Abstract
Background
The role of fomites and the environment in nosocomial infections is becoming widely recognized. In this paper we discuss the use of Cupron copper oxide impregnated non-porous solid surface in the hospital setting and present in vitro testing data via USA Environmental Protection Agency (EPA) approved testing protocols that demonstrate the efficacy of these products to assist in reduction in environmental contamination and potentially nosocomial infections.
Results
The two countertops tested passed all the acceptance criteria by the EPA (>99.9% kill within 2 hours of exposure) killing a range of bacterial pathogens on the surface of the countertops even after repeated exposure of the countertops to the pathogen, and multiple wet and dry abrasion cycles.
Conclusions
Cupron enhanced EOS countertops thus may be an important adjunct to be used in hospital settings to reduce environmental bioburden and potentially nosocomial infections.
Keywords: Copper oxide, EPA registered, Antimicrobial surface, Environmental bioburden
Background
Hospital Acquired Infections (HAI) have exacted a heavy toll worldwide with over 2 million patients annually contracting an infection in the US [1], being one of the leading causes of death in the US behind cancer and strokes [2]. In Europe, out of 3 million HAI [3] approximately 50,000 resulted in death [4], and in Australia more than 177,000 HAI occur per year [5] whilst in the province of Quebec, Canada the rate of HAI are estimated to be around 11% [6]. The HAI rates in developing countries are significantly higher [7-9]. According to the USA Center for Disease Control (CDC) some of the predominant HAI organisms are Staphylococcus aureus, Pseudomonas aeruginosa, and Enterobacter species [10]. Methicillin resistant S. aureus accounts for 50% of HAI associated with multidrug resistant pathogens [10]. The Extended Prevalence of Infection in Intensive Care (EPIC II) study demonstrated a 50% HAI rate in ICU patients sampled from over 75 countries and two of the most predominant organisms were resistant Staphylococci and P. aeruginosa[11]. HAI are associated with considerable mortality, morbidity and costs [2,12]. Recent intervention efforts including improvement of national surveillance, use of aggressive antibiotic control programs, healthcare staff education for improved hygiene, isolation of infected patients, use of disposable equipment, cleaning and disinfection of environmental surfaces and equipment, improvement of cleaning equipment and sanitary facilities, increase in nursing and janitorial resources and better nutrition [13-17], have been shown to reduce HAI rates. However further supplemental interventions are required. The link between contaminated hard surfaces to HAI has been demonstrated [18-28] and an antimicrobial protected touch surface would assist in reduction of pathogen buildup upon touch surfaces as long as that activity can be indisputably demonstrated.
Thus, our objective was to demonstrate the efficacy of two solid surface countertops incorporating copper (I) oxide in killing a range of pathogens according to protocols that were approved by the USA Environmental Protection Agency (EPA).
Methods
Bacterial strains
The following bacterial strains were tested: Staphylococcus aureus (ATCC 6538); Enterobacter aerogenes (ATCC 13048); Pseudomonas aeruginosa (ATCC 15442); Methicillin resistant Staphylococcus aureus (MRSA)(ATCC 33592); and Escherichia coli 0157:H7 (ATCC 35150).
Materials
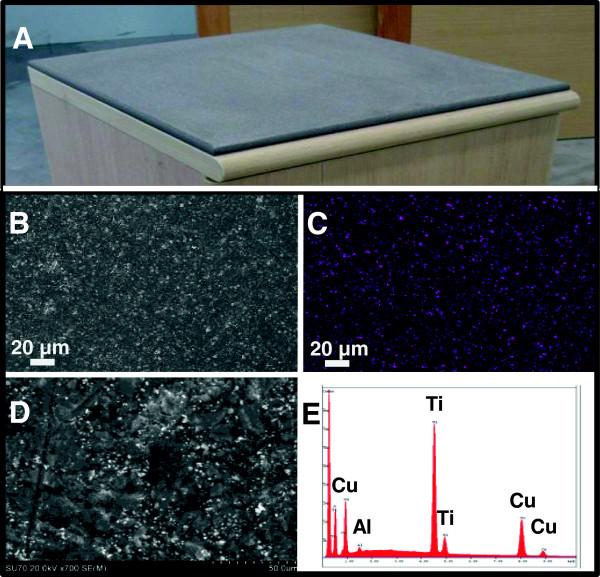
The studied countertops were composed of homogenous blends of polyester, acrylic alloys and fillers, inert pigment and dyes, with (test samples) or without (control samples) Cupron’s 16% copper (I) oxide weight/weight. Three and two separate manufacturing lots of the test and control countertop samples were tested, respectively. A total of 1500 pieces, cut into one inch by one inch squares (Figure 1), 300 per each manufacturing lot, were tested. The countertops were examined by Scanning Electron Microscopy (SEM) and Energy-dispersive X-ray spectroscopy (EDS) by using Hitachi FE-SEM SU-70. The Cupron Enhanced EOS Surface is a novel polymeric solid surface that has all the properties of a solid surface including hardness, firmness, and the ability to be easily cleaned and shaped or fashioned with the antimicrobial ability of copper. The surface can be easily refinished and repaired in the event of damage or aesthetic appeal. The surfaces are currently available in two color choices due to the addition of pigments to alter the color of the surfaces at the time of manufacture. The surface is produced by mixing a blend of acrylic and polyester resins with copper oxide and pigments, which is then heated until liquified and poured into casting molds. The material is allowed to cure allowing the polymerization of the material to produce a solid surface which can then be cut and shaped to produce a final product or installed surface.
Figure 1.
SEM pictures and EDS analysis of a representative countertop containing copper oxide particles. A. A representative picture of a tested countertop impregnated with copper oxide; B. A SEM imaging of the Countertop (white dots indicating copper oxide particles; C. EDS imaging of the Countertop (purple dots indicating the copper oxide); D. cut through SEM imaging of the Countertop (white dots indicating copper oxide particles); and E. corresponding EDS spectra of D, showing a peak corresponding to copper.
Biocidal testing protocols
The biocidal testing of the countertops was conducted by an independent laboratory, MicroBiotest, a division of Microbac Laboratories, Inc. Sterling, VA, using Good Laboratory Practice (GLP) according to protocols pre-approved by the USA EPA.
Protocol 1- sanitizer activity
The carriers were cleaned with 70% isopropyl alcohol, rinsed with deionized water, and allowed to air dry. After steam sterilization for 15 minutes at 121°C, each carrier was placed into a plastic Petri dish matted with two pieces of filter paper using sterile forceps. Two or three lots of manufacturing of each test material were tested per microorganism. Five replicates of each material were used in each test. Organisms from stock cultures were transferred to Tryptic Soy Broth and incubated for 24 hours at 35-37°C (25-30°C for ATCC 13048). Two loopfuls of culture were transferred consecutively daily for three days for the inoculation stocks and the pellicle of bacteria were aspirated. Daily transfers were done for at least 3 consecutive days but for no more than 10 days. To this culture 0.25 ml of heat-inactivated fetal bovine serum (FBS) and 0.05 ml of Triton X-100 were added to 4.7 ml bacteria suspension to yield 5% FBS and 0.01% Triton X-100 organic soil load. The challenge microorganism titer was determined by serially diluting a final 48 hour culture using phosphate buffered solution (PBS) and selected dilutions were plated in duplicate using Tryptic Soy Agar (TSA) pour plates. Carriers were inoculated with 0.02 ml of the 48 hour culture. The bacterial inoculum per experiment is detailed in Table 1. All control plates were incubated in parallel to the test plates. The inoculum was spread to within ~1/8 inch of the control or test carrier before air drying for 20–40 minutes at 35-37°C and 38-42% relative humidity. After 120 minutes exposure at 21°C, the carriers were transferred to 20 ml neutralizer solution (2x Letheen broth [29]) and sonicated for 5 minutes and rotated to mix. Within one hour serial dilutions (10−1 to 10−4) were made in PBS and plated using TSA and incubated for 48 hours at 35-37°C for colony observation and enumeration, taking into account also the 20 fold dilution used to retrieve the bacteria from the carriers. The following controls were performed: culture purity control - each prepared culture was streaked using TSA for purity control; organic soil purity control - duplicate 1 ml aliquots of organic soil were plated in TSA pour plates for sterility control; neutralizer sterility control – a jar containing the neutralizer was incubated with the test plates and observed for growth or no growth; carrier sterility control – an uninoculated test (per lot) and control carrier was put in independent jars containing the neutralizer, incubated and observed for growth or no growth; carrier viability control – for each challenge microorganism, a single inoculated control carrier was subcultured in a jar containing the neutralizer, incubated and the neutralizer observed for growth or no growth; and neutralization confirmation control – for each challenge microorganisms, per lot of the test article, a single sterile test carrier was put in individuals jars containing 20 ml of the neutralizer. To each jar a 1 ml aliquot of the diluted inoculum was added to reach ~100 colony forming units (CFU)/ml in the neutralizer. The jar was mixed and the 1 ml inoculum was removed and plated in duplicate.
Table 1.
CFU recovered from control samples
|
Microorganism |
Protocol |
Replicate |
Inoculum |
Titers recovered |
Average |
|---|---|---|---|---|---|
| CFU/carrier | CFU/carrier | CFU/carrier | |||
|
S. aureus |
1 |
1 |
0.22 × 108 |
8.4 × 105 |
7.5 × 105 |
| |
|
2 |
|
7.0 × 105 |
|
| |
|
3 |
|
7.2 × 105 |
|
| |
2 - initial |
1 |
0.42 × 108 |
1.1 × 106 |
1.3 × 106 |
| |
|
2 |
|
1.6 × 106 |
|
| |
|
3 |
|
1.2 × 106 |
|
| |
|
4 |
|
1.3 × 106 |
|
| |
2 - Final |
1 |
0.46 × 108 |
9.9 × 105 |
1.1 × 106 |
| |
|
2 |
|
1.1 × 106 |
|
| |
|
3 |
|
1.3 × 106 |
|
| |
|
4 |
|
1.0 × 106 |
|
| |
3 - 2 hours |
1 |
0.38 × 108 |
8.2 × 105 |
9.3 × 105 |
| |
|
2 |
|
1.0 × 106 |
|
| |
|
3 |
|
9.4 × 105 |
|
| |
3 - 6 hours |
1 |
|
2.0 × 106 |
1.8 × 106 |
| |
|
2 |
|
1.8 × 106 |
|
| |
|
3 |
|
1.7 × 106 |
|
| |
3 - 12 hours |
1 |
|
2.5 × 106 |
2.5 × 106 |
| |
|
2 |
|
2.4 × 106 |
|
| |
|
3 |
|
3.7 × 106 |
|
| |
3 - 18 hours |
1 |
|
3.7 × 106 |
3.6 × 106 |
| |
|
2 |
|
3.6 × 106 |
|
| |
|
3 |
|
3.5 × 106 |
|
| |
3 - 24 hours |
1 |
|
4.6 × 106 |
4.6 × 106 |
| |
|
2 |
|
4.6 × 106 |
|
| |
|
3 |
|
4.5 × 106 |
|
|
E. aerogenes |
1 |
1 |
0.38 × 109 |
7.4 × 106 |
7.9 x106 |
| |
|
2 |
|
8.8 × 106 |
|
| |
|
3 |
|
7.6 × 106 |
|
| |
2 - initial |
1 |
0.22 × 109 |
1.1 × 106 |
1.1 × 106 |
| |
|
2 |
|
1.0 × 106 |
|
| |
|
3 |
|
1.2 × 106 |
|
| |
|
4 |
|
1.1 × 106 |
|
| |
2 - Final |
1 |
0.4 × 109 |
1.5 × 106 |
1.2 × 106 |
| |
|
2 |
|
1.4 × 106 |
|
| |
|
3 |
|
8.3 × 105 |
|
| |
|
4 |
|
1.1 × 106 |
|
| |
3 - 2 hours |
1 |
0.38 × 109 |
2.0 × 106 |
2.0 × 106 |
| |
|
2 |
|
2.1 × 106 |
|
| |
|
3 |
|
2.0 × 106 |
|
| |
3 - 6 hours |
1 |
|
3.8 × 106 |
3.9 × 106 |
| |
|
2 |
|
3.9 × 106 |
|
| |
|
3 |
|
3.9 × 106 |
|
| |
3 - 12 hours |
1 |
|
5.1 × 106 |
4.7 × 106 |
| |
|
2 |
|
5.4 × 106 |
|
| |
|
3 |
|
3.6 × 106 |
|
| |
3 - 18 hours |
1 |
|
4.8 × 106 |
5.6 × 106 |
| |
|
2 |
|
6.8 × 106 |
|
| |
|
3 |
|
5.2 × 106 |
|
| |
3 - 24 hours |
1 |
|
8.5 × 106 |
7.9 × 106 |
| |
|
2 |
|
8.4 × 106 |
|
| |
|
3 |
|
6.8 × 106 |
|
| MRSA |
1 |
1 |
0.38 × 109 |
7.4 × 105 |
8.5 × 105 |
| |
|
2 |
|
9.8 × 105 |
|
| |
|
3 |
|
8.2 × 105 |
|
| |
2 - initial |
1 |
0.36 × 108 |
7.3 × 105 |
7.5 × 105 |
| |
|
2 |
|
9.5 × 105 |
|
| |
|
3 |
|
6.6 × 105 |
|
| |
|
4 |
|
6.5 × 105 |
|
| |
2 - Final |
1 |
0.32 × 108 |
5.6 × 105 |
6.9 × 105 |
| |
|
2 |
|
5.7 × 105 |
|
| |
|
3 |
|
8.0 × 105 |
|
| |
|
4 |
|
8.2 × 105 |
|
| |
3 - 2 hours |
1 |
0.26 × 108 |
4.0 × 105 |
4.0 × 105 |
| |
|
2 |
|
3.8 × 105 |
|
| |
|
3 |
|
4.2 × 105 |
|
| |
3 - 6 hours |
1 |
|
8.6 × 105 |
8.8 × 105 |
| |
|
2 |
|
9.8 × 105 |
|
| |
|
3 |
|
7.9 × 105 |
|
| |
3 - 12 hours |
1 |
|
9.9 × 105 |
1.0 × 106 |
| |
|
2 |
|
1.2 × 106 |
|
| |
|
3 |
|
9.1 × 105 |
|
| |
3 - 18 hours |
1 |
|
1.8 × 106 |
1.7 × 106 |
| |
|
2 |
|
1.6 × 106 |
|
| |
|
3 |
|
1.7 × 106 |
|
| |
3 - 24 hours |
1 |
|
1.8 × 106 |
1.8 × 106 |
| |
|
2 |
|
1.8 × 106 |
|
| |
|
3 |
|
1.7 × 106 |
|
|
P. aeruginosa |
1 |
1 |
0.2 × 108 |
6.8 × 106 |
7.0 × 106 |
| |
|
2 |
|
7.4 × 106 |
|
| |
|
3 |
|
6.9 × 106 |
|
| |
2 - initial |
1 |
0.2 × 109 |
1.0 × 106 |
1.3 × 106 |
| |
|
2 |
|
1.4 × 106 |
|
| |
|
3 |
|
1.4 × 106 |
|
| |
|
4 |
|
1.5 × 106 |
|
| |
2 - Final |
1 |
0.34 × 109 |
2.4 × 106 |
2.0 × 106 |
| |
|
2 |
|
1.9 × 106 |
|
| |
|
3 |
|
1.6 × 106 |
|
| |
|
4 |
|
2.0 × 106 |
|
| |
3 - 2 hours |
1 |
0.3 × 109 |
2.6 × 105 |
2.5 × 105 |
| |
|
2 |
|
2.5 × 105 |
|
| |
|
3 |
|
2.5 × 105 |
|
| |
3 - 6 hours |
1 |
|
5.2 × 105 |
5.2 × 105 |
| |
|
2 |
|
5.3 × 105 |
|
| |
|
3 |
|
5.3 × 105 |
|
| |
3 - 12 hours |
1 |
|
7.2 × 105 |
7.2 × 105 |
| |
|
2 |
|
7.1 × 105 |
|
| |
|
3 |
|
7.4 × 105 |
|
| |
3 - 18 hours |
1 |
|
9.8 × 105 |
9.6 × 105 |
| |
|
2 |
|
9.5 × 105 |
|
| |
|
3 |
|
9.6 × 105 |
|
| |
3 - 24 hours |
1 |
|
9.8 × 105 |
9.7 × 105 |
| |
|
2 |
|
9.2 × 105 |
|
| |
|
3 |
|
1.0 × 106 |
|
|
E. coli O157:H7 |
1 |
1 |
0.36 × 109 |
6.4 × 106 |
6.6 × 106 |
| |
|
2 |
|
6.7 × 106 |
|
| |
|
3 |
|
6.6 × 106 |
|
| |
2 - initial |
1 |
0.24 × 109 |
1.5 × 106 |
1.1 × 106 |
| |
|
2 |
|
8.3 × 106 |
|
| |
|
3 |
|
1.1 × 106 |
|
| |
|
4 |
|
1.1 × 106 |
|
| |
2 - Final |
1 |
0.22 × 109 |
7.8 × 105 |
9.4 × 105 |
| |
|
2 |
|
8.0 × 105 |
|
| |
|
3 |
|
1.2 × 106 |
|
| |
|
4 |
|
9.9 × 105 |
|
| |
3 - 2 hours |
1 |
0.36 × 109 |
2.5 × 105 |
2.6 × 105 |
| |
|
2 |
|
2.6 × 105 |
|
| |
|
3 |
|
2.7 × 105 |
|
| |
3 - 6 hours |
1 |
|
5.2 × 105 |
5.3 × 105 |
| |
|
2 |
|
5.2 × 105 |
|
| |
|
3 |
|
5.4 × 105 |
|
| |
3 - 12 hours |
1 |
|
7.9 × 105 |
7.7 × 105 |
| |
|
2 |
|
7.7 × 105 |
|
| |
|
3 |
|
7.6 × 105 |
|
| |
3 - 18 hours |
1 |
|
1.0 × 106 |
1.0 × 106 |
| |
|
2 |
|
1.1 × 106 |
|
| |
|
3 |
|
1.0 × 106 |
|
| |
3 - 24 hours |
1 |
|
1.2 × 106 |
1.2 × 106 |
| |
|
2 |
|
1.2 × 106 |
|
| 3 | 1.2 × 106 |
Protocol 2- residual sanitizer activity
A sanitization test was followed as described above (Protocol 1) using 4 replicates per material. Post this initial test a Gardner apparatus was used to simulate surface wear of the test and control samples. The abrasion tester was used at a speed of 2.25 to 2.5 for a total contact time of 4–5 seconds for one complete cycle. A wear cycle equals one pass to the left and a return pass to the right. After a minimum of 15 minutes after the wear cycle each carrier was reinoculated as described above and dried for a minimum of 30 minutes. After each set of surface wear, absolute ethanol was used to sterilize the apparatus and the foam liner and cotton cloth were changed after each wear test. Wet cycles and dry cycles were alternated and for wet wear cycles the boat assembly included a new foam liner and dry cotton cloth sprayed with sterile deionized water using a preval sprayer from a distance of 75±1 cm for not more than one second. At least 24 hours passed between the initial inoculation and final sanitizer. Overall 12 wear cycles were completed before sanitizer activity was assessed using the method outlined above. All the controls as outlined for Protocol 1 were performed.
Protocol 3- continuous bacterial reduction
A sanitization test was followed as described above (Protocol 1) using 5 replicates per each material tested. The carriers were consecutively inoculated for 8 times by adding the challenge microorganism at 0, 3, 6, 9, 12, 15, 18 and 21 hours. Efficacy was assessed at 2, 6, 12, 18 and 24 hours, which corresponds to 1, 2, 4, 6, and 8 inoculations. After exposure the carriers were transferred to a neutralizer solution and sonicated and rotated to mix. Within one hour, serial dilutions (10−1 to 10−4) were spread on plates using appropriate media and incubated for 48 hours for colony observation and enumeration. All the controls as outlined for Protocol 1 were performed.
Results
The challenge microorganisms were confirmed for purity by Gram stain and colony morphology. Controls demonstrated that the organic soil, carrier and neutralizing medium were sterile. The neutralizing solution itself did not show any bacterial inhibition. The bacterial titers (actual CFU after taking into consideration the relevant dilutions) recovered from the control samples following the different protocols, which included air drying, sonication, and recovering the bacteria from the exposed carrier, are summarized in Table 1. Tables 2, 3 and 4 show the bacterial titers recovered from the Test samples following the various protocols and the percent reduction in the bacterial titers recovered as compared to the mean bacterial titers recovered from the control samples. All experiments conducted with the copper oxide impregnated countertops demonstrated over a 3 log (>99.9%) reduction against all organisms tested, as compared to the control countertops without copper (Tables 2, 3 and 4). Out of the 192 data points obtained (average of 4 or 5 replicates each) for the test countertops, there were only two exceptions for the continuous sanitizer activity test - with a 99.8% and 99.2% reductions against Pseudomonas aeruginosa (Table 4), which exceeds the 99% reduction requirement set up by the EPA for continuous efficacy kill rates. As determined by SEM and EDS analysis, copper oxide particles are homogenously distributed within (Figure 1D and E) and throughout the surface (Figure 1B and C) of the test countertops.
Table 2.
Results from Protocol 1- Sanitizer Activity
| Countertop | Organism | CFU/ Recovered from control samples* | Lot | CFU recovered from test samples | % reduction ** |
|---|---|---|---|---|---|
| Test 1 |
S. aureus |
7.5 × 105 |
1 |
<1;<1;5;<1;<1 |
>99.9 |
| 2 |
18;<1;11;<1;22 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
7.9 × 106 |
1 |
1200;790;50;1200;<1 |
>99.9 |
|
| 2 |
760;840;1200;800;620 |
>99.9 |
|||
| 3 |
<1;620;<1;500;<1 |
>99.9 |
|||
|
MRSA |
8.5 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
7 × 106 |
1 |
90;580;<1;50;160 |
>99.9 |
|
| 2 |
440;<1;400;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
6.6 × 106 |
1 |
470;690;450;480;380 |
>99.9 |
|
| 2 |
560;360;320;390;360 |
>99.9 |
|||
| Test 2 |
S. aureus |
7.5 × 105 |
1 |
<1;50;<1;80;<1 |
>99.9 |
| 2 |
280;<1;70;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
7.9 × 106 |
1 |
70;540;140;650;120 |
>99.9 |
|
| 2 |
240;750;240;460;410 |
>99.9 |
|||
| 3 |
770;610;410;230;450 |
>99.9 |
|||
|
MRSA |
8.5 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
7.0 × 106 |
1 |
820;740;600;880;890 |
>99.9 |
|
| 2 |
930;840;730;870;990 |
>99.9 |
|||
| E. coli 0157:H7 | 6.6 × 106 | 1 |
640;720;300;700;700 |
>99.9 |
|
| 2 | 660;540;490;760;300 | >99.9 |
*Values taken from Table 1.
**Compared to control, each number represents an average of 5 replicates per manufacturing lot. Either 2 or 3 lots were examined per organism.
Table 3.
Results from protocol 2- residual sanitizer efficacy
| Countertop | Organism | CFU recovered from control samples* | Lot | CFU recovered from test samples | % reduction** |
|---|---|---|---|---|---|
| Test 1- Initial |
S. aureus |
1.3 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
E. aerogenes |
1.1 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| 3 |
390;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
MRSA |
7.5 × 105 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
P. aeruginosa |
1.3 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;90 |
>99.9 |
|||
|
E. coli 0157:H7 |
1.1 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| Test 1- Final |
S. aureus |
1.1 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
E. aerogenes |
1.2 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
MRSA |
6.9 × 105 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
P. aeruginosa |
2.0x106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
E. coli 0157:H7 |
9.4 × 105 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| Test 2- Initial |
S. aureus |
1.3 × 106 |
1 |
4.5;<1.5;<1.5;<1.5 |
>99.9 |
| 2 |
<1.5;<1.5;<1.5;200 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;240 |
>99.9 |
|||
|
E. aerogenes |
1.1 × 106 |
1 |
<1.5;60;180;<1.5 |
>99.9 |
|
| 2 |
9;150;420;<1.5 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
MRSA |
7.6 × 105 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
P. aeruginosa |
1.3 × 106 |
1 |
150;<1.5;9;230 |
>99.9 |
|
| 2 |
450;570;<1.5;<1.5 |
>99.9 |
|||
|
E. coli 0157:H7 |
1.1 × 106 |
1 |
<1.5;60;180;<1.5 |
>99.9 |
|
| 2 |
90;150;420;<1.5 |
>99.9 |
|||
| Test 2- Final |
S. aureus |
1.1 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
| 2 |
<1.5;330;<1.5;<1.5 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
E. aerogenes |
1.2 × 106 |
1 |
380;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;320 |
>99.9 |
|||
| 3 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
MRSA |
6.9 × 105 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
|
P. aeruginosa |
2.0 × 106 |
1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|||
| E. coli 0157:H7 | 9.4 × 105 | 1 |
<1.5;<1.5;<1.5;<1.5 |
>99.9 |
|
| 2 | <1.5;<1.5;<1.5;<1.5 | >99.9 |
*Values taken from Table 1.
**Compared to control, each number represents an average of 4 replicates per manufacturing lot. Either 2 or 3 lots were examined per organism.
Table 4.
Results from protocol 3- continuous self sanitizing activity
| Countertop | Organism | CFU recovered from control samples | Lot | CFU recovered from test samples | % reduction** |
|---|---|---|---|---|---|
| Test 1–2 hours |
S. aureus |
9.3 × 105 |
1 |
220;340;500;670;290 |
>99.9 |
| 2 |
420;270;290;320;220 |
>99.9 |
|||
| 3 |
380;420;340;290;270 |
>99.9 |
|||
|
E. aerogenes |
2.0 × 106 |
1 |
11;220;<1<1<1 |
>99.9 |
|
| 2 |
<1;100;220;<1;<1 |
>99.9 |
|||
| 3 |
80;40;170;80 |
>99.9 |
|||
|
MRSA |
4.0 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
2.5 × 105 |
1 |
480;370;480;180;120 |
99.9 |
|
| 2 |
420;480;240;450;360 |
99.8 |
|||
|
E. coli 0157:H7 |
2.6 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
140;<1;<1;<1;150 |
99.9 |
|||
| Test 1–6 hours |
S. aureus |
1.8 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
3.9 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
8.8 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
5.2 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;170;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
5.3 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 1–12 hours |
S. aureus |
2.5 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
4.7 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
1.0 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
7.2 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
7.7 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 1–18 hours |
S. aureus |
3.6 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
5.6 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
1.7 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
9.6 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
1.0 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 1–24 hours |
S. aureus |
4.6 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
7.9 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
1.8 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
9.7 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
1.2 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 2–2 hours |
S. aureus |
9.3 × 105 |
1 |
760;580;770;730;550 |
>99.9 |
| 2 |
780;770;520;540;460 |
>99.9 |
|||
| 3 |
480;420;420;450;410 |
>99.9 |
|||
|
E. aerogenes |
2.0 × 106 |
1 |
250;240;460;250;280 |
>99.9 |
|
| 2 |
620;640;330;340;260 |
>99.9 |
|||
| 3 |
360;240;280;220;270 |
>99.9 |
|||
|
MRSA |
4.0 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
2.5 × 105 |
1 |
260;200;540;200;400 |
99.9 |
|
| 2 |
200;410;560;280;680 |
99.2 |
|||
|
E. coli 0157:H7 |
2.6 × 105 |
1 |
<1;130;210;<1;30 |
>99.9 |
|
| 2 |
440;250;170;390;130 |
>99.9 |
|||
| Test 2–6 hours |
S. aureus |
1.8 × 106 |
1 |
280;260;330;230;700 |
>99.9 |
| 2 |
320;300;220;260;200 |
>99.9 |
|||
| 3 |
160;120;100;140;180 |
>99.9 |
|||
|
E. aerogenes |
3.9 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
8.8 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
5.2 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
5.3 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 2–12 hours |
S. aureus |
2.5 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
4.7 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
1.0 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
7.2 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
7.7 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 2–18 hours |
S. aureus |
3.6 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
5.6 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
1.7 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
9.6 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. coli 0157:H7 |
1.0 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| Test 2–24 hours |
S. aureus |
4.6 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
E. aerogenes |
7.9 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| 3 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
MRSA |
1.8 × 106 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
|
P. aeruginosa |
9.7 × 105 |
1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 |
<1;<1;<1;<1;<1 |
>99.9 |
|||
| E. coli 0157:H7 | 1.2 × 106 | 1 |
<1;<1;<1;<1;<1 |
>99.9 |
|
| 2 | <1;<1;<1;<1;<1 | >99.9 |
*Values taken from Table 1.
**Compared to control, each number represents an average of 5 replicates per manufacturing lot. Either 2 or 3 lots were examined per organism.
Discussion
Bacteria can persist on inanimate surfaces for months [30] and can be a potential source for outbreaks of nosocomial infections [18,19,27]. Thus using self-sanitizing surfaces can be a very important adjunct in the fight against nosocomial pathogens [31]. The data collected under GLP independent testing using a predefined concentration of cultivated ATCC referenced bacterial strains, demonstrated the antimicrobial properties of Cupron copper oxide impregnated countertops. Protocol number 1 tested the capacity of copper oxide infused countertops to kill a number of cultivated pathogens (Table 2) under conditions prescribed by the US EPA for the in vitro testing of the antimicrobial efficacy of copper oxide particles suspended in a plastic matrix. The organisms tested constitute a broad representation of current HAI organisms, and with over a three log reduction (>99.9%) achieved within 2 hours of exposure the authors conclude that these copper oxide infused countertops can be an additional tool for bioburden reduction and potentially reducing the risk of HAI. Importantly, as demonstrated by using Protocol 2, simulating prolonged surface wear, the countertops continue to be highly efficacious even after 12 consecutive wet and dry wear and inoculation cycles (Table 3), simulating surface abrasion that occurs due to cleaning and use. Despite the erosion of the countertops’ surface, there was no reduction in biocidal efficacy. This is explained by the distribution of the copper oxide particles throughout the matrix, on and within the surface (Figure 1), and the appearance of “new” particles on the surface as the countertop surface is eroded. This property of the countertops practically endows them with biocidal properties for the life of the product. Protocol 3 demonstrated that the countertops are efficacious to consecutive bacterial inoculations (Table 4) in the same exact spot, indicating that the countertops do not lose their biocidal efficacy following bacterial kill, but maintain this biocidal property continuously.
Copper has a long history as an antimicrobial and preventative measure and metallic copper countertops have previously been approved for EPA public health claims [32]. Field trials of these countertops have demonstrated the reduction in bioburden in a variety of clinical settings [33-37] and a reduction in the risk of infections [38,39].
Based on the data presented in this publication, Cupron Enhanced EOS Surfaces infused with copper have been approved for public health claims relating to their anti bacterial efficacy. Some of the approved health claims are a) “This surface continuously reduces bacterial* contamination achieving a 99.9% reduction within two hours of exposure.”; b) “This surface kills greater than 99.9% of Gram negative and Gram positive bacteria* within two hours of exposure.”; c) “This surface kills greater than 99.9% of bacteria* within two hours and continues to kill 99% of bacteria* even after repeated contamination.”; and d) “This surface helps inhibit the buildup and growth of bacteria* within two hours of exposure between routine cleaning and sanitizing steps”. *Testing demonstrates effective antibacterial activity against Staphylococcus aureus (ATCC 6538), Enterobacter aerogenes (ATCC 13048), MRSA (ATCC 33592), Escherichia coli O157:H7 (ATCC 35150) and Pseudomonas aeruginosa (ATCC 15442).
These non-porous Cupron copper oxide containing solid surfaces possess comparable efficacy to the metallic hard surfaces, however represent a much more feasible alternative to the metallic surfaces as they are expected to be significantly more affordable and aesthetically more pleasing. Field trials are ongoing at the time of publication to demonstrate the efficacy of these countertops in a “real world” setting (Borkow and Monk, unpublished). The biocidal properties of copper oxide against a range of organisms have also been previously demonstrated [40-43].
Limitations of this study include that the data is based upon ATCC laboratory strains and conducted in a controlled setting such as a laboratory, however further work is ongoing utilizing field trials of the surface to demonstrate the efficacy in real world applications.
Bacterial resistance to biocidal control agents is of concern in infection prevention and can be exemplified by highly antibiotic resistant bacteria (with up to 2200-fold decreased sensitivity to the antibiotic (e.g. [44]) that have evolved in less than 50 years of antibiotic usage, making infected patient treatment extremely difficult (e.g. [45]). Consequently, the possibility of development of resistance to biocides is a real concern [46,47]. Importantly, as opposed to antibiotics, despite evolving in the continued presence of copper, no microorganisms that are highly resistant to copper have been found, but only microorganisms with increased copper tolerance [31]. Importantly, no resistant bacteria evolved in vitro when repeatedly and consecutively exposed to fabrics containing copper oxide particles [42]. The reason why no resistance to copper is found in microorganisms exposed to constant relatively high doses of copper, is because copper exerts its biocidal/antimicrobial activity not through one mechanism (as most antibiotics), but through several parallel non-specific mechanisms [48,49]. Both metallic copper and copper oxide particles, in the presence of humidity, even that present in air, release copper ions. The released copper ions can migrate and reach the microorganisms even though they may not be in direct contact with the copper oxide particles. These ions can cause plasma membrane permeabilization, membrane lipid peroxidation, alteration of proteins and inhibition of their biological assembly and activity, and denaturation of nucleic acids [48,49]. It is likely that the first site that copper ions damage is the microorganisms’ envelope via electrostatic forces [50], altering the membrane integrity and permeability [51,52]. Copper ions can also cause conformational changes in the structure of intracellular or membrane proteins or in the proteins active site also by direct interaction or by displacing essential metals from their native binding sites in the proteins (e.g. [53,54]). Furthermore, the redox cycling between Cu2+ and Cu1+, which can catalyze the production of highly reactive hydroxyl radicals, can subsequently damage lipids, proteins, DNA and other biomolecules [48,55]. In addition, metallic copper, as well as cuprous oxide particles, in the absence of humidity, cause massive membrane damage and kill microorganisms within minutes via direct “contact killing” [56-58]. Apparently, the metal-bacterial contact damages the cell envelope, which, in turn, makes the cells susceptible to further damage by copper ions [58].
Microorganisms cannot cope when exposed to high concentrations of copper and are irreversibly damaged, as demonstrated also in this study. Thus, the development of resistant bacteria to copper due to the introduction of the copper containing countertops to the hospital environment is not a concern.
With the ongoing HAI problem and the role of fomites and the environment being more clearly defined, the role of antimicrobial products with EPA approved public health claims, above and beyond the treated article claims and with clinical data supporting their role in HAI prevention, will become more important.
Conclusion
The tested Cupron Enhanced EOS Surfaces containing copper oxide kill above 99.9% of a wide range of bacteria within two hours of exposure and continue to do so even after repeated contamination and multiple wet and dry abrasion cycles, passing all the acceptance criteria required by the EPA. These biocidal surfaces thus may be an important adjunct to be used in hospital settings to reduce environmental bioburden and potentially nosocomial infections.
Abbreviations
CFU: Colony forming units; EDS: Energy-dispersive X-ray spectroscopy; EPA: Environmental Protection Agency; FBS: Fetal bovine serum; GLP: Good Laboratory Practice; HAI: Hospital Acquired Infections; MRSA: Methicillin resistant Staphylococcus aureus; PBS: Phosphate buffered solution; SEM: Scanning Electron Microscopy; TSA: Tryptic Soy Agar.
Competing interests
KT is an employee of EOS Surfaces. ABM, VK and GB are employees of Cupron Inc. This study was funded by Cupron Inc. and EOS Surfaces that developed the antimicrobial surfaces.
Authors’ contributions
ABM and GB made substantial contributions to conception, design, analysis and interpretation of data of the study, and writing the manuscript; VK and KT were key in designing and developing the test materials studied, and revising the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Contributor Information
Alastair B Monk, Email: amonk@cupron.com.
Vikram Kanmukhla, Email: vik@cupron.com.
Ken Trinder, Email: kgt@eos-surfaces.com.
Gadi Borkow, Email: gadi@cupron.com.
Acknowledgements
Cupron and EOS would like to acknowledge MicroBioTest, a division of MicroBac for providing the GLP compliant independent test data.
References
- Valles J, Ferrer R. Bloodstream infection in the ICU. Infect Dis Clin North Am. 2009;23:557–569. doi: 10.1016/j.idc.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, Pollock DA, Cardo DM. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control. Annual epidemiological report on communicable diseases in Europe. Stockholm; 2010. [PubMed] [Google Scholar]
- Kock R, Becker K, Cookson B, Gemert-Pijnen JE, Harbarth S, Kluytmans J, Mielke M, Peters G, Skov RL, Struelens MJ, Tacconelli E, Navarro TA, Witte W. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010;15:19688. doi: 10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- Ferguson JK. Preventing healthcare-associated infection: risks, healthcare systems and behaviour. Intern Med J. 2009;39:574–581. doi: 10.1111/j.1445-5994.2009.02004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvernement du Québec. Loi modifiant la Loi sur les services de santé et les services sociaux concernant la prestation sécuritaire de services de santé et de services sociaux. 2009.
- Hughes AJ, Ariffin N, Huat TL, Abdul MH, Hashim S, Sarijo J, Hughes AJ, Ariffin N, Huat TL, Abdul Molok H, Hashim S, Sarijo J, Abd Latif NH, Abu Hanifah Y, Kamarulzaman A. Prevalence of nosocomial infection and antibiotic use at a university medical center in Malaysia. Infect Control Hosp Epidemiol. 2005;26:100–104. doi: 10.1086/502494. [DOI] [PubMed] [Google Scholar]
- Rosenthal VD, Maki DG, Salomao R, Moreno CA, Mehta Y, Higuera F, Cuellar LE, Arikan OA, Abouqal R, Leblebicioglu H. Device-associated nosocomial infections in 55 intensive care units of 8 developing countries. Ann Intern Med. 2006;145:582–591. doi: 10.7326/0003-4819-145-8-200610170-00007. [DOI] [PubMed] [Google Scholar]
- Rosenthal VD. Device-associated nosocomial infections in limited-resources countries: findings of the International Nosocomial Infection Control Consortium (INICC) Am J Infect Control. 2008;36:S171–12. doi: 10.1016/j.ajic.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- Roberts RR, Scott RD, Hota B, Kampe LM, Abbasi F, Schabowski S, Ahmad I, Ciavarella GG, Cordell R, Solomon SL, Hagtvedt R, Weinstein RA. Costs attributable to healthcare-acquired infection in hospitalized adults and a comparison of economic methods. Med Care. 2010;48:1026–1035. doi: 10.1097/MLR.0b013e3181ef60a2. [DOI] [PubMed] [Google Scholar]
- Curtis LT. Prevention of hospital-acquired infections: review of non-pharmacological interventions. J Hosp Infect. 2008;69:204–219. doi: 10.1016/j.jhin.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer SJ, White LF, Lamb J, Girvan EK, Robertson C. Measuring the effect of enhanced cleaning in a UK hospital: a prospective cross-over study. BMC Med. 2009;7:28. doi: 10.1186/1741-7015-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton D, Foster A, Ballantyne L, Kingsmore P, Bedwell D, Hall TJ, Hickok SS, Jeanes A, Coen PG, Gant VA. Performance of ultramicrofibre cleaning technology with or without addition of a novel copper-based biocide. J Hosp Infect. 2010;74:62–71. doi: 10.1016/j.jhin.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Pratt RJ, Pellowe CM, Wilson JA, Loveday HP, Harper PJ, Jones SR, McDougall C, Wilcox MH. epic2: national evidence-based guidelines for preventing healthcare-associated infections in NHS hospitals in England. J Hosp Infect. 2007;65(Suppl 1):S1–S64. doi: 10.1016/S0195-6701(07)60002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren MW, Rollins MS, Jeanes A, Hall TJ, Coen PG, Gant VA. Removing bacteria from hospital surfaces: a laboratory comparison of ultramicrofibre and standard cloths. J Hosp Infect. 2008;70:265–271. doi: 10.1016/j.jhin.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Bhalla A, Pultz NJ, Gries DM, Ray AJ, Eckstein EC, Aron DC, Donskey CJ. Acquisition of nosocomial pathogens on hands after contact with environmental surfaces near hospitalized patients. Infect Control Hosp Epidemiol. 2004;25:164–167. doi: 10.1086/502369. [DOI] [PubMed] [Google Scholar]
- Boyce JM, Potter-Bynoe G, Chenevert C, King T. Environmental contamination due to methicillin-resistant Staphylococcus aureus: possible infection control implications. Infect Control Hosp Epidemiol. 1997;18:622–627. doi: 10.2307/30141488. [DOI] [PubMed] [Google Scholar]
- Eckstein BC, Adams DA, Eckstein EC, Rao A, Sethi AK, Yadavalli GK, Donskey CJ. Reduction of Clostridium Difficile and vancomycin-resistant Enterococcus contamination of environmental surfaces after an intervention to improve cleaning methods. BMC Infect Dis. 2007;7:61. doi: 10.1186/1471-2334-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman ER, Platt R, Bass R, Onderdonk AB, Yokoe DS, Huang SS. Impact of an environmental cleaning intervention on the presence of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci on surfaces in intensive care unit rooms. Infect Control Hosp Epidemiol. 2008;29:593–599. doi: 10.1086/588566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MK, Bonten MJ, Blom DW, Lyle EA, van de Vijver DA, Weinstein RA. Reduction in acquisition of vancomycin-resistant enterococcus after enforcement of routine environmental cleaning measures. Clin Infect Dis. 2006;42:1552–1560. doi: 10.1086/503845. [DOI] [PubMed] [Google Scholar]
- Hota B. Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin Infect Dis. 2004;39:1182–1189. doi: 10.1086/424667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Siu LK, Chen TC, Ma L, Chiang WG, Chen YH, Lin SF, Chen TP. Methicillin-resistant Staphylococcus aureus and Acinetobacter baumannii on computer interface surfaces of hospital wards and association with clinical isolates. BMC Infect Dis. 2009;9:164. doi: 10.1186/1471-2334-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutters R, Nonnenmacher C, Susin C, Albrecht U, Kropatsch R, Schumacher S. Quantitative detection of Clostridium difficile in hospital environmental samples by real-time polymerase chain reaction. J Hosp Infect. 2009;71:43–48. doi: 10.1016/j.jhin.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Sabino R, Sampaio P, Carneiro C, Rosado L, Pais C. Isolates from hospital environments are the most virulent of the Candida parapsilosis complex. BMC Microbiol. 2011;11:180. doi: 10.1186/1471-2180-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38:S25–S33. doi: 10.1016/j.ajic.2010.04.196. [DOI] [PubMed] [Google Scholar]
- Young JM, Naqvi M, Richards L. Microbial contamination of hospital bed handsets. Am J Infect Control. 2005;33:170–174. doi: 10.1016/j.ajic.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Champagne VK, Helfritch DJ. A demonstration of the antimicrobial effectiveness of various copper surfaces. J Biol Eng. 2013;7:8. doi: 10.1186/1754-1611-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130–138. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkow G, Monk AB. Fighting nosocomial infections with biocidal non-intrusive hard and soft surfaces. World J Clin Infect Dis. 2012;12:77–90. [Google Scholar]
- Gould SWJ, Fielder MD, Kelly AF, Morgan M, Kenny J, Naughton DP. The antimicrobial properties of copper surfaces against a range of important nosocomial pathogens. Anal Microbiol. 2009;59:151–156. doi: 10.1007/BF03175613. [DOI] [Google Scholar]
- Rai S, Hirsch BE, Attaway HH, Nadan R, Fairey S, Hardy J, Miller G, Armellino D, Moran WR, Sharpe P, Estelle A, Michel JH, Michels HT, Schmidt MG. Evaluation of the antimicrobial properties of copper surfaces in an outpatient infectious disease practice. Infect Control Hosp Epidemiol. 2012;33:200–201. doi: 10.1086/663701. [DOI] [PubMed] [Google Scholar]
- Casey AL, Adams D, Karpanen TJ, Lambert PA, Cookson BD, Nightingale P, Nightingale P, Miruszenko L, Shillam R, Christian P, Elliott TS. Role of copper in reducing hospital environment contamination. J Hosp Infect. 2010;74:72–77. doi: 10.1016/j.jhin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Karpanen TJ, Casey AL, Lambert PA, Cookson BD, Nightingale P, Miruszenko L, Elliott TS. The antimicrobial efficacy of copper alloy furnishing in the clinical environment: a crossover study. Infect Control Hosp Epidemiol. 2012;33:3–9. doi: 10.1086/663644. [DOI] [PubMed] [Google Scholar]
- Marais F, Mehtar S, Chalkley L. Antimicrobial efficacy of copper touch surfaces in reducing environmental bioburden in a South African community healthcare facility. J Hosp Infect. 2010;74:80–82. doi: 10.1016/j.jhin.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Schmidt MG, Attaway HH, Sharpe PA, John J Jr, Sepkowitz KA, Morgan A, Fairey SE, Singh S, Steed LL, Cantey JR, Freeman KD, Michels HT, Salgado CD. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J Clin Microbiol. 2012;50:2217–2223. doi: 10.1128/JCM.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou PA. The role of antimicrobial copper surfaces in reducing healthcare associated infections. Eur Infect Dis. 2011;5:125–128. [Google Scholar]
- Salgado CD, Sepkowitz KA, John JF, Cantey JR, Attaway HH, Freeman KD, Sharpe PA, Michels HT, Schmidt MG. Copper surfaces reduce the rate of healthcare-acquired infections in the intensive care unit. Infect Control Hosp Epidemiol. 2013;34:479–486. doi: 10.1086/670207. [DOI] [PubMed] [Google Scholar]
- Borkow G, Gabbay J. Putting copper into action: copper-impregnated products with potent biocidal activities. FASEB J. 2004;18:1728–1730. doi: 10.1096/fj.04-2029fje. [DOI] [PubMed] [Google Scholar]
- Borkow G, Sidwell RW, Smee DF, Barnard DL, Morrey JD, Lara-Villegas HH, Shemer-Avni Y, Gabbay J. Neutralizing viruses in suspensions by copper oxide based filters. Antimicrob Agents Chemother. 2007;51:2605–2607. doi: 10.1128/AAC.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkow G, Okon-Levy N, Gabbay J. Copper oxide impregnated wound dressings: biocidal and safety studies. Wounds. 2010;22:310–316. [PubMed] [Google Scholar]
- Borkow G. Using copper to fight microorganisms. Curr Chem Biol. 2012;6:93–103. doi: 10.2174/187231312801254723. [DOI] [Google Scholar]
- Goto H, Shimada K, Ikemoto H, Oguri T. Antimicrobial susceptibility of pathogens isolated from more than 10,000 patients with infectious respiratory diseases: a 25-year longitudinal study. J Infect Chemother. 2009;15:347–360. doi: 10.1007/s10156-009-0719-3. [DOI] [PubMed] [Google Scholar]
- Durai R, Ng PC, Hoque H. Methicillin-resistant Staphylococcus aureus: an update. AORN J. 2010;91:599–606. doi: 10.1016/j.aorn.2009.11.065. [DOI] [PubMed] [Google Scholar]
- Maillard JY. Bacterial resistance to biocides in the healthcare environment: should it be of genuine concern? J Hosp Infect. 2007;65(Suppl 2):60–72. doi: 10.1016/S0195-6701(07)60018-8. [DOI] [PubMed] [Google Scholar]
- Maillard JY. Antimicrobial biocides in the healthcare environment: efficacy, usage, policies, and perceived problems. Ther Clin Risk Manag. 2005;1:307–320. [PMC free article] [PubMed] [Google Scholar]
- Borkow G, Gabbay J. Copper as a biocidal tool. Curr Med Chem. 2005;12:2163–2175. doi: 10.2174/0929867054637617. [DOI] [PubMed] [Google Scholar]
- Borkow G, Gabbay J. An ancient remedy returning to fight microbial, fungal and viral infections. Curr Chem Biol. 2009;3:272–278. [Google Scholar]
- Nan L, Liu Y, Lu M, Yang K. Study on antibacterial mechanism of copper-bearing austenitic antibacterial stainless steel by atomic force microscopy. J Mater Sci Mater Med. 2008;19:3057–3062. doi: 10.1007/s10856-008-3444-z. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Kitamoto K, Anraku Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J Bacteriol. 1988;170:2676–2682. doi: 10.1128/jb.170.6.2676-2682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery SV, Howlett NG, Radice S. Copper toxicity towards Saccharomyces cerevisiae: dependence on plasma membrane fatty acid composition. Appl Environ Microbiol. 1996;62:3960–3966. doi: 10.1128/aem.62.11.3960-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstrom AR, Levine RL. Copper inhibits the protease from human immunodeficiency virus 1 by both cysteine-dependent and cysteine-independent mechanisms. Proc Natl Acad Sci U S A. 1991;88:5552–5556. doi: 10.1073/pnas.88.13.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstrom AR, Shames BD, Levine RL. Reactivity of cysteine residues in the protease from human immunodeficiency virus: identification of a surface-exposed region which affects enzyme function. Arch Biochem Biophys. 1993;304:163–169. doi: 10.1006/abbi.1993.1334. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Espirito SC, Lam EW, Elowsky CG, Quaranta D, Domaille DW, Chang CJ. et al. Bacterial killing by dry metallic copper surfaces. Appl Environ Microbiol. 2011;77:794–802. doi: 10.1128/AEM.01599-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans M, Erbe A, Mathews S, Chen Y, Solioz M, Mucklich F. Role of copper oxides in contact killing of bacteria. Langmuir. 2013;29:16160–16166. doi: 10.1021/la404091z. [DOI] [PubMed] [Google Scholar]
- Mathews S, Hans M, Mucklich F, Solioz M. Contact killing of bacteria on copper is suppressed if bacterial-metal contact is prevented and is induced on iron by copper ions. Appl Environ Microbiol. 2013;79:2605–2611. doi: 10.1128/AEM.03608-12. [DOI] [PMC free article] [PubMed] [Google Scholar]