Abstract
Small interfering RNAs (siRNAs) potently silence expression of target genes. In principle siRNA libraries can be used to perform effective genome-scale functional genetic screens in mammalian cells, but their development has been hampered by the need to chemically synthesize thousands of oligonucleotides and to incorporate them into expression vectors. We have developed a technology to efficiently convert a double-stranded cDNA library into a retroviral siRNA library in which 21-base siRNAs are produced in infected cells at high levels and efficiently block expression of their target genes. The key steps are the generation of random cDNA fragments that are fused to a hairpin linker, cleavage with the MmeI endonuclease that creates 20- to 21-bp cDNA fragments, conversion to a double-stranded DNA that contains two copies of the cDNA insert in a head-to-head palindrome, and insertion of the construct downstream of a polymerase III promoter. We constructed a siRNA library with 3 × 106 clones from a mouse embryo cDNA library; siRNAs were found against many different genes; and multiple siRNAs can be generated from a single mRNA. We further showed that specific siRNAs were efficiently produced in stably infected mammalian cells and resulted in significant and specific reduction of their target mRNAs. Because no prior knowledge about target transcripts is needed, a cDNA-derived siRNA library will generate siRNAs against unknown transcripts and genes. Finally, cDNA-derived siRNA libraries can be readily generated from any cell type or species, enabling genome-wide functional screens in many biological systems.
RNA interference (RNAi) is a powerful experimental technique for silencing gene expression both in cultured eukaryotic cells and living organisms. It is initiated by the introduction of double-stranded RNA (dsRNA) into cells, which is processed into double-stranded small interfering RNAs (siRNAs) 21–23 bases in length (reviewed in refs. 1–4). siRNAs in turn stimulate the RNA-induced silencing complex (RISC) and result in the degradation of mRNAs homologous in sequence to the siRNAs. In Caenorhabditis elegans and Drosophila, RNAi is an extraordinarily powerful tool for elucidating and manipulating gene function in somatic cells (5–10).
The original RNAi technology, which predominantly employs dsRNAs of >30 bp, is not effective in mammalian cells because their introduction activates an interferon response, resulting in nonspecific mRNA degradation and inhibition of protein synthesis. However, smaller synthetic 20- to 23-bp double-stranded siRNAs do not induce an interferon response yet remain potent and specific inhibitors of endogenous gene expression (11). Several vectors have been developed to enable transient as well as stable expression of siRNAs. In general, these employ an RNA Pol III promoter to express short hairpin RNAs (shRNAs) consisting of 19–29 bp of double-stranded stem separated by a loop of 4–9 bases. Subsequent processing of the shRNAs into siRNAs (12–16) and incorporation of one strand into the RISC directs the degradation of the homologous mRNA target.
In principle, one should be able to use siRNA libraries to perform effective genome-scale functional genetic screens in mammalian cells. However, their development has been hampered by the need to chemically synthesize thousands of oligonucleotides and to incorporate them into viral or other types of vectors. Additionally, because of the base pairing specificity required for siRNA function, a new library must be generated for each species. Moreover, construction of a siRNA expression library with oligonucleotides requires knowledge of the sequence of all potential target messenger RNAs, information that is presently available for a handful of organisms. For the sequenced organisms, existing algorithms for predicting gene structure are not accurate (17), and gene coding sequences are not always recognized. Finally, high-throughput oligonucleotide synthesis currently used for construction of siRNA expression libraries has a high error rate (about one error in every 200 bases) and thus sequence validation of each siRNA expression construct is necessary (D. Root, personal communication).
Here we describe siRNA production by enzymatic engineering of DNA (SPEED), a simple, effective, and inexpensive strategy to construct genome-wide siRNA expression libraries from populations of double-stranded cDNA. This method negates the need for expensive chemical synthesis of oligonucleotides and facilitates the generation of siRNA libraries that encompass all expressed genes, including those with unknown structure and functions, from diverse mRNA sources in many species.
Materials and Methods
Construction of siRNA Library from cDNA Library. Steps refer to those in Fig. 1.
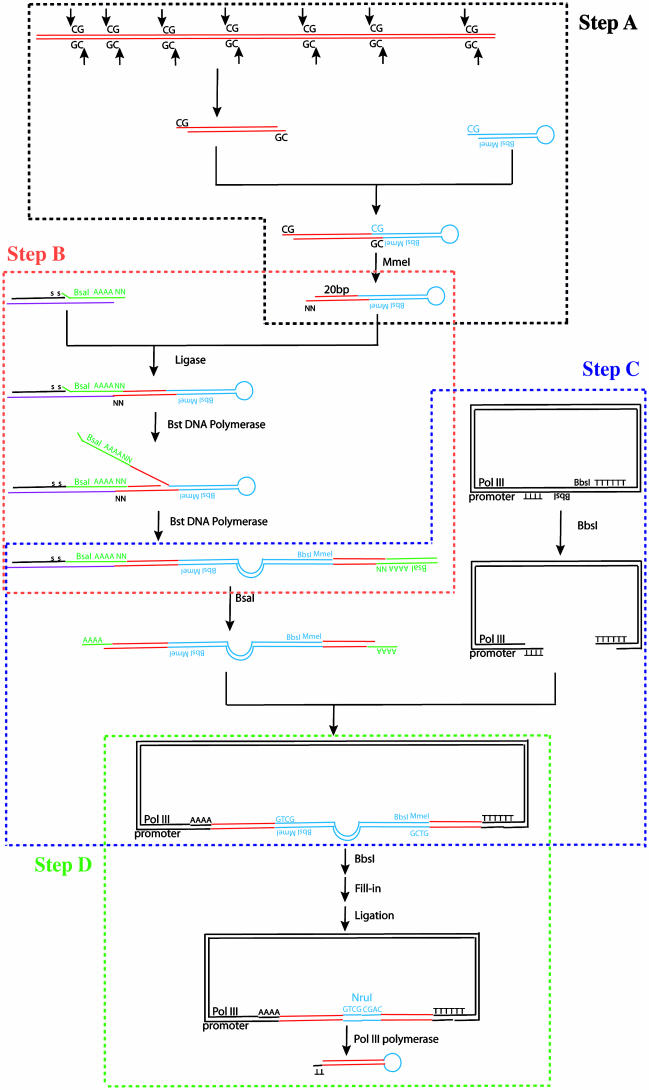
Fig. 1.
Schematic outline for construction of a siRNA library. Step A, hairpin linker attachment; step B, conversion of extended hairpins into palindromic dsDNAs; step C, cloning into the retrovirus expression vector; step D, creation of a hairpin loop. Details are given in Materials and Methods and Results.
Step A: Hairpin Linker Attachment. Inserts from a cDNA library were digested with AciI, HpaII, HpyCH4IV, HinP1 I, and TaqI endonucleases and ligated to a self-annealed PAGE-purified hairpin linker, 5′-pCGGTCGGAGTCTTCATGATTCAAGAGATCATGAAGACTCCGAC-3′. The hairpin linker has recognition sequences for MmeI (TCCGAC) and BbsI (GAAGAC) at its open end. The ligation product was digested with MmeI, and the resulting extended hairpins were purified as ≈40-bp species by PAGE.
Step B: Conversion of Extended Hairpins into Palindromic Double-Stranded DNAs. The extended hairpins were ligated with an extension linker comprised of three oligonucleotides: an extension priming oligo, 5′-GACTCTGATACACTGT-s-C-s-T-3′ [black (colors are references to Fig. 1); the last two bases at the 3′ end are thiophosphate-modified]; an extension sense primer, 5′-CAGAGTCGGTCTCAAAAANN-3′ (green); and an extension antisense primer, 5′-TTTTTGAGACCGACTGACAGACAGTGTATCAGAGTCGGG-3′. The extension sense primer contains the recognition sequence GGTCTC for BsaI. The ligation products were then converted into double-stranded DNA (dsDNA) by the polymerase and strand displacement activity of the Bst DNA polymerase large subunit in the presence of single-stranded DNA binding protein.
Step C: Cloning into the Retrovirus Expression Vector. The extension products from Step B were digested with BsaI, leaving a 5′-AAAA overhang on both ends. The Pol III promoter expression vector was constructed by PCR amplification of the human U6 promoter (14), using the primers 5′-TGACAAGCTTGCTAGCAAAAA ACTGTCT TCGA AGACTCTTTTTT TCGTCCTTTCCACAAGATATATAAAGCCA-3′ and 5′-GGCCTCTAGACCCGAGTCCAACACCCGTGGGAATCCCATG-3′ and resulting in the placement of tandem BbsI sites just downstream from the U6 promoter. Digestion with BbsI left two 5′-TTTT overhangs that were then ligated to the 5′-AAAA overhangs of the BsaI-digested palindromic cDNA extension products. The ligation reaction was transformed into bacteria, and plasmid DNA was purified.
Step D: Creation of a Hairpin Loop. DNA was digested with BbsI to remove most of the original hairpin linker sequence, leaving CGAC single-stranded 5′ overhangs at each end. These overhangs were filled in with the Klenow fragment of DNA Pol I, and subsequent blunt-end self-ligation generated an 8-bp sequence (GTCGCGAC) containing an NruI site between the two 20- to 21-bp cDNA palindrome sequences.
Construction of siRNA Expression Vectors. The viral backbone of our retroviral expression constructs is MSCV-pgk-EGFP (MSCV, murine stem cell virus) derived from pMSCVhyg (BD Clontech); these vectors express GFP from the pgk promoter. It was modified by inserting a Pol III promoter driving a hairpin siRNA between the NheI and XbaI sites in the U3 region of the 3′ LTR with the Pol III promoter in the opposite orientation to the LTR promoter. H1-lucB has an H1 promoter (18) driving expression of a siRNA targeting the B site (5′-GTGCGCTGCTGGTGCCAAC-3′) in the luciferase gene (lucB site). H1-PU718 has an H1 promoter driving expression of a siRNA targeting nucleotides 718–737 (5′-GAAGCTCACCTACCAGTTC-3′) of the human PU.1 gene (PU718 site). U6-PU718 has a U6 promoter driving expression of a siRNA targeting the PU718 site. U6-PU646 has a U6 promoter driving expression of a siRNA targeting nucleotides 646–665 (5′-GATGACCTACCAGAAGATG-3′) of the human PU.1 gene (PU646 site). H1-AAAA-PU718 has a modified H1 promoter with the last four nucleotides changed to AAAA and driving expression of a siRNA targeting the PU718 site. U6-AAAA-PU718 has a modified U6 promoter with the last four nucleotides changed to AAAA and driving expression of a siRNA targeting the PU718 site. In the vectors above, the loop of the siRNA hairpin is 5′-TTCAAGAGA-3′. U6-AAAA-PU718-0, 2, 4, 6, and 8 express PU718 siRNA with the hairpin containing palindrome loops of 0, 2 (5′-GC-3′), 4 (5′GTAC-3′), 6 (5′-GTCGAC-3′), and 8 (5′-GTCGCGAC-3′) nucleotides, respectively, from a U6 promoter with AAAA as the last bases. U6-AAAA-CD53-8 encodes siRNA targeting nucleotides 310–329 (5′-GTGTCTGCTTATGTCGTTCT-3′) of human CD53, and U6-AAAA-CD45-8 encodes siRNA targeting nucleotides 1377–1396 (5′-GATTGCCTCAATCTGGATAA-3′) of human CD45 from U6 promoter with the AAAA as the last bases and the eight-base palindrome loop (5′-GTCGCGAC-3′).
Retroviral Infection. Retroviral vectors were transfected into BOSC23 cells (19) to produce virus. UT7 cells expressing the ecotropic receptor (20) were infected by centrifuging UT7 cells with virus in the presence of 4 μg/ml polybrene for 2 h then incubating at 37°C for 5 h, followed by removal of the virus. UT7 cells were cultured with alpha minimum essential medium with 20% FCS and 5 ng/ml human granulocyte–macrophage colony-stimulating factor (GM-CSF). NIH 3T3 cells were cultured in DMEM with 10% calf serum and infected without spinning.
Western and Northern Blot Analysis. Western blotting was performed as described (21) with PU.1 antibody from Santa Cruz Biotechnology and Stat3 antibody from Cell Signaling Technology (Beverly, MA). Northern blotting for siRNA products was performed as described (22).
Fluorescence-Activated Cell Sorting (FACS) Staining and Analysis. UT7 cells were infected with U6-AAAA-CD53-8 or U6-AAAA-CD45-8 retroviruses, and infected GFP positive cells were sorted by FACS on a MoFlo machine (DakoCytomation, Carpinteria, CA) and propagated. Cells were stained with phycoerythrin (PE)-labeled CD53 or CD45 or a PE-labeled rat IgG2a control antibody. Cells were stained for 1 h at 4°C and washed twice before being analyzed on a FACScan (Becton Dickinson). All antibodies were from BD Pharmingen.
Real-Time RT-PCR. Total RNA was purified from FACS-sorted NIH 3T3 cells with TRIzol reagent (Invitrogen). cDNA was synthesized with SuperScript II (Invitrogen). Real-time PCR was performed on ABI Prism 7700 with SYBR Green (Perkin–Elmer Biosystems). The clone 548 target was amplified with primers 5′-GCGCTCCAAGAGCTCCAAGCCGCAT-3′ and 5′-CCTCCTCTGCGGCCTGCTGCAGGAT-3′. The clone 550 target was amplified with primers 5′-TGCAGACCTCTGACAAGGATGAGAGC-3′ and 5′-TGGAAGTTGCCGTTGCGGTCACAGT-3′. The absolute amount of the transcript targeted by the siRNA, as well as the GAPDH control, was deduced from the standard curve for amplification of the respective genes.
Results
Fig. 1 outlines our procedure (SPEED) for converting a population of double-stranded cDNAs into a siRNA library that is inserted into a retrovirus vector. It is convenient to divide the process into four steps (Fig. 1).
Step A: Hairpin Linker Attachment. First a dsDNA or a double-stranded cDNA library (red) is cleaved with a mixture of five restriction endonucleases, each of which leaves a 5′-CG overhang but has a different 4-bp recognition sequence. This combination covers 6 of 256 possible 4-bp sequences, resulting in one cleavage every 43 bp. These fragmented cDNAs are ligated to a hairpin linker (blue) with a complementary 5′-CG overhang. This hairpin has a recognition site for MmeI, a type IIs restriction endonuclease that cleaves 20–21 bp away from its recognition sequence and leaves a two-base 5′ overhang. Type IIs endonucleases have been used to capture small segments of DNA in the SAGE (serial analysis of gene expression) protocol (23). Note that the hairpin linker can be ligated to either the 5′ or 3′ end of the double-stranded cDNA fragment. Subsequent digestion by MmeI generates uniformly sized extended hairpins, each of which contains one 20- to 21-bp cDNA fragment.
Step B: Conversion of Extended Hairpins into Palindromic dsDNAs. Next an “extension linker” comprised of three different oligonucleotides is ligated to the cDNA-extended hairpin. As a result the 5′ and 3′ ends of the extended hairpin become ligated to the extension “sense” oligonucleotide (green) and the extension “antisense” oligonucleotide (purple), respectively. Note that the 3′ end of the sense oligonucleotide has two exposed random nucleotides that pair with the two random nucleotides at the 3′ end of the extended hairpin. Next, the extension priming oligonucleotide (black) serves as primer for DNA elongation by the highly processive Bst DNA polymerase large fragment, which lacks a 5′–3′ exonuclease domain. This reaction converts the hairpin into a dsDNA that contains palindromic copies of the 20- to 21-bp cDNA insert (red) as well as a palindrome derived from the original hairpin linker (blue). Also, a BsaI restriction endonuclease site placed near the 3′ end of the extension “sense” oligonucleotide is duplicated by elongation with the Bst DNA polymerase.
Step C: Cloning into the Retrovirus Expression Vector. Digestion of the resulting dsDNA with BsaI generates 5′AAAA overhangs on each end. In our murine stem cell virus (MSCV) retrovirus expression vector the hairpin siRNAs are transcribed from a Pol III promoter in the opposite orientation to the retroviral LTRs. The Pol III promoter-siRNA cassette is inserted into the 3′ LTR, which contains a self-inactivating deletion. When the retrovirus RNA is copied by reverse transcriptase, two copies of the Pol III promoter-siRNA cassette are created, one in the resulting 5′ LTR and the other in the 3′ LTR, and thus infected cells contain two copies of the cassette.
We engineered tandem BbsI endonuclease sites, in a back-to-back orientation, at the 3′ end of the Pol III promoter. Thus, when digested with BbsI, TTTT overhangs are created on both 5′ ends of the vector DNA. These ends pair with the 5′-AAAA ends of the double-stranded BsaI-digested palindromic cDNA extension products to be ligated to form a completely double-stranded molecule. Note that the 5′-TTTT overhang distal to Pol III promoter in the vector, together with the two adjacent T residues in the vector DNA, generates the Pol III termination sequence. The four A bases at the 5′ end of the BsaI-digested palindromic cDNA extension product become the last four bases of the Pol III promoter.
Step D: Creation of a Hairpin Loop. Preliminary studies showed that dsDNAs resulting from step C could be used to generate packaged retroviruses. However, the levels of mRNAs homologous to the cDNA portion of the hairpin were not affected in infected cells (data not shown). We surmised that the original hairpin linker sequence (blue) somehow prevented proper RNA processing, and thus we removed the majority of this sequence from the vector–insert complex by digesting with BbsI. This enzyme leaves a four-base overhang at each end that was then filled in by Klenow fragment of DNA Pol I. Subsequent blunt-end ligation generated an 8-bp sequence, 5′-GTCGCGAC, containing a NruI recognition site (blue) between the palindromic copies of the cDNA insert. After transcription by RNA Pol III, these eight bases form the loop connecting the two strands of the hairpin.
Effect of Promoter Modification and Palindrome Loop for siRNA Production. Our protocol introduces two major modifications to established methods for Pol III-driven production of siRNA hairpins (12–16). First, the last four bases of the Pol III promoter are changed from the wild-type sequence to AAAA. Second, the “loop” we created in the RNA hairpin is an eight-base palindromic sequence, whereas normally it is a nonpalindromic sequence. Neither of these modifications significantly affects expression of the small hairpin RNA and its subsequent processing to a functional 21-base siRNA, as we show in Figs. 2 and 3.
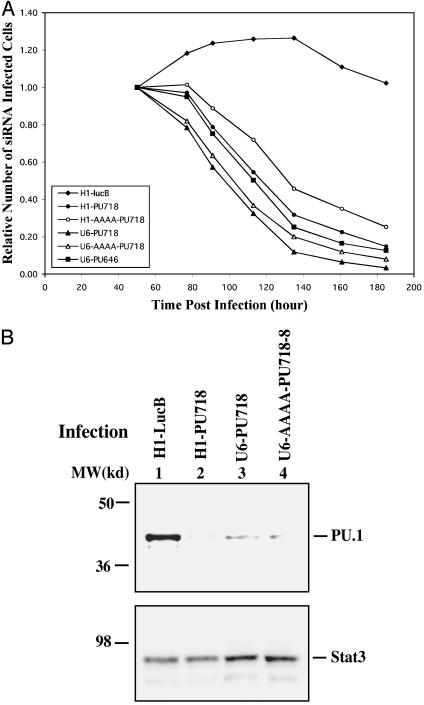
Fig. 2.
Effects of promoter modification. (A) Expression of PU.1 siRNA causes loss of infected cells. UT7 cells were infected with the indicated retroviruses, and the percentage of infected GFP-positive cells, among the total propidium iodide-negative live cells, was measured by FACS at various times after infection. Plotted is the ratio of GFP-positive to -negative cells normalized to the ratio at 50 h postinfection. (B) Specific reduction of PU.1 protein. UT7 cells were infected with the indicated viruses. Two days after infection, GFP-positive cells were sorted and total cell proteins were used for Western blots; the same membrane was blotted with PU.1 and Stat3 antibodies. U6-AAAA-PU718-8 expresses PU718 siRNA with an eight-base palindrome hairpin from a U6 promoter with AAAA as the last bases.
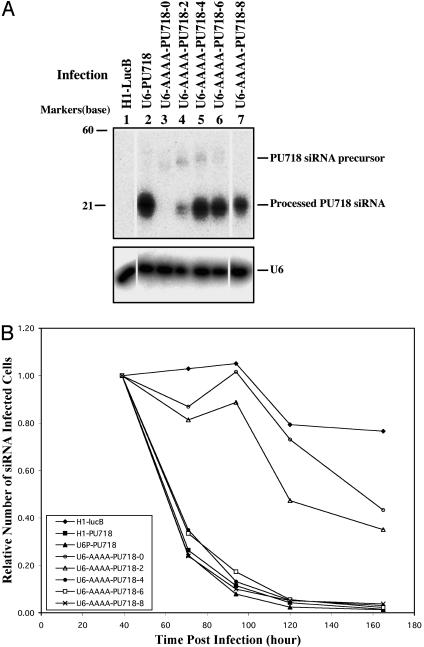
Fig. 3.
Effects of palindrome loop lengths on siRNA function. (A) Expression in NIH 3T3 cells. Cells were infected with the indicated viruses, and after 2 days GFP-positive cells were sorted by FACS and propagated. Northern blots were performed with total cellular RNA (22); the membrane was sequentially hybridized with probes for siRNAs targeting the PU718 site (5′-GAAGCTCACCTACCAGTTC-3′) and for endogenous U6 RNA. (B) Functional expression in UT7 cells. UT7 cells were infected with the indicated viruses, and the relative number of siRNA-infected cells were calculated as described in Fig. 2, except that the ratio of GFP-positive cells to GFP-negative cells was normalized to the ratio at 39 h postinfection.
Our preliminary studies examining functional expression of siRNAs took advantage of the fact that the PU.1 transcription factor is essential for survival of UT7 cells (data not shown). After infection of these cells with an MSCV-pgk-GFP retroviral vector expressing either siRNA or cDNA, the proportion of infected cells reaches its highest level after ≈2 days, as determined by the proportion of GFP-positive cells. If the siRNA does not affect cell growth or survival, this proportion remains constant thereafter as the cells continue to proliferate (Fig. 2 A; H1-LucB). In contrast, expression of a siRNA (termed PU718) targeting site 718 in PU.1 mRNA caused the proportion of infected cells to decrease markedly after the first 2 days. This was the case whether the PU718 siRNA was expressed from a U6 or H1 promoter (H1-PU718 and U6-PU718) or the AAAA sequence (H1-AAAA-PU718 and U6-AAAA-PU718) was present at the 3′ end of the promoter (Fig. 2 A). The ability of siRNAs expressed from promoters with the AAAA sequence to block PU.1 function was only marginally less than those expressed from the wild-type promoters (Fig. 2 A). Expression of a second siRNA, targeting site 646 in the PU.1 mRNA (PU646), also resulted in a rapid decrease of infected cells (Fig. 2 A), establishing that the loss of infected cells was likely due specifically to knockdown of PU.1 and not an effect of the siRNA on expression of other genes.
The Western blots in Fig. 2B extend these observations by showing a specific reduction of PU.1 protein in cells in which the PU718 siRNA was expressed from wild-type H1 or U6 promoters or from a U6 promoter with the additional AAAA sequence. This effect was specific for PU.1 because the level of the control STAT3 protein was unaffected (Fig. 2B), as was the total profile of UT7 cell proteins (determined by Ponceau S staining of the membrane; data not shown).
Next we systematically investigated the effects of different sized palindrome loops on siRNA expression and processing (Fig. 3). Northern blot analysis of cells infected with virus expressing PU718 siRNA with four-, six-, or eight-base palindrome loops from the AAAA-modified U6 promoter showed that all had high levels of the processed 21-base PU718 siRNA (Fig. 3A). Expression of functional PU718 siRNAs with a four-, six-, or eight-base palindrome loop was also demonstrated by the rapid elimination of infected cells (U6-AAAA-PU718-4, -6, and -8; Fig. 3B). Note that the configuration of the U6-AAAA-PU718-8 vector closely resembles that of our siRNA library clones in that it expresses PU718 siRNA from the AAAA-modified U6 promoter and there is an eight-base palindrome loop. As shown in Fig. 2B, infection of UT7 cells by the U6-AAAA-PU718-8 virus reduced the level of PU.1 protein to that achieved by the U6-PU718 vector.
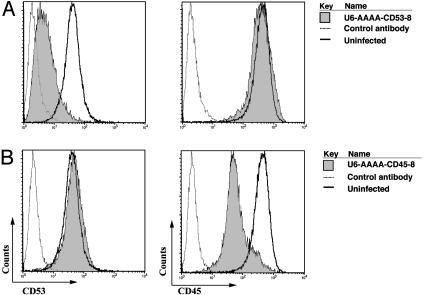
Next we demonstrated that many other siRNAs could be efficiently and functionally generated from our modified U6 expression cassette. As example siRNA sequences were created to target CD53 and CD45 mRNAs, these siRNA sequences were inserted into the modified U6-AAAA promoter, and the construct contained the same eight-base palindrome loop as did the clones from the siRNA library. Fig. 4A Left showed that expression of the siRNA specific for CD53 caused an ≈10-fold reduction in the level of surface CD53 expression without affecting the level of the control CD45 protein (Fig. 4A Right). Conversely, expression of a CD45 siRNA lowered CD45 expression ≈10-fold without affecting the level of the control CD53 protein (Fig. 4B).
Fig. 4.
Specific inhibition of CD53 and CD45 expression by siRNAs. Viral vectors expressing a CD45- or CD53-specific siRNA contained a U6 promoter with the AAAA as the last bases and the eight-base palindrome loop (5′-GTCGCGAC-3′). UT7 cells were infected with the indicated viruses, and infected GFP-positive cells were sorted by FACS and propagated. U6-AAAA-CD53-8-infected (A) or U6-AAAA-CD45-8-infected (B) cells and uninfected cells (solid line in each panel) were stained with phycoerythrin (PE)-labeled CD53 (Left) or CD45 (Right) or a PE-labeled rat IgG2a control antibody (dotted line in all panels).
Taken together, these experiments demonstrate that our retrovirus vectors express high levels of functional siRNAs. In particular, our data suggest that there is some flexibility in the identity of the last four bases of the Pol III promoter and in the size and the sequence of the hairpin loop. Thus, our modifications of the promoter and palindrome loop were well tolerated by the cellular machinery in producing and processing functional siRNA.
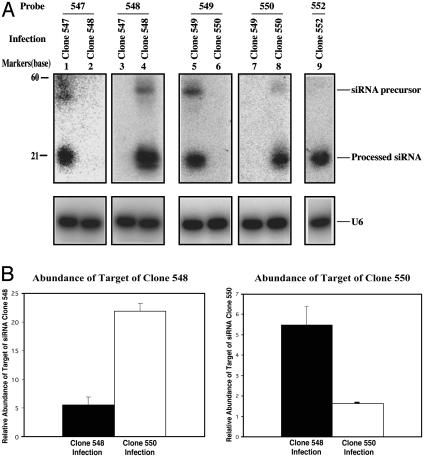
Validation of SPEED. After these preliminary studies, we applied the SPEED process detailed in Fig. 1 to construction of a siRNA library from a mouse embryo cDNA library. We obtained three million independent clones, all of which contained inserts. Twenty-seven clones were chosen at random for sequencing, all of which revealed matches against known mouse genes (Table 1, which is published as supporting information on the PNAS web site). Five random clones were used to generate stable NIH 3T3 cell lines. Northern blotting with probes corresponding to their mRNA targets demonstrated that each of the cDNA-derived inserts produced the expected processed ≈21-base siRNAs (Fig. 5A). More importantly, these clones caused specific reductions in their corresponding target mRNA. For example, real-time PCR analysis showed that expression of clone 548 caused a marked reduction of the corresponding 548 clone target mRNA, compared to cells expressing clone 550 siRNA (Fig. 5B). Conversely, cells expressing clone 550 siRNA exhibited marked reduction of the corresponding clone 550 target mRNA, but there was no effect on expression of clone 548 mRNA (Fig. 5B). These data demonstrate that clones in our siRNA library efficiently express the encoded siRNA and result in potent and highly specific inhibition of expression of the corresponding endogenous genes.
Fig. 5.
Functional expression of random siRNA clones created from the mouse embryo library. (A) Expression of siRNA clones in NIH 3T3 cells. The indicated clones from the mouse embryo siRNA library were used to infect NIH 3T3 cells. GFP-positive infected cells were purified and cultured. Northern blot analysis was performed on total RNA from purified cells by using probes identical to the 5′ strands of the respective siRNAs (see Table 1 for sequence information). The U6 probe was used as a control. (B) Knockdown of endogenous genes by clones 548 and 550. Real-time PCR was performed on RNA from populations of infected NIH 3T3 cells infected with clones 548 or 550 (see Table 1 for sequence information). The absolute amount of the transcript targeted by the siRNA as well as the GAPDH control were deduced from the standard curve for amplification of the respective genes. The relative abundance of target of siRNA clone 548 (Left) or clone 550 (Right) was normalized to that of GAPDH.
Discussion
We have developed the SPEED process to generate siRNA libraries from double-stranded cDNA. Multiple siRNAs are produced from each cDNA, thus increasing the chances of obtaining a functional siRNA for any desired gene; a typical ≈2,000-bp cDNA would be expected to generate ≈100 different siRNA clones. Analyzing many siRNAs against each target is particularly advantageous for genes that exhibit alternative exon splicing, because siRNAs specific for individual exons will enable investigation of the function of individual spliced isoforms. Another potential application of our technology is “expression knock-down cloning.” Here a population of cells, each of which expresses a unique siRNA, is screened for a physiological or biochemical property (e.g., loss of a surface protein or expression of a reporter gene) that results from inhibition of a possibly unknown gene in a signaling or biochemical pathway.
Because no prior knowledge about transcripts is needed to generate a cDNA-derived siRNA library, libraries from many tissues, species, and cell lines can be readily generated to enrich for siRNAs targeting relevant genes. A library of siRNA clones generated from a nonredundant cDNA collection, e.g., a mammalian gene collection (MGC) (24, 25) full-length library or a UniGene library (National Center for Biotechnology Information), may be the most cost-effective method for production of arrayed clones against most expressed genes. A further advantage of SPEED is that the use of a high-fidelity polymerase reduces the error rate in clones by at least three orders of magnitude below that of chemically synthesized siRNA expression constructs.
In addition to cDNA libraries, other DNAs can be converted into a siRNA library by SPEED. For example, SPEED can be used to provide a library of siRNAs that target different segments of a viral genome such as HIV. Screening of such a library for siRNAs that potently inhibit virus replication or other viral-encoded functions could identify potential siRNA-based therapeutics. Finally, the basic design principles of SPEED can be applied to convert any DNA into inverted repeats encoding dsRNAs that can be used for genome-wide screening with RNAi. SPEED will enable the application of the powerful RNAi technology to many organisms at a genome-wide level.
Supplementary Material
Acknowledgments
We thank Xiaowu Zhang for the mouse embryo cDNA library, Changzheng Chen for help with Northern blotting, members of the Lodish lab, David Sabatini, David Root, Nir Hacohen, William Hahn, David Bartel, and Eric Lander for discussion and critical reading of the manuscript, and Ann Oakenfull for preparation of the manuscript. This work was supported by grants from the National Science Foundation and the Singapore–Massachusetts Institute of Technology Alliance (to H.F.L.). B.L. was supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society.
Abbreviations: dsDNA, double-stranded DNA; FACS, fluorescence-activated cell sorting; Pol, polymerase; RNAi, RNA interference; siRNA, small interfering RNA; SPEED, siRNA production by enzymatic engineering of DNA.
See Commentary on page 5313.
References
- 1.Fire, A., Xu, S. Q., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 3.McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 4.Paddison, P. J. & Hannon, G. J. (2003) Curr. Opin. Mol. Ther. 5, 217–224. [PubMed] [Google Scholar]
- 5.Ashrafi, K., Chang, F. Y., Watts, J. L., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nature 421, 268–272. [DOI] [PubMed] [Google Scholar]
- 6.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 7.Lee, S. S., Lee, R. Y. N., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nat. Genet. 33, 40–48. [DOI] [PubMed] [Google Scholar]
- 8.Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S. J. M., Copley, R. R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. (2000) Nature 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. & Ahringer, J. (2000) Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 10.Lum, L., Yao, S. Q., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M. & Beachy, P. A. (2003) Science 299, 2039–2045. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 13.Miyagishi, M. & Taira, K. (2002) Nat. Biotechnol. 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 14.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul, C. P., Good, P. D., Winer, I. & Engelke, D. R. (2002) Nat. Biotechnol. 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 16.Sui, G. C., Soohoo, C., Affar, E., Gay, F., Shi, Y. J., Forrester, W. C. & Shi, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn, J. L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N. M., Hartman, S., Harrison, P. M., Nelson, F. K., Miller, P., Gerstein, M., et al. (2003) Genes Dev. 17, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Cancer Cell 2, 243–247. [DOI] [PubMed] [Google Scholar]
- 19.Pear, W. S., Nolan, G. P., Scott, M. L. & Baltimore, D. (1993) Proc. Natl. Acad. Sci. USA 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albritton, L. M., Tseng, L., Scadden, D. & Cunningham, J. M. (1989) Cell 57, 659–666. [DOI] [PubMed] [Google Scholar]
- 21.Luo, B., Aster, J. C., Hasserjian, R. P., Kuo, F. & Sklar, J. (1997) Mol. Cell. Biol. 17, 6057–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 23.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 24.Strausberg, R. L., Feingold, E. A., Klausner, R. D. & Collins, F. S. (1999) Science 286, 455–457. [DOI] [PubMed] [Google Scholar]
- 25.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.