Abstract
It has been previously described that elevation of endogenous spermine levels in Arabidopsis could be achieved by transgenic overexpression of S-Adenosylmethionine decarboxylase (SAMDC) or Spermine synthase (SPMS). In both cases, spermine accumulation had an impact on the plant transcriptome, with up-regulation of a set of genes enriched in functional categories involved in defense-related processes against both biotic and abiotic stresses. In this work, the response of SAMDC1-overexpressing plants against bacterial and oomycete pathogens has been tested. The expression of several pathogen defense-related genes was induced in these plants as well as in wild type plants exposed to an exogenous supply of spermine. SAMDC1-overexpressing plants showed an increased tolerance to infection by Pseudomonas syringae and by Hyaloperonospora arabidopsidis. Both results add more evidence to the hypothesis that spermine plays a key role in plant resistance to biotic stress.
Keywords: biotic stress, spermine, jasmonate, polyamines, stress response and stress tolerance
Introduction
Polyamines (PAs) constitute a group of low molecular weight aliphatic amines, whose most widespread forms in living organisms are the diamine putrescine (Put), the triamine spermidine (Spd) and the tetraamine spermine (Spm). In plants, PAs have been implicated as key players in growth and development processes, as well as in the response to biotic and abiotic stresses (Kusano et al., 2008; Alcázar et al., 2010; Takahashi and Kakehi, 2010). Intracellular PA levels in plants are mostly regulated by anabolic and catabolic processes, as well as by their conjugation to hydroxycinnamic acids and macromolecules like proteins and DNA.
PA biosynthesis pathway has been well characterized in A. thaliana (Alcazar et al., 2006). In this species, PA synthesis is initiated with Put synthesis from aminoacid arginine by the sequential action of arginine decarboxylase (ADC; EC 4.1.1.19), agmatine iminohydrolase (AIH; EC 3.4.3.12), and N-carbamoylputrescine amidohydrolase (CPA; EC 3.5.1.53). Spd and Spm are the result of sequential additions of aminopropyl moieties to Put and Spd by the enzymes Spd synthase (SPDS; EC 2.5.1.16) and Spm synthase (SPMS; EC 2.5.1.22), respectively. Decarboxylated S-adenosylmethionine (dcSAM) is used by both enzymes as donor molecule of aminopropyl groups, and is synthesized from the decarboxylation of S-adenosylmethionine (SAM) in a reaction catalyzed by SAM decarboxylase (SAMDC; EC 4.1.1.50). Spd and dcSAM can also form a structural isomer of Spm, known as thermospermine (tSpm), in a reaction catalyzed by tSpm synthase (tSPMS; EC 2.5.1.79). Characterization of the Arabidopsis genome has allowed to identify two genes encoding ADC (ADC1 and ADC2) (Watson and Malmberg, 1996; Watson et al., 1997) and one for each AIH and CPA (Janowitz et al., 2003; Piotrowski et al., 2003). The Arabidopsis genome also carries two genes encoding SPDS (SPDS1 and SPDS2) (Hanzawa et al., 2002), one coding for SPMS (SPMS) (Panicot et al., 2002), another one coding for tSPMS (ACL5) (Knott et al., 2007; Kakehi et al., 2008), and at least four coding for SAMDC (SAMDC1-4) (Urano et al., 2003).
PAs are catabolized through the activity of diamine oxidases (DAO; EC 1.4.3.6) and polyamine oxidases (PAO; EC 1.5.3.3). DAOs display high affinity for diamines, like Put, producing Δ1-pyrroline, hydrogen peroxide (H2O2) and ammonia, while PAOs oxidize secondary amine groups from Spd and Spm leading to the formation of 4-aminobutanal or (3-aminopropyl)-4-aminobutanal, along with 1,3-diaminopropane (DAP) and H2O2. PAOs are also able to catalyze the back-conversion of Spm to Spd, producing 3-aminopropanal and H2O2. Some PAO isoforms are also involved in back-conversion processes of tSpm to Spm, and Spm to Put, that lead to the production of H2O2 (Moschou et al., 2012). At least five genes encoding putative PAOs (Alcazar et al., 2006; Takahashi et al., 2010) and 10 genes encoding putative DAOs (Planas-Portell et al., 2013) are present in the Arabidopsis genome.
PA metabolism is altered in a variety of plant hosts in response to several pathogens (Walters, 2003a,b), suggesting a role for PAs in the biotic defense response. However, the precise mechanism(s) of action by which PAs could exert this defensive role remains unclear, although some possible mechanisms of action have been proposed. An up-regulation of PA biosynthesis and catabolism has been observed during hypersensitive response (HR) induced by the powdery mildew fungus Blumeria graminis f. sp. hordei in barley (Cowley and Walters, 2002), as well as in tobacco plants exposed to tobacco mosaic virus (TMV) (Marini et al., 2001). PA catabolism produces H2O2, a reactive oxygen species (ROS) that could have an antimicrobial effect as well as participate in host defense mechanisms, including cell wall modifications, or act as a signal molecule triggering processes like HR (Walters, 2003a). PA oxidation has also been observed in compatible interactions between plant hosts and different types of pathogens. Spm accumulates in the apoplast of tobacco plants infected by Pseudomonas syringae pv. tabaci, and its oxidation by PAO leads to the production of H2O2 (Moschou et al., 2009). Moreover, PAO overexpression enhances tobacco tolerance to biotrophic bacteria P. syringae as well as to the hemibiotrophic oomycete Phytophtora parasitica var nicotianae (Moschou et al., 2009). Apoplastic PA accumulation and further oxidation has also been observed in tobacco leaves exposed to the biotrophic bacterium Pseudomonas viridiflava (Burkholder) Dowson, restricting bacterial growth in the host (Marina et al., 2008).
A role for Spm in defense signaling has been pointed out in A. thaliana, in which exogenous Spm induces a set of genes that are also expressed in response to the cucumber mosaic virus (CMV) infection (Mitsuya et al., 2009). Changes in Spm metabolism and subcellular localization have been associated with plant host responses to pathogenic attack. Induction of acidic pathogenesis-related proteins (PR) observed during TMV infection in tobacco is produced by Spm accumulation in the leaf apoplast, and this induction is not dependent on Salicylic acid (SA) (Yamakawa et al., 1998). Moreover, Spm accumulation, which can also be mimicked by exogenous Spm application on tobacco leaves, triggers a “Spm-signaling pathway” that causes mitochondrial dysfunction by activation of mitogen-activated protein kinases and increase of the expression of a set of HR-specific genes, leading tobacco leaf cells to develop defense responses and HR-like cell death (Takahashi et al., 2003, 2004; Mitsuya et al., 2007).
More recently, the manipulation of PA levels by transgenic approaches and the use of loss or gain-of-function mutations have emerged as new tools to gain knowledge about the role of PAs in plant stress responses (Alcázar et al., 2010; Gill and Tuteja, 2010). Consistent with the hypothesis that Spm could perform a key role in defense signaling, transgenic 35S::SPMS Arabidopsis plants accumulate Spm and are more resistant to infection by P. viridiflava than the wild type (WT). On the contrary, Spm-deficient spms mutant lines are more susceptible to infection (Gonzalez et al., 2011). Comparison of the transcriptomes of Spm-accumulating and Spm-deficient mutants showed that many genes only overexpressed in 35S::SPMS lines participate in pathogen perception and defense responses, including several families of disease resistance genes, transcription factors, kinases, and nucleotide- and DNA/RNA-binding proteins (Gonzalez et al., 2011). At the same time, most of those genes appear also induced in other Spm–accumulating lines obtained by overexpression of SAMDC1 gene (Marco et al., 2011).
In this work we have compared the expression levels of some disease resistance genes between SAMDC1-overexpressing lines and WT plants. Furthermore, we have also compared the effect of Spm accumulation on the susceptibility to the bacteria P. syringae, and to the oomycete Hyaloperonospora arabidopsidis. In both cases, SAMDC1-overexpressing lines were more resistant to infection than WT plants.
Materials and methods
Plant growth conditions
Infection by the bacterial pathogen P. syringae and by the oomycete H. arabidopsidis was tested in three Arabidopsis transgenic lines (pBISDCs-S3', pBISDCs-S9', pBISDCs-S15) overexpressing the SAMDC1 gene under the control of CaMV35S constitutive promoter (Marco et al., 2011) and in the ecotype Col-0, WT obtained from the Nottingham Arabidopsis Stock Center (University of Nottingham, Loughborough, UK). pBISDCs lines exhibit a similar phenotype to WT plants in terms of growth and development.
Arabidopsis seeds were sown in pots with a 1:1:1 mixture of soil, vermiculite and sand, stratified for 2 days at 4°C, and transferred to growth chambers. Plants were grown in Sanyo MLR-350 (Sanyo Electric Co., Japan) chambers, either under long day conditions (illumination at 23°C for 16 h, darkness at 16°C for 8 h), or short day conditions (illumination at 18°C for 10 h, darkness at 16°C for 14 h), and watered with mild nutrient solution (recipe from Arabidopsis Biological Resource Center, The Ohio State University, USA, handling plants and seeds guide, http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/handling.htm).
Seedlings were also grown on plates under long day conditions. Seeds surface was sterilized by washing in 30% (v/v) commercial bleach, 0.01% (v/v) Triton X-100 by 10 min and rinsed three times with sterile distilled water. Sterile seeds were plated on 4% agar plates containing one half strength MS medium (½MS) (Murashige and Skoog, 1962). When required, seedlings were also grown in ½MS plates supplemented with 0.1, 0.5, and 1 mM of Put, Spd, or Spm. Whole seedling samples were taken after 5 days of growth and immediately frozen in liquid Nitrogen and stored at −80°C for RNA extraction.
P. syringae infection conditions and disease evaluation
P. syringae pv. maculicola ES4326 and pv. tomato DC3000, kindly supplied by Dr. Jürgen Zeier (Julius-von-Sachs-Institute of Biological Sciences, University of Würzburg, Germany) and Dr. John Stavrinides (Department of Botany, University of Toronto, Canada), respectively, were used to infect Arabidopsis WT plants, as well as pBISDCs SAMDC1-overexpressing lines. Both bacterial strains were cultivated at 28°C in King's B medium (King et al., 1954). Streptomycin (100 μg/ml) or rifampicin (50 μg/ml) were added to select growth of strains pv. maculicola ES4326 and pv. tomato DC3000, respectively. For plant inoculation, bacterial cells were grown until cultures reached an OD600 of 0.1, collected by centrifugation, washed, and resuspended in 10 mM MgCl2 to a final concentration of 3 × 106 CFU/ml.
Leaves of 15 day-old plants grown in long day conditions were inoculated with P. syringae according to Zeier et al. (2004). Briefly, bacterial suspension was inoculated on the abaxial surface of leaves, using a 1-mL syringe without a needle. Control inoculations were performed with 10 mM MgCl2 pH 7.0. 10 different plants were inoculated with each bacterial strain and disease extension was quantified by two approaches. Image analysis was used to evaluate chlorosis in the diseased plants. Leaves were scanned at 600 dpi and images were processed using the Image Processing Tool Kit 5.0 (Reindeer Graphics Inc.) and Photoshop 7.0 softwares (Adobe Systems, Inc) to measure leaf area. Since yellow generates a lighter shade of gray than green, chlorosis-induced yellowing was estimated by the conversion of leaf images to gray tones and the determination of their luminance percentages. On the other hand, severity of infection was also estimated by measuring in planta bacterial growth. Foliar extracts were obtained by cutting three leaf discs from each plant with a 0.5 cm-diameter borer, homogenizing them with 10 mM MgCl2. Then, serial dilutions of the extracts thus obtained were plated on King's B agar medium supplemented with the appropriate antibiotic. The number of colony forming units (CFU) was determined after 24 h incubation at 28°C. Both, images and plant extracts were obtained 2 and 3 days after infection. Samples were also harvested, frozen in liquid Nitrogen and stored at −80°C for RNA extraction.
H. arabidopsidis infection conditions and disease evaluation
H. arabidopsidis isolate Noks1, was used to infect Col-0 WT Arabidopsis plants by using a dry powder containing pathogen oospores, obtained from previously infected leaves kindly supplied by Dr. Mahmut Tör (Warwick HRI, University of Warwick, UK). Methods for subculturing H. arabidopsidis and preparing inoculum for experiments were modified from Tör et al. (2002). Powder was sprinkled on pots containing Arabidopsis seeds, which were then stratified and left to germinate under short day conditions as described above. Presence of sporangiophores on cotyledons was checked daily and cotyledons with abundant sporulation were selected. H. arabidopsidis conidiospores were released from infected cotyledons by rinsing infected tissues with distilled water followed by centrifugation. Sedimented conidiospores were resuspended in distilled water, counted with a Neubauer chamber (Hauser Scientific Partnership, HORSHAM, PA 19044) and diluted to a final concentration of 5 × 104 conidiospores/ml.
Inocula consisting of 2 μl of this conidiospores suspension were applied on cotyledons of 14 day-old WT plants and pBISDCs transgenic lines. After inoculation, plants were grown under short day conditions and periodically sprinkled with water to maintain moisture. A control batch of plants was inoculated with distilled water and grown in parallel. 10 days after inoculation, 25 leaves were sampled for each line and the developed sporangiophores were counted.
RNA extraction
Total RNA was extracted from plant tissues using Total Quick RNA Cells and Tissues Kit (Talent SRL, Italy), following the protocol established by manufacturer. RNA was quantified by measuring the absorbance at 260 nm, and their integrity was checked by denaturing agarose gel electrophoresis.
Quantitative RT-PCR
RNA was treated with RNase free-DNAse (Roche diagnostics, Spain) to remove contaminating genomic DNA. A total of 1 μg of DNA free-total RNA was reverse transcribed to first-strand complementary DNA (cDNA) with random hexamers using SuperScript®III First-Strand Synthesis System 1st (Invitrogen, Spain) according to manufacturer's instructions. Quantitative real time PCR (qRT-PCR) was performed on GeneAmp®5700 Sequence Detection System (PE Applied Biosystems, Japan), using Power SYBR®Green PCR Master Mix (PE Applied Biosystems). 20 μl reactions contained 1 μl of cDNA, 100 nM of each pair of target primers (FW and REV) and 10 μl of SYBR Green PCR Master Mix. PCR conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 1 min. Three technical replicates from three independent biological experiments were performed for qRT-PCR analyses. Primers used for real-time PCR are described in Table 1. The efficiency of primers and the data were analyzed according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). Gene coding for actin-2 (ACT-2; AT3G18780; An et al., 1996) was used as a reference gene.
Table 1.
Primers used in qRT-PCR analyses.
| Gene | AGI locus | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| PR-1 | AT2G14610 | CCACAAGATTATCTAAGGGGTC | TTCCACTGCATGGGACCTA |
| PR-2 | AT3G57260 | CATCCTCGACGTTCCCAGTT | TGTCGGCCTCCGTTTGA |
| PR-5 | AT1G75050 | AACGGCGGCGGAGTTC | GCCGCCATCGCCTACTAGA |
| CYP79F1 | AT1G16410 | CATCCGTGCCATCACCATAA | CAAATCTGCGTCCCGCTCTCT |
| WAK1 | AT1G21245 | TGCTCTCAGGTCAAAAGGCATT | CGCAAAGTAACTCACCAGATGT |
| FLS2 | AT5G46330 | CCTGGACCTGTCTCACAACCA | ACGTAAGATTCATCCTTCCGAA |
| LOX2 | AT3G45140 | CAACGACAACAAGGATAAGA | CTGGCGACTCATAGAACT |
| AOC1 | AT3G25760 | CGTCCCATTCACAAACAACTC | CAGAGACCAGCCGTGATTCC |
| AOC2 | AT3G25780 | ACTGGAACGGCGGTTACG | GGCTCCATCGCCTTAGCTT |
| AOS | AT5G42650 | CGGGCGGGTCATCAAG | GCCGTTGGATTTAATCACAGAT |
| DAD1 | AT2G44810 | GGAGACGCCGTGGGTTT | GGCGAGTCACGGCTCA |
| JMT | AT1G19640 | CCAACATCACTTACTATATTCAT | GAAGAACTCGCATTACCT |
| ACT-2 | AT3G18780 | GATTCAGATGCCCAGAAAGTCTTG | TGGATTCCAGCAGCTTCCAT |
Statistical analyses
Data was analyzed by One-Way Analysis of Variance (ANOVA) followed by post-hoc comparisons by Tukey's HSD or Dunnet's T3 t-test. A probability level < 0.05 was considered statistically significant. Calculations were performed using IBM®SPSS®Statistics v20.0 Software.
Results
pBISDCs transgenic lines show constitutive elevated expression levels of disease response and jasmonic acid metabolism genes
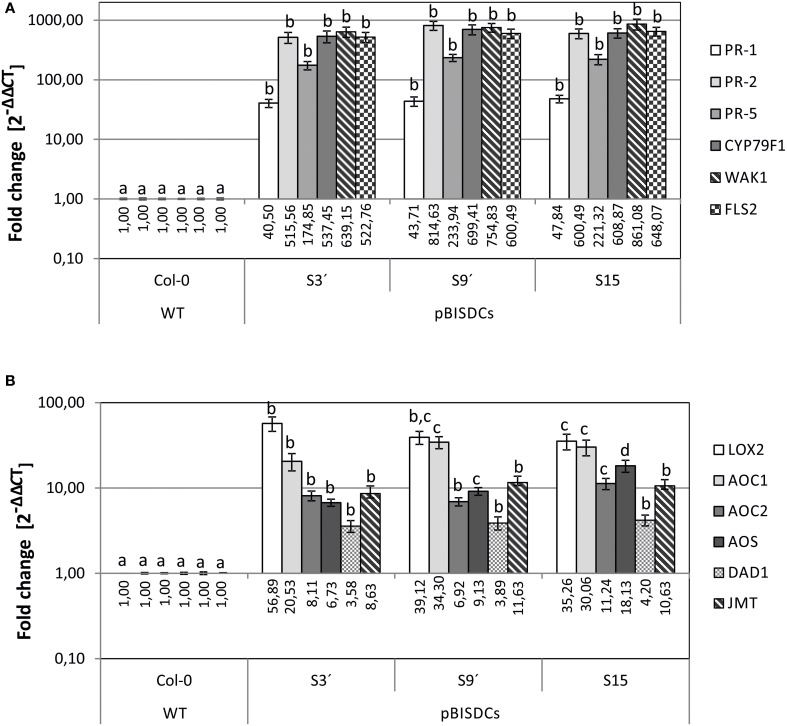
Previous comparison of the transcriptomes of WT and pBISDCs transgenic lines showed that overexpression of SAMDC1 gene in Arabidopsis leads to higher Spm levels and to the induction of a set of genes enriched in functional categories involved in defense-related processes against both biotic and abiotic stresses (Marco et al., 2011). Some of these genes were selected and their expression checked by qRT-PCR to confirm the results obtained in transcriptome studies. Expression levels of genes that encode for pathogenesis-related proteins PR-1 (AT2G14610), PR-2 (AT3G57260) and PR-5 (AT3G57260) (Uknes et al., 1992; Van Loon et al., 2006); CYP79F1 (AT1G16410), a cytochrome P450 involved in the biosynthesis of aliphatic glucosinolates (Hansen et al., 2001; Chen et al., 2003); Cell-Wall associated kinase WAK1 (AT1G21245; Verica and He, 2002) as well as the flagellin receptor FLS2 (AT5G46330; Gomez-Gomez and Boller, 2000) were determined by qRT-PCR (Figure 1A). Our results demonstrate that all disease response genes tested showed a dramatic increase in mRNA abundance in transgenic lines overexpressing SAMDC1. The expression level of these genes was very similar in all the pBISDCs lines selected, ranging from 100 to 800-fold higher than WT, except for PR-1, whose expression increased less than 100-fold (Figure 1A).
Figure 1.
qRT-PCR analysis of biotic stress-related genes in leaves of 4-week old A. thaliana WT and pBISDCs transgenic lines overexpressing SAMDC1 (S3', S9, and S15). Expression levels were determined for a set of biotic stress defense-related genes (A), as well as jasmonate and methyl-jasmonate biosynthesis genes (B). For each gene, data is expressed as fold change relative to the level measured in WT plants (2−ΔΔCT). Graph show the mean of three biological replicates ± standard deviation. Significant differences between plant lines are indicated with letters (ANOVA, Tukey HSD test, p < 0.05).
Previous transcriptome analysis showed that another set of overexpressed genes in SAMDC1-overexpressing lines were enriched in the category of genes related to jasmonic acid biosynthesis and response (Marco et al., 2011). Expression levels of genes that encode jasmonate (JA) and methyljasmonate (MeJA) biosynthesis enzymes such as Chloroplast lipoxygenase LOX2 (AT3G45140; Bell et al., 1995); Allene oxide cyclase, AOC1 (AT3G25760) and AOC2 (AT3G25780) (Stenzel et al., 2003); Allene oxide synthase AOS (AT5G42650; Kubigsteltig et al., 1999); chloroplastic phospholipase A1 DAD1 (AT2G44810; Ishiguro et al., 2001), and jasmonate O-methyltransferase JMT (AT1G19640; Seo et al., 2001), were also tested by qRT-PCR in WT and pBISDCs lines (Figure 1B). Higher levels of expression were observed for all JA and MeJA biosynthesis genes in SAMDC1-overexpressing lines when compared to WT plants, with LOX2 showing the most pronounced increase, with a mean of 43.76-fold higher than WT (Figure 1B).
Spm treatment raises the expression of defense-related genes PR-1, PR-5, as well as jasmonic acid biosynthesis AOS and AOC1 genes
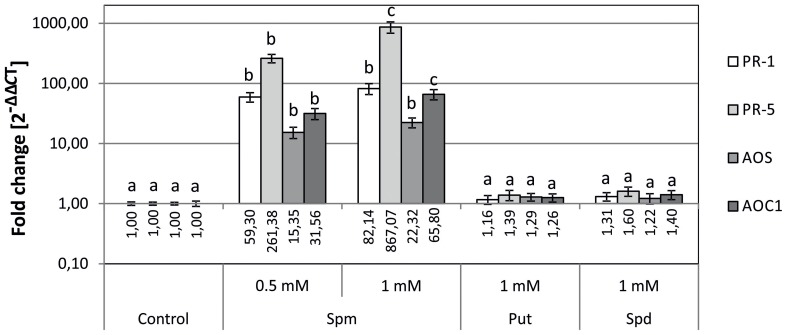
To determine whether the changes in gene expression observed in SAMDC1-overexpressing plants (Figure 1) were due to their higher Spm levels than WT plants (Marco et al., 2011), the effect of exogenous applied PAs on expression of defense response and JA biosynthesis genes was tested. WT seeds were sown on MS media containing exogenously supplied PAs and expression levels of PR-1, PR-5, AOS and AOC1 genes were determined by qRT-PCR 5 days after germination. A positive correlation between exogenous Spm concentration and transcript level was observed for the set of defense-related and JA biosynthesis genes analyzed (Figure 2). Conversely, exogenous addition of Put or Spd did not produce significant changes in gene expression (Figure 2).
Figure 2.
Effects of external polyamine treatment on the expression of biotic stress defense-related genes (PR-1 and PR5) and jasmonate biosynthesis genes (AOS and AOC1). Plants were grown for 5 days in plates supplemented with Put, Spd, or Spm, as well as in control plates without the amendment of PAs. Expression levels were determined by qRT-PCR. For each gene, data is expressed as fold change relative to the level measured in WT plants in control conditions (2−ΔΔCT). Graph show the mean of three biological replicates ± standard deviation. Significant differences between treatments are indicated with letters (ANOVA, Tukey HSD test, p < 0.05).
Overexpression of SAMDC1 enhances resistance to the oomycete H. arabidopsidis and to the bacteria P. syringae
The qRT-PCR analyses shown above (Figure 1), as well as previous transcriptome analysis of pBISDCs plants (Marco et al., 2011), suggested that SAMDC1-overexpressing plants, with elevated Spm levels, have constitutively activated a set of genes related to the defense response of plants to pathogenic microorganisms. Therefore, the response of the transgenic and WT lines against infection by bacterial or oomycete pathogens was studied.
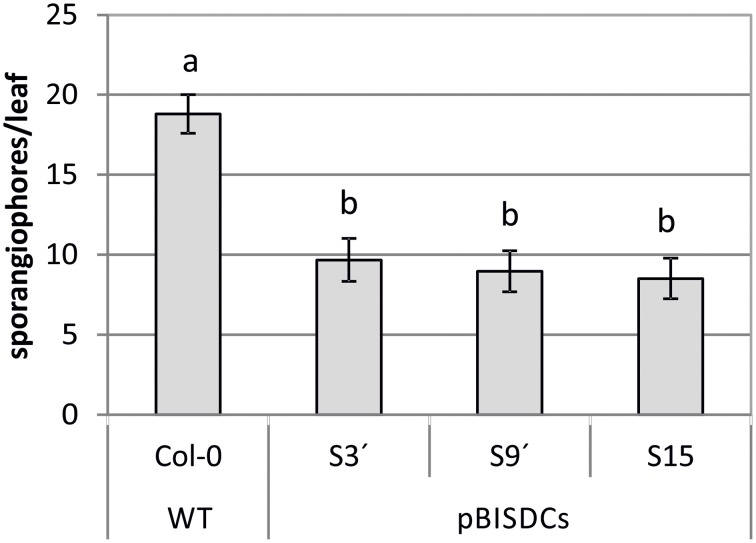
WT and PBISDCs lines were infected with H. arabidopsidis isolate Noks1. Two-week-old plants were exposed to a suspension containing H. arabidopsidis conidiospores and extension of infection was evaluated after 10 days by counting the number of sporangiophores by leaf. The number of sporangiophores by leaf observed in pBISDCs lines was approximately a half of the number obtained in WT plants, suggesting a lower propagation of H. arabidopsidis (Figure 3).
Figure 3.
Effects of H. arabidopsidis infection in Arabidopsis WT and pBISDCs transgenic lines overexpressing SAMDC1 (S3', S9', and S15). Fourteen-day old plants were inoculated with a conidiosporium suspension of H. arabidopsidis isolate Noks1. Disease extension was estimated 10 days after inoculation by counting the number of sporangiophores developed in each leaf. Twenty-five leaves were sampled for each line. Graph shows the mean ± standard deviation. Significant differences between plant lines are indicated with letters (ANOVA, Tukey HSD test, p < 0.05).
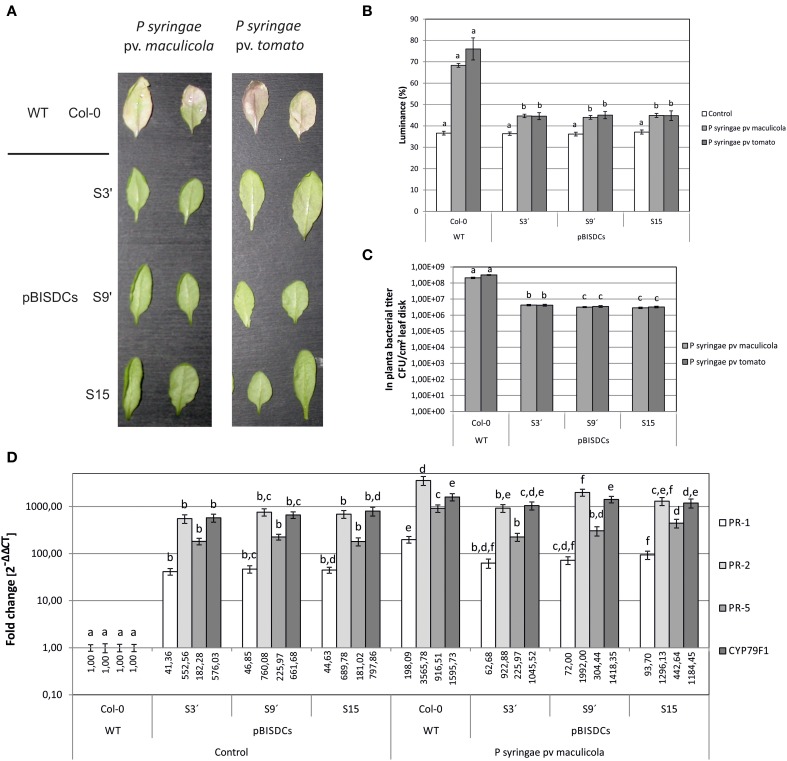
Additionally, two strains of the bacterial pathogen P. syringae, pv. maculicola ES4326 and pv. tomato DC3000, were also inoculated in 2-week old leaves of WT and pBISDCs transgenic lines. Both strains infect Arabidopsis leaves and cause initial chlorosis followed by appearance of dark spots. Progress of infection was followed by visual estimation of chlorosis appearance (Figure 4A) or by quantifying leaf luminance percentage after conversion to grayscale and image analysis (Figure 4B). Three days after inoculation, visual symptoms of chlorosis were observed in WT plants inoculated with either P. syringae strains (Figure 4A) along with an increase in leaf luminance (Figure 4B). However, SAMDC1-overexpressing lines did not show yellowing symptoms 3 days after inoculation (Figure 4A), with luminance levels close to control conditions (Figure 4B), suggesting at least a delay in infection. P. syringae propagation in planta was also estimated by the determination of CFU in foliar disks 3 days after inoculation. Compared to WT plants, pBISDCs lines showed a 10-fold reduction in the propagation of both strains of P. syringae (Figure 4C).
Figure 4.
Effects of P. syringae infection in Arabidopsis WT and pBISDCs transgenic lines overexpressing SAMDC1 (S3', S9, and S15). Leaves of 15 day-old plants were inoculated with a suspension of P. syringae strains pv. maculicola ES4326 or pv. tomato DC3000. Ten different plants for each line were inoculated. Disease extension was evaluated for each strain 3 days after inoculation visually (A), as well as by quantifying leaf luminance percentage by image analysis using the Image Processing Tool Kit 5.0 and Photoshop 7.0 softwares (B). Also, propagation of P. syringae in planta was estimated by determination of the number of CFU/cm2 of leaf disk (C). Ten different plants for each line were inoculated. Results show the mean ± standard deviation obtained for each combination of line and bacterial strain. For each condition, significant differences between lines are indicated with letters [ANOVA, Tukey HSD test (luminance data) or Dunnet's T3 test (CFU data), p < 0.05]. Plant response to P. syringae pv. maculicola infection was also studied by comparison of the expression levels for a set of biotic stress defense-related genes 3 days after inoculation with the bacterial strain or control inoculums (D). For each gene, data is expressed as fold change relative to the level measured in WT plants inoculated with control inoculum (2−ΔΔCT). Graph show the mean of three biological replicates ± standard deviation. Significant differences between plant lines are indicated with letters (ANOVA, Tukey HSD test, p < 0.05).
In addition, leaves of plants of each line infected with P. syringae pv. maculicola or exposed to control inoculum were collected 3 days after infection and expression of defense-related genes PR-1, PR-5, CYP79F1, and WAK1 was determined by qRT-PCR (Figure 4D). A dramatic induction of the expression of the four genes was observed in WT leaves, ranging from 200-fold in the case of PR-1 or 1000 to 2000-fold in the case of PR-2, PR-5 or CYP79F1. In turn, the expression levels of those genes in uninfected leaves of SAMDC-1 transgenic plants were higher than in uninfected WT plants (Figure 4D) and similar to the levels reached in them after infection (Figure 4D).
Discussion
Previous studies have shown that overexpression of SAMDC1 or SPMS genes in Arabidopsis leads to plants with higher Spm content than WT plants (Gonzalez et al., 2011; Marco et al., 2011). Transcriptome studies have pointed out that a common set of 233 genes is induced in those Spm-accumulating lines. This set of genes is enriched in functional categories involved in defense-related processes during both biotic and abiotic stresses as well as JA biosynthesis and response (Marco et al., 2011). These results suggest a connection between the increase of Spm levels and the induction of biotic stress responses. The connection among Spm levels and biotic stress responses is confirmed when the set of induced genes found at SAMDC1-overexpressing lines is compared to a set of 312 ESTs differentially expressed during Systemic Acquired Resistance (SAR) in Arabidopsis (Maleck et al., 2000). When this comparison is made, a set of 71 common genes emerges, including transcripts coding for pathogenesis-related proteins PR-1, PR-2 as well as the JA biosynthesis enzyme LOX2, which appear overexpressed in both Arabidopsis SAR and pBISDCs transcriptomes (Maleck et al., 2000; Marco et al., 2011).
The gene expression analysis made in the present work confirmed that expression levels of PR-1, PR-2, and PR-5 genes, are higher in Spm-accumulating pBISDCs lines than in WT plants (Figure 1A), as previously observed by transcriptome studies (Marco et al., 2011). Exogenously applied Spm also produces PR-1 and PR-5 induction in WT plants (Figure 2), suggesting that their expression levels in pBISDCs lines could be related to the higher Spm levels found in these plants. As expected, those genes are also induced when WT plants are infected by P. syringae pv. maculicola (Figure 4D). Induction of acidic PR proteins in response to abiotic stress and Spm treatment has been previously described in tobacco (Yamakawa et al., 1998). Moreover, PR-1 induction by Spm in Arabidopsis has been also reported by Mitsuya et al. (2009). Additionally, overexpression of SPDS in Citrus sinensis Osbeck (sweet orange) leads to plants with higher Spm levels, as well as to the overexpression of genes that code for putative PR proteins, like PR-4A and PR-10A (Fu et al., 2011; Fu and Liu, 2013).
Our qRT-PCR analysis also confirmed the up-regulation of the defense-related genes CYP79F1, WAK1 and FLS2 in SAMDC1-overexpressing lines (Figure 1A). Selection of these genes was made based on previous transcriptome data (Marco et al., 2011) and the different roles played by them during plant pathogenic responses. CYP79F1, a member of the cytochrome P450 (CYP) superfamily, is a key enzyme in the biosynthesis pathway of aliphatic glucosinolates from methionine (Hansen et al., 2001; Chen et al., 2003). Glucosinolates constitute an essential part of plant defense secondary metabolites (Halkier and Gershenzon, 2006). Diverse defense pathways control glucosinolate biosynthesis by activation of different subsets of biosynthetic enzymes (CYP among them), leading to the accumulation of specific glucosinolate profiles (Mikkelsen et al., 2003). In this trend, CYP79F1 is induced by MeJA (Mikkelsen et al., 2003; Guo et al., 2013) and our results demonstrate that it is also induced by P. syringae pv. maculicola infection (Figure 4D). WAK1 is the most studied member of a five-member family of Cell-Wall associated protein kinases (WAK1-5) (Verica and He, 2002). WAK1 is induced by P. syringae pv. maculicola infection and by SA treatment (Schenk et al., 2000). Expression of WAK1 is required by the plant to survive against lethal accumulation of SA during plant–pathogen interactions (He et al., 1998), and its ectopic overexpression confers resistance to Botrytis cinerea (Brutus et al., 2010). WAK1 is also induced by MeJA and ethylene (Schenk et al., 2000). In addition, another member of WAK family, WAK2, appears up-regulated in pBISDCs and Arabidopsis SAR transcriptomes (Maleck et al., 2000; Marco et al., 2011). FLS2 is a receptor kinase essential in the perception of flagellin, a potent elicitor of the defense response to bacterial infection (Gomez-Gomez and Boller, 2000). Flagellin perception initiates a battery of downstream defense pathways that leads to stomatal closure to avoid bacterial invasion, as well as to the activation of mechanisms inhibiting bacterial multiplication in the plant apoplast (Zipfel et al., 2004; Melotto et al., 2006).
Furthermore, levels of expression of genes coding for JA and MeJA biosynthesis enzymes, including LOX2, were also checked by qRT-PCR (Figures 1B, 2). Again, Spm was the unique PA able to induce AOS and AOC1 genes by external treatment (Figure 2), suggesting that the induction of JA and MeJA biosynthesis genes observed in SAMDC1-overexpressing plants (Figure 1B) could be promoted by their modified Spm levels. This induction could lead to the rise of JA levels in pBISDCs plants and promote JA-mediated defense mechanisms. It has been previously described that Spm treatment induces JA biosynthesis in lima bean, promoting the production of herbivore-induced volatile terpenoids that attract predatory mites (Ozawa et al., 2009). Previous studies have suggested possible interactions between PAs and JA in disease response. MeJA treatment increases PA levels and renders an improved disease response in barley seedlings exposed to powdery mildew (Walters et al., 2002), as well as in wheat plants infected with leaf rust (Haggag and Abd-El-Kareem, 2009) or in loquat fruits inoculated with Colletotrichum acutatum spores (Cao et al., 2014).
SAMDC1-ovexexpressing plants show an enhanced tolerance when infected by any of the two strains of P. syringae assayed (Figure 4A) and by the oomycete H. arabidopsidis (Figure 3), in terms of in planta pathogen propagation. Enhanced tolerance to biotrophic bacteria P. viridiflava has also been observed in Arabidopsis Spm-accumulating lines obtained by overexpression of SPMS, as well as in WT plants treated with exogenous Spm (Gonzalez et al., 2011). In addition, sweet orange SPDS-overexpressing plants are also less susceptible to Xanthomonas axonopodis pv. citri, the bacterial agent that causes citrus canker (Fu et al., 2011).
In summary, qRT-PCR studies conducted in this work confirmed that pBISDCs lines have an up-regulated expression of genes that code for members of the pathogen defense system, as suggested by previous transcriptome studies (Marco et al., 2011). This constitutive activation of the defense-response mechanisms has also a positive impact on the susceptibility of pBISDCs lines against bacterial (Figure 4) and oomycete infection (Figure 3). Results obtained in this study add more evidence to the role of Spm in plant response to biotic stress, and reinforce the hypothesis that, among the different mechanisms postulated by which Spm could exert their protective action, transcriptional changes of defense genes might play an important role. It remains to be determined which of the changes in gene expression observed in the transcriptome of Spm accumulating plants are the result of the direct action of Spm or the consequence of intricate cross-talking between Spm and other biotic defense-signaling pathways, including JA and MeJA.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research has been supported during these last years by the following grants: CICYT-BIO2002-04459-C02-02, MEC-BIO2005-09252-C02-02 and GV2005-070. We thank Jane Moreno for revising the English language.
References
- Alcázar R., Altabella T., Marco F., Bortolotti C., Reymond M., Koncz C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249 10.1007/s00425-010-1130-0 [DOI] [PubMed] [Google Scholar]
- Alcazar R., Marco F., Cuevas J. C., Patron M., Ferrando A., Carrasco P., et al. (2006). Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 28, 1867–1876 10.1007/s10529-006-9179-3 [DOI] [PubMed] [Google Scholar]
- An Y. Q., McDowell J. M., Huang S., McKinney E. C., Chambliss S., Meagher R. B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121 10.1046/j.1365-313X.1996.10010107.x [DOI] [PubMed] [Google Scholar]
- Bell E., Creelman R. A., Mullet J. E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 92, 8675–8679 10.1073/pnas.92.19.8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457 10.1073/pnas.1000675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Cai Y., Yang Z., Joyce D. C., Zheng Y. (2014). Effect of MeJA treatment on polyamine, energy status and anthracnose rot of loquat fruit. Food Chem. 145, 86–89 10.1016/j.foodchem.2013.08.019 [DOI] [PubMed] [Google Scholar]
- Chen S., Glawischnig E., Jorgensen K., Naur P., Jorgensen B., Olsen C. E., et al. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33, 923–937 10.1046/j.1365-313X.2003.01679.x [DOI] [PubMed] [Google Scholar]
- Cowley T., Walters D. R. (2002). Polyamine metabolism in barley reacting hypersensitively to the powdery mildew fungus Blumeria graminis f. sp. hordei. Plant Cell Environ. 25, 461–468 10.1046/j.0016-8025.2001.00819.x [DOI] [Google Scholar]
- Fu X. Z., Chen C. W., Wang Y., Liu J. H., Moriguchi T. (2011). Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: involvement of H(2)O(2) production and transcriptional alteration. BMC Plant Biol. 11:55 10.1186/1471-2229-11-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Z., Liu J. H. (2013). Transcriptional profiling of canker-resistant transgenic sweet orange (Citrus sinensis Osbeck) constitutively overexpressing a spermidine synthase gene. Biomed. Res. Int. 2013:918136 10.1155/2013/918136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S., Tuteja N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 5, 26–33 10.4161/psb.5.1.10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- Gonzalez M. E., Marco F., Minguet E. G., Carrasco Sorli P., Blázquez M. A., Carbonell J., et al. (2011). Perturbation of spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis thaliana defense against Pseudomonas viridiflava. Plant Physiol. 156, 2266–2277 10.1104/pp.110.171413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Shen W., Qian H., Zhang M., Liu L., Wang Q. (2013). Jasmonic acid and glucose synergistically modulate the accumulation of glucosinolates in Arabidopsis thaliana. J. Exp. Bot. 64, 5707–5719 10.1093/jxb/ert348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggag W., Abd-El-Kareem F. (2009). Methyl jasmonate stimulates polyamines biosynthesis and resistance against leaf rust in wheat plants. Arch. Phytopathol. Plant Protect. 41, 16–31 10.1080/03235400600914355 [DOI] [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Hansen C. H., Wittstock U., Olsen C. E., Hick A. J., Pickett J. A., Halkier B. A. (2001). Cytochrome p450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276, 11078–11085 10.1074/jbc.M010123200 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y., Imai A., Michael A. J., Komeda Y., Takahashi T. (2002). Characterization of the spermidine synthase-related gene family in Arabidopsis thaliana. FEBS Lett. 527, 176–180 10.1016/s0014-5793(02)03217-9 [DOI] [PubMed] [Google Scholar]
- He Z. H., He D., Kohorn B. D. (1998). Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–63 10.1046/j.1365-313X.1998.00092.x [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209 10.1105/tpc.010192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowitz T., Kneifel H., Piotrowski M. (2003). Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett. 544, 258–261 10.1016/s0014-5793(03)00515-5 [DOI] [PubMed] [Google Scholar]
- Kakehi J.-I., Kuwashiro Y., Niitsu M., Takahashi T. (2008). Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant Cell Physiol. 49, 1342–1349 10.1093/pcp/pcn109 [DOI] [PubMed] [Google Scholar]
- King E. O., Ward M. K., Raney D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307 [PubMed] [Google Scholar]
- Knott J. M., Römer P., Sumper M. (2007). Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 581, 3081–3086 10.1016/j.febslet.2007.05.074 [DOI] [PubMed] [Google Scholar]
- Kubigsteltig I., Laudert D., Weiler E. W. (1999). Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 208, 463–471 10.1007/s004250050583 [DOI] [PubMed] [Google Scholar]
- Kusano T., Berberich T., Tateda C., Takahashi Y. (2008). Polyamines: essential factors for growth and survival. Planta 228, 367–381 10.1007/s00425-008-0772-7 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K. A., et al. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410 10.1038/82521 [DOI] [PubMed] [Google Scholar]
- Marco F., Alcazar R., Tiburcio A. F., Carrasco P. (2011). Interactions between polyamines and abiotic stress pathway responses unraveled by transcriptome analysis of polyamine overproducers. OMICS 15, 775–781 10.1089/omi.2011.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina M., Maiale S. J., Rossi F. R., Romero M. F., Rivas E. I., Gárriz A., et al. (2008). Apoplastic polyamine oxidation plays different roles in local responses of tobacco to infection by the necrotrophic fungus Sclerotinia sclerotiorum and the biotrophic bacterium Pseudomonas viridiflava. Plant Physiol. 147, 2164–2178 10.1104/pp.108.122614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F., Betti L., Scaramagli S., Biondi S., Torrigiani P. (2001). Polyamine metabolism is upregulated in response to tobacco mosaic virus in hypersensitive, but not in susceptible, tobacco. New Phytol. 149, 301–309 10.1046/j.1469-8137.2001.00017.x [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980 10.1016/j.cell.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M. D., Petersen B. L., Glawischnig E., Jensen A. B., Andreasson E., Halkier B. A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131, 298–308 10.1104/pp.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya Y., Takahashi Y., Berberich T., Miyazaki A., Matsumura H., Takahashi H., et al. (2009). Spermine signaling plays a significant role in the defense response of Arabidopsis thaliana to cucumber mosaic virus. J. Plant Physiol. 166, 626–643 10.1016/j.jplph.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Mitsuya Y., Takahashi Y., Uehara Y., Berberich T., Miyazaki A., Takahashi H., et al. (2007). Identification of a novel Cys2/His2-type zinc-finger protein as a component of a spermine-signaling pathway in tobacco. J. Plant Physiol. 164, 785–793 10.1016/j.jplph.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Moschou P. N., Sarris P. F., Skandalis N., Andriopoulou A. H., Paschalidis K. A., Panopoulos N. J., et al. (2009). Engineered polyamine catabolism preinduces tolerance of tobacco to bacteria and oomycetes. Plant Physiol. 149, 1970–1981 10.1104/pp.108.134932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou P. N., Wu J., Cona A., Tavladoraki P., Angelini R., Roubelakis-Angelakis K. A. (2012). The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J. Exp. Bot. 63, 5003–5015 10.1093/jxb/ers202 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Ozawa R., Bertea C. M., Foti M., Narayana R., Arimura G., Muroi A., et al. (2009). Exogenous polyamines elicit herbivore-induced volatiles in lima bean leaves: involvement of calcium, H2O2 and Jasmonic acid. Plant Cell Physiol. 50, 2183–2199 10.1093/pcp/pcp153 [DOI] [PubMed] [Google Scholar]
- Panicot M., Minguet E. G., Ferrando A., Alcazar R., Blazquez M. A., Carbonell J., et al. (2002). A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell 14, 2539–2551 10.1105/tpc.004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M., Janowitz T., Kneifel H. (2003). Plant C-N hydrolases and the identification of a plant N-carbamoylputrescine amidohydrolase involved in polyamine biosynthesis. J. Biol. Chem. 278, 1708–1712 10.1074/jbc.M205699200 [DOI] [PubMed] [Google Scholar]
- Planas-Portell J., Gallart M., Tiburcio A. F., Altabella T. (2013). Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol. 13:109 10.1186/1471-2229-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P. M., Kazan K., Wilson I., Anderson J. P., Richmond T., Somerville S. C., et al. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 97, 11655–11660 10.1073/pnas.97.21.11655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H. S., Song J. T., Cheong J. J., Lee Y. H., Lee Y. W., Hwang I., et al. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. U.S.A. 98, 4788–4793 10.1073/pnas.081557298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I., Hause B., Miersch O., Kurz T., Maucher H., Weichert H., et al. (2003). Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 51, 895–911 10.1023/A:1023049319723 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Kakehi J.-I. (2010). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 105, 1–6 10.1093/aob/mcp259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Berberich T., Miyazaki A., Seo S., Ohashi Y., Kusano T. (2003). Spermine signalling in tobacco: activation of mitogen-activated protein kinases by spermine is mediated through mitochondrial dysfunction. Plant J. 36, 820–829 10.1046/j.1365-313X.2003.01923.x [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Berberich T., Yamashita K., Uehara Y., Miyazaki A., Kusano T. (2004). Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the tobacco mosaic virus-induced hypersensitive response and during leaf- and flower-senescence. Plant Mol. Biol. 54, 613–622 10.1023/B:PLAN.0000038276.95539.39 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Cong R., Sagor G. H., Niitsu M., Berberich T., Kusano T. (2010). Characterization of five polyamine oxidase isoforms in Arabidopsis thaliana. Plant Cell Rep. 29, 955–965 10.1007/s00299-010-0881-1 [DOI] [PubMed] [Google Scholar]
- Tör M., Gordon P., Cuzick A., Eulgem T., Sinapidou E., Mert-Türk F., et al. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003 10.1105/tpc.001123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., et al. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656 10.1105/tpc.4.6.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K., Yoshiba Y., Nanjo T., Igarashi Y., Seki M., Sekiguchi F., et al. (2003). Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ. 26, 1917–1926 10.1046/j.1365-3040.2003.01108.x [DOI] [Google Scholar]
- Van Loon L. C., Rep M., Pieterse C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- Verica J. A., He Z. H. (2002). The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129, 455–459 10.1104/pp.011028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D. (2003a). Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol. 159, 109–115 10.1046/j.1469-8137.2003.00802.x [DOI] [PubMed] [Google Scholar]
- Walters D., Cowley T., Mitchell A. (2002). Methyl jasmonate alters polyamine metabolism and induces systemic protection against powdery mildew infection in barley seedlings. J. Exp. Bot. 53, 747–756 10.1093/jexbot/53.369.747 [DOI] [PubMed] [Google Scholar]
- Walters D. R. (2003b). Polyamines and plant disease. Phytochemistry 64, 97–107 10.1016/S0031-9422(03)00329-7 [DOI] [PubMed] [Google Scholar]
- Watson M. B., Malmberg R. L. (1996). Regulation of Arabidopsis thaliana (L.) Heynh arginine decarboxylase by potassium deficiency stress. Plant Physiol. 111, 1077–1083 10.1104/pp.111.4.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. W., Yu W., Galloway G. L., Malmberg R. L. (1997). Isolation and characterization of a second arginine decarboxylase cDNA from Arabidopsis (Accession No AF009647). Plant Physiol. 114, 1569 10.1104/pp.114.4.1567 [DOI] [Google Scholar]
- Yamakawa H., Kamada H., Satoh M., Ohashi Y. (1998). Spermine is a salicylate-independent endogenous inducer for both tobacco acidic pathogenesis-related proteins and resistance against tobacco mosaic virus infection. Plant Physiol. 118, 1213–1222 10.1104/pp.118.4.1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J., Delledonne M., Mishina T., Severi E., Sonoda M., Lamb C. (2004). Genetic elucidation of nitric oxide signaling in incompatible plant-pathogen interactions. Plant Physiol. 136, 2875–2886 10.1104/pp.104.042499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., Felix G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]