect, as it appears that survivors, where all telomeres are engaged in recombination, arise earlier in the 7L-Gal tiTEL strain and also in an Exo1-dependent manner.
It is not entirely clear why highly transcribed telomeres are particularly prone to Exo1 activity. It was suggested that excess TERRA may lead to an inhibition of Ku70/80 dimer as TERRA could be found to co-precipitate with Ku80 in cross-linked yeast extracts.37 Indeed, since Ku70/80 protects telomeres from Exo1-dependent degradation, this is an attractive possibility. An alternative possibility is that TERRA binds to and inhibits the CST complex, another telomere protective protein assembly. Finally, it was shown that increased telomere transcription is able to disrupt the fold-back structure on yeast chromosomes,38 which renders the telomeres more accessible to Exo1.9 It will be interesting to determine which protective structures are perturbed when high levels of TERRA transcription are induced.
Telomeric RNA–DNA Hybrids Lead to Exo1-Independent Telomeric Processing
We postulated that TERRA might be able to form RNA–DNA hybrids with the template strand from which it gets transcribed due to its repetitive and G-rich nature. Co-transcriptionally formed RNA–DNA hybrids are three-stranded nucleic acid structures, which are often referred to as R-loops due to the displaced DNA strand39 (see Fig. 2). It is assumed that R-loops or the RNA polymerase II complex itself can hinder DNA replication by causing stalled and collapsed replication forks.40 These can lead to DNA double strand breaks (DSB) and nucleolytic resection. Moreover, R-loops and replication impairment have been implicated in Rad52-dependent transcription-associated recombination (TAR).41 We and others have recently reported that RNA–DNA hybrids do indeed exist at yeast telomeres.33,37 Furthermore, in the absence of RNase H activity, an enzyme that resolves R-loops by degrading the RNA moiety in an RNA–DNA hybrid, the hybrid levels were significantly increased. Telomeric RNA–DNA hybrid levels were also increased in mutants of the THO complex, which has been implicated in co-transcriptional hybrid removal.33,37 In the complete absence of TMMs (telomerase and HDR), telomere shortening and progressive senescence were enhanced in RNase H and THO mutants. Indeed, this outcome is similar to what we had previously observed for 7L-Gal tiTEL cells in the absence of a TMM (Fig. 1A).30 Strikingly, the accelerated senescence resulting from increased hybrid accumulation at telomeres was not dependent on Exo1-mediated resection,33 as it was seen for the accelerated senescence caused by induced telomere transcription (Fig. 1A).32 These data suggested that high levels of TERRA transcription and increased accumulation of telomeric RNA–DNA hybrids lead to different responses at chromosome ends. Consistently, TERRA levels were not increased in RNase H mutants33 or mutants of the THO complex.37 To determine whether or not a nuclease other than Exo1 may be processing telomeres in RNase H mutants, we performed non-denaturing dot blot experiments to measure single-stranded 3′ telomeric DNA overhangs (Fig. 3). RNase H mutants showed 3-fold increased levels of telomeric ssDNA, which were not reduced if EXO1 was deleted in addition, hinting toward either an Exo1-independent role of resection or incomplete telomeric replication. It will be interesting to determine if resection occurs in RNase H mutants and if it is a result of processing stalled DNA replication forks that have encountered R-loops. Together, these data demonstrate that in the absence of TMMs, increased TERRA transcription and increased R-loops cause telomere dysfunction in mechanistically distinct manners.
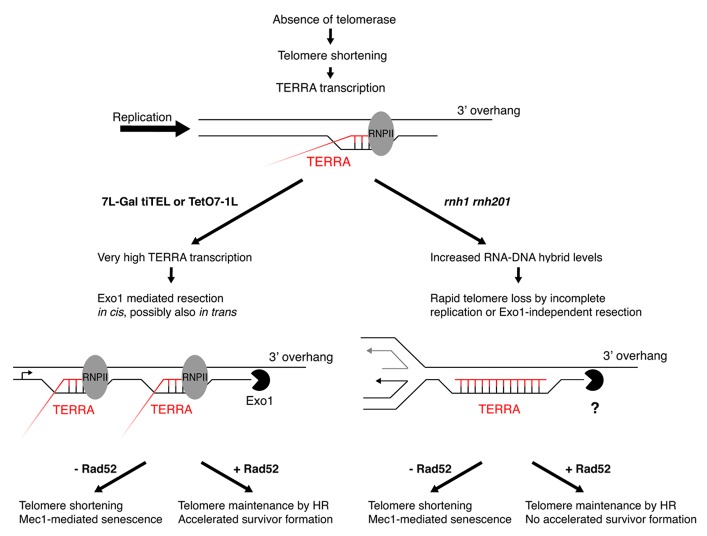
Figure 2. Model: Increased telomeric transcription or RNA–DNA hybrid accumulation cause distinct processing of telomeres. On the one hand (left side), induced TERRA transcription of a single telomere results in very high TERRA transcription that allows Exo1-mediated resection in cis. In the absence of recombination-dependent repair and telomerase, resection leads to rapid telomere shortening and triggers senescence. In recombination-proficient cells, however, the fast telomere shortening can be compensated by recombination processes resulting in telomere maintenance (Fig. 1C and D). Interestingly, strong TERRA transcription might also have an effect in trans, as survivor formation is accelerated if telomere transcription is induced at the tiTEL (Fig. 1C). In absence of recombination, this leads to telomere shortening and senescence, but in presence of recombination, telomere maintenance occurs. In the presence of increased telomeric R-loops (right side) an Exo1-independent overhang is generated, leading to similar phenotypic outcomes as the tiTELs, however, no accelerated survivor formation was detected.33
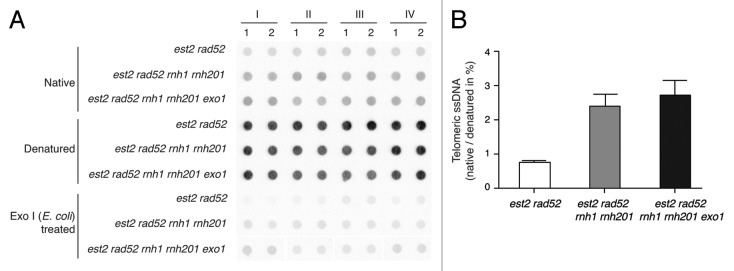
Figure 3. Accumulation of telomeric RNA–DNA hybrids results in an Exo1-independent increase in telomeric 3′ ssDNA overhangs. (A) Genomic DNA was spotted onto a nylon membrane and incubated with a telomeric C-probe to detect ssDNA 3′ telomeric overhang (native) and total telomeric DNA (denatured) as previously described.9 Native DNA was treated with E. coli-derived Exo I to confirmed the 3′ nature of the ssDNA. (B) Single-stranded telomeric DNA is represented as the 3′ telomeric ssDNA signal (native) divided by total telomeric DNA signal (denatured) from two technical (1, 2) and four biological (I–VI) replicates.
In recombination-proficient cells (RAD52+), the accumulation of RNA–DNA hybrids had the completely opposite effect upon telomerase deletion, leading to a delay in senescence and slightly elongated average telomere length, which was due to increased telomere recombination.33 Especially the shortest telomeres were prone to recombination, suggesting that RNA–DNA hybrids promote recombination at short telomeres. Interestingly, short telomeres show increased levels of TERRA transcription, which in the presence of telomerase, promotes its recruitment to these telomeres.34 Our data suggest that in the absence of telomerase, increased TERRA at short telomeres also promotes telomere elongation, but through HDR rather than telomerase. It will be important to determine the physiological relevance of RNA–DNA hybrids and Exo1-mediated resection during HDR- and telomerase-mediated telomere elongation. Indeed, we demonstrated that the overexpression of RNase H was able to promote premature senescence in telomerase-negative cells through the inhibition of HDR. This strongly suggests that hybrids counteract telomere shortening in pre-senescent cells by promoting HDR.
Increased Telomeric Transcription or RNA-DNA Hybrid Accumulation Cause Distinct Processing of Telomeres
In summary, we demonstrate that telomere shortening by induced tiTEL transcription is dependent on Exo1-mediated resection, whereas Exo1 activity did not contribute to the accelerated senescence in RNase H mutants. Moreover, when transcription at the tiTEL (TetO7-1L) was induced, RNA–DNA hybrid levels were not increased significantly.37 One explanation for this would be that hybrids are continuously removed by the next incoming transcription complex at the heavily transcribing tiTEL. Alternatively, extensive resection of the 5′ end by Exo1 at these short telomeres might interfere with hybrid formation on the C- strand.
We propose a model (Fig. 2) in which tiTEL transcription and impaired TERRA hybrid displacement from the telomere by RNase H have similar phenotypic outcomes but the underlying mechanisms differ. The results described above have focused on telomerase-negative senescing cells, but it was also shown that TERRA has a role in telomerase-positive cells by helping to recruit telomerase to the telomere from which TERRA originated.34 Therefore, TERRA seems to promote telomere elongation in both telomerase-negative and telomerase-positive cells via HDR and telomerase-mediated elongation, respectively. Deciphering how TERRA influences telomere processing in these different genetic contexts will be an important future goal.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge C. Azzalin and A. Maicher for critical reading of this manuscript. Balk B was supported by a Landesgraduiertenförderung fellowship. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 1036) to Luke B and the Netzwerk Alters-Forschung (Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg) to Luke B.
Glossary
Abbreviations:
- TERRA
telomeric repeat-containing RNA
- HDR
homology-directed repair
- TMM
telomere maintenance mechanism
- ALT
alternative lengthening of telomeres
- HR
homologous recombination
- RNAPII
RNA polymerase II
- tiTEL
transcriptionally inducible telomere
- UAS
upstream activating sequence
- PD
population doubling
- DSB
double strand break
- TAR
transcription associated recombination
- ssDNA
single-stranded DNA
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27798
References
- 1.Sfeir A. Telomeres at a glance. J Cell Sci. 2012;125:4173–8. doi: 10.1242/jcs.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–52. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–25. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 4.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 5.Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–4. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 6.Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-V. [DOI] [PubMed] [Google Scholar]
- 7.Wright JH, Gottschling DE, Zakian VA. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 8.de Bruin D, Zaman Z, Liberatore RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–13. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- 9.Poschke H, Dees M, Chang M, Amberkar S, Kaderali L, Rothstein R, Luke B. Rif2 promotes a telomere fold-back structure through Rpd3L recruitment in budding yeast. PLoS Genet. 2012;8:e1002960. doi: 10.1371/journal.pgen.1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdallah P, Luciano P, Runge KW, Lisby M, Géli V, Gilson E, Teixeira MT. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11:988–93. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enomoto S, Glowczewski L, Berman J. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2626–38. doi: 10.1091/mbc.02-02-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–35. doi: 10.1016/S0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 14.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–4. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 15.Reddel RR. The role of senescence and immortalization in carcinogenesis. Carcinogenesis. 2000;21:477–84. doi: 10.1093/carcin/21.3.477. [DOI] [PubMed] [Google Scholar]
- 16.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 17.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–9. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 18.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–43. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 19.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–10. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 22.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–77. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol. 2010;30:4808–17. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–94. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–89. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 26.Iglesias N, Redon S, Pfeiffer V, Dees M, Lingner J, Luke B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011;12:587–93. doi: 10.1038/embor.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat Struct Mol Biol. 2012;19:948–56. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- 28.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–13. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maicher A, Kastner L, Dees M, Luke B. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012;40:6649–59. doi: 10.1093/nar/gks358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farnung BO, Brun CM, Arora R, Lorenzi LE, Azzalin CM. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS One. 2012;7:e35714. doi: 10.1371/journal.pone.0035714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer V, Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet. 2012;8:e1002747. doi: 10.1371/journal.pgen.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 34.Cusanelli E, Romero CA, Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell. 2013;51:780–91. doi: 10.1016/j.molcel.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci U S A. 1994;91:12061–5. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Géli V, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol. 2009;11:980–7. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
- 37.Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013;32:2861–71. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol. 2000;20:7991–8000. doi: 10.1128/MCB.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Gómez-González B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, Foiani M, Aguilera A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–19. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaillard H, Herrera-Moyano E, Aguilera A. Transcription-associated genome instability. Chem Rev. 2013;113:8638–61. doi: 10.1021/cr400017y. [DOI] [PubMed] [Google Scholar]