Abstract
Zinc is an essential micronutrient that can also be toxic. An intricate mechanism exists in yeast that maintains cellular zinc within an optimal range. The centerpiece of this mechanism is the Zap1p protein, a transcription factor that senses zinc deficiency and responds by up-regulating genes involved in zinc metabolism. A microarray screen for novel Zap1p target genes suggested a role in zinc homeostasis for four homologous yeast genes. The expression of two of these genes, YDR492w and YOL002c, suggested direct regulation by Zap1p, whereas the expression of YOL002c and a third homologous gene, YOL101c, was induced by high zinc. YDR492w and YOL002c are confirmed to be direct Zap1p target genes. The induction of YOL002c and YOL101c by toxic metal ion exposure is shown to be mediated by the Mga2p hypoxia sensor. Furthermore, YOL101c is induced by deletion of the Aft1p iron-responsive transcription factor. These three genes, along with a fourth yeast homolog, YLR023c, have phenotypic effects on zinc tolerance and Zap1p activity. Because of their metalloregulation, zinc-related phenotypes, and highly conserved motifs containing potential metal-binding residues, this family has been renamed the IZH gene family (Implicated in Zinc Homeostasis). Furthermore, these genes are regulated by exogenous fatty acids, suggesting a dual role in lipid metabolism. The IZH genes encode membrane proteins that belong to a ubiquitous protein family that includes hemolysin III and vertebrate membrane steroid receptors. We propose that the IZH genes affect zinc homeostasis either directly or indirectly by altering sterol metabolism.
Zinc is an essential micronutrient and many of the proteins involved in maintaining zinc homeostasis are highly conserved throughout evolution. For example, the ZIP and CDF families of zinc transporter proteins can be found in organisms ranging from bacteria to humans (1). The coordinate induction of ZIP (ZRT1, ZRT2, and ZRT3) and CDF (ZRC1) gene transcription in zinc-limited Saccharomyces cerevisiae is administered by the zinc-sensing transcription factor, Zap1p (2). Zap1p binds to an 11-bp site in its target promoters called a zinc responsive element (ZRE) with the consensus sequence ACCTTNAAGGT. In a previous study, DNA microarrays were used to define the Zap1p regulon as well as to identify genes induced by zinc excess (2). Herein, we describe the initial characterization of a previously unrecognized family of four yeast genes discovered in the course of that study. Two of these genes are induced by zinc-limitation in a Zap1p-dependent manner and two respond to excess metals via the hypoxia sensor Mga2p. All four genes encode membrane proteins with mutant phenotypic effects on zinc tolerance and homeostasis. Therefore, we have designated these genes IZH1-4 (Implicated in Zinc Homeostasis).
Two observations suggested other roles for the Izh proteins unrelated to zinc metabolism. First, some IZH genes are transcriptionally regulated by fatty acids (3). Second, the proteins they encode are homologs of vertebrate membrane steroid receptors (mSRs) that mediate rapid, posttranslational (also referred to as nongenomic) effects of steroids (4).
This report demonstrates metalloregulation of three of the four IZH genes as well as zinc-dependent mutant phenotypes for all four genes. Increased IZH gene dosage is also shown to affect zinc homeostasis. Lipid- and oxygen-dependent expression of IZH2 and IZH4 is confirmed, and analysis of related genes link IZH gene function with sterol metabolism. We propose that the Izh proteins affect zinc metabolism either by altering membrane sterol content or by directly altering cellular zinc levels.
Materials and Methods
Yeast Strains. The strains used in this study are described in Table 2, which is published as supporting information on the PNAS web site. kanMX4::IZH deletion strains were either purchased from EUROSCARF or generated by PCR-based gene disruption using short flanking homology (5). Multiple mutants were generated from a heterozygous quadruple knockout strain engineered by successive rounds of mating and sporulation. The kanMX4 markers in the izh2, izh3, and izh4 strains were replaced with the hphMX4 (hygromycin), natMX4 (nourseothricin), and ura3MX4 (URA+) cassettes, respectively (6, 7). Nourseothricin was obtained from WERNER BioAgents (www.webioage.com).
DNA Manipulations. A complete list of primers is given in Data Set 1, which is published as supporting information on the PNAS web site. All PCR products were cloned into their respective plasmids by gap repair (8). IZH-lacZ fusions were generated as described (2). Only the fusion of the second in-frame ATG in the IZH2 ORF to lacZ resulted in a functional promoter construct (2, 3). IZH1mutZRE-lacZ and IZH2mutZRE-lacZ constructs in which each position in the ZREs was altered by transversion mutation (mutZRE) were generated by overlap extension PCR (9). HIS4-lacZ (10), CYC1-lacZ (11), FET3-lacZ (gift of A. Dancis, University of Pennsylvania, Philadelphia), and OLE1-lacZ (p62::934) (12) reporters were used as controls. pNB404 contains lacZ driven by a minimal CYC1 promoter lacking upstream activating sequences (CYC1ΔUAS-lacZ) (13). Insertion of putative regulatory elements between the XbaI and XhoI sites of this plasmid was used to test element function. An IZH1 ZRE insert was generated by overlap extension PCR. LORE-lacZ (pAM6) and ZRE-lacZ (pDg2 URA3/pDg2 LEU2) were made previously (12, 14). The inserts for these constructs contain the OLE1 low-oxygen response element (LORE, ACTCAACAA) and the ZRT1 ZRE (ACCCTCAAGGT), respectively.
Analysis of the promoter regions of zinc-inducible genes was performed by using rsa-tools (http://rsat.ulb.ac.be/rsat) (15). The OLE1 LORE and the FET3 FeRE (iron response element, TGCACCCA) were used as seed sequences for the program.
Single- and multicopy plasmids were generated by the insertion of PCR-amplified genomic fragments (from ≈1,000 bp upstream of ATG to ≈500 bp downstream of STOP) into the centromeric pRS315 and episomal YEp353 vectors. Fragments were inserted into YEp353 between the EcoRI and BamHI sites and into pRS315 between the SacI and SalI sites. For GAL1-driven overexpression, each ORF was amplified by PCR and inserted between the SacI and SalI sites of the pRS316-GAL1 plasmid (16). For IZH2 the GAL1 promoter was fused to both the first and second in-frame ATG codons to make GAL1-IZH2.1 and GAL1-IZH2.2. GAL1-hZip1 and GAL1-ZRT1Δ2 express the human hZip1 and mutant yeast Zrt1p zinc transporter proteins, respectively. Overexpression was achieved by using an estradiol-inducible system (17) or by 2% galactose.
Yeast Growth and Assays. Yeast and bacterial transformations were performed by standard methods. Microarrays were previously published (2). It was necessary to culture yeast for 4 days in selectable minimal media before re-inoculation in high-zinc media to ensure consistent results. Protocol for the preparation of chelexed synthetic media (CSD) (2) and low-zinc media (14) are published. Zinc, as ZnCl2, was added back to CSD to a final concentration of 50 nM (deficiency), 10 μM (repletion), and 3 mM (excess) for microarrays and lacZ assays. To induce zinc toxicity, 6 mM zinc was added to liquid media and 12 mM zinc was added to agar plates. A 37.5% wt/vol stock of sodium myristate was first dissolved in 50% EtOH/25% Tween-40 and then added to CSD to a final concentration of 0.375% myristate. Doubling time was determined by dividing ln 2 by the slope of a line generated by plotting growth time (log phase only) versus ln OD600. β-galactosidase activity was measured by using published procedures (2) and is expressed in Miller units.
Results
Microarray Analysis. Previously published microarray data showed higher mRNA levels for two genes, YDR492w and YOL002c, in zinc-deficient vs. zinc-replete wild-type cells and greater expression in a zinc-deficient wild-type strain vs. a zinc-limited zap1 strain. These genes also showed decreased expression in zinc-deficient vs. zinc-replete zap1 cells (Fig. 1). YDR492w and YOL002c are highly similar in sequence and analysis of the S. cerevisiae genome revealed two other related genes, YLR023c and YOL101c. YOL002c and YOL101c showed higher expression in cells exposed to excess zinc. YLR023c did not show significantly altered expression in response to zinc status. These genes are herein named IZH1 (YDR492w), IZH2 (YOL002c), IZH3 (YLR023c), and IZH4 (YOL101c).
Fig. 1.
IZH gene expression response to Zn2+. Comparative expression of IZH genes across four experimental regimes (n = 2 for each). In this and all subsequent figures, –, +, and ++ indicate 50 nM, 10 μM, and 3 mM Zn2+ in the growth medium, respectively. Data are presented as log2 of the ratio of expression. Dotted lines demarcate a 2-fold difference in expression.
IZH1 and IZH2 Are Zap1p Target Genes. IZH1 and IZH2 possess putative ZREs in their promoter regions, located at –416 (ACCTTTAGGGT) and –225 (TCCTCTAGGGT), respectively. Confirmation of the functionality of the IZH2 ZRE was reported previously (2). The IZH1 ZRE-lacZ construct yielded 30-fold more activity under zinc deficiency than under zinc repletion (455.8 ± 12.2 vs. 15.3 ± 0.7). In a zap1 strain, this effect disappeared (9.3 ± 1.4 vs. 17.6 ± 6.6).
IZH1- and IZH2-lacZ reporters were inducible by zinc deficiency in a wild-type strain, but not in a zap1 strain (Fig. 2 A and B). IZH3-lacZ, CYC1-lacZ, and HIS4-lacZ controls were not induced by zinc deficiency, demonstrating that general changes in the levels of transcription or translation are not responsible for the observed induction of IZH1 and IZH2 (Fig. 2C). Moreover, IZH1mutZRE-lacZ and IZH2mutZRE-lacZ reporters, in which the ZREs were mutated, were not inducible by zinc limitation (Fig. 2 A and B).
Fig. 2.
Zap1p-dependent regulation of IZH1 and IZH2. IZH1-lacZ and IZH1mutZRE-lacZ (A) and IZH2-lacZ and IZH2mutZRE-lacZ (B) reporter fusion constructs show Zap1p-dependent induction during Zn2+-limitation. (C) A similar analysis of the IZH3-lacZ, HIS4-lacZ, and CYC1-lacZ control constructs. (D) The IZH2-lacZ reporter responds to both Zn2+ and exogenous myristate (C14:0). Black bars show reporter activity in – Zn2+, and gray bars show reporter activity in + Zn2+. In this and all other figures showing lacZ data, a representative experiment performed in triplicate is shown and the error bars represent ±1 SD.
IZH2 Is Regulated by Fatty Acids. A previous report showed that IZH1, IZH2, and IZH4 expression is induced by fatty acids (3). We have confirmed that IZH2-lacZ responds independently to both zinc and the addition of exogenous myristate (Fig. 2D). Fatty acids can induce gene expression via the Oaf1p/Pip2p complex that binds to oleate response elements (OREs). Putative OREs are present in the IZH2 (–159 to –167 bp), IZH1 (–302 to –328 bp), and IZH4 (–240 to –263 bp) promoters (3).
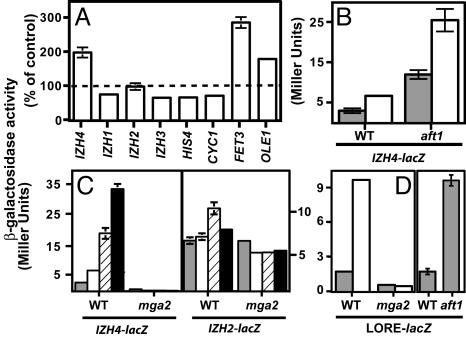
IZH4 Is Induced by Excess Zinc. The IZH4-lacZ reporter showed elevated activity (2-fold) when grown in elevated zinc, whereas a general inhibitory effect of high zinc on lacZ activity is demonstrated by a significant decrease in IZH1-lacZ, IZH3-lacZ, HIS4-lacZ, and CYC1-lacZ activities in cells exposed to 3 mM zinc (Fig. 3A). Indeed, high levels of zinc have been shown to inhibit both protein and RNA synthesis by up to 70% in certain yeast species (18). Unlike the control constructs, the activity of IZH2-lacZ did not decrease in high zinc, suggesting the existence of some factor that maintains elevated expression.
Fig. 3.
Metal induction of IZH2 and IZH4.(A) Relative activities of promoter-lacZ reporters shown as % change in activity in cells exposed to ++ zinc relative to activity in + zinc. (B) Effect of aft1 mutation on IZH4-lacZ activity. (C) Mga2p dependence of IZH4-lacZ and IZH2-lacZ activity in cells exposed to 3 mM Zn2+ (white bars), 400 μM Co2+ (hatched bars), or 400 μM Ni2+ (black bars). Control treatment has no metals added (gray bars). (D) Effect of Zn2+, Mga2p, and Aft1p on LORE-lacZ activity. In B and D, gray bars show activity in + zinc, and white bars show activity in ++ zinc.
IZH2 and IZH4 Are Part of the Hypoxic Response. Table 1 lists genes with an average induction of >2-fold in cells exposed to 3 mM zinc (n = 2) and includes known targets of the Mga2p hypoxia sensor, OLE1 (12) and Ty1 elements (19). The remaining genes are known to be induced by either low pO2, by deletion of the Ssn6p O2-activated repressor complex (20, 21) or by iron deficiency via the Aft1p iron-responsive transcription factor (22). Indeed, Fig. 3A confirms zinc induction for the OLE1-lacZ and FET3-lacZ reporters. A screen for regulatory elements in the promoters of the O2-regulated genes by using rsa-tools generated a probability-based consensus matrix that matched the LORE (low-oxygen response element). A similar screen of the promoters of the Aft1p-target genes generated a consensus matrix that matched the FeRE. With these matrices, we scanned 750 bp of the promoters of all genes in Table 1 and found that most of the O2-regulated promoters contained putative LOREs and that all of the Aft1p-target promoters contained putative FeREs (Data Set 2, which is published as supporting information on the PNAS web site).
Table 1. Genes induced >2-fold by zinc excess.
| Group | Gene name | Fold induction |
|---|---|---|
| Induced by SSN6 deletion or by low oxygen | IZH4* | 6.6 |
| OLE1* | 3.4 | |
| HSP26* | 3.4 | |
| YGL039w* | 2.9 | |
| ERG3* | 2.8 | |
| PIR3 | 2.6 | |
| YOR338w* | 2.3 | |
| HSP30* | 2.3 | |
| COS10* | 2.2 | |
| IZH2* | 2.1 | |
| NCE103* | 2.0 | |
| AHP1* | 2.0 | |
| YGR161c* | 2.0 | |
| HSP104 | 2.0 | |
| YOL106w† | 2.8 | |
| Miscellaneous | PDR3* | 2.6 |
| MGA2* | 2.3 | |
| UBS1 | 2.1 | |
| HSP150 | 2.0 | |
| Iron metabolism | FIT3† | 7.6 |
| FIT2† | 7.2 | |
| TAF1† | 6.2 | |
| TIS11† | 3.6 | |
| ENB1† | 3.4 | |
| ARN1† | 3.0 | |
| FTR1*† | 2.4 | |
| FRE1† | 2.3 | |
| SIT1† | 2.2 | |
| FET3† | 2.0 | |
| HMX1† | 2.0 | |
| Ty retrotransposons | YBL005w-A (YBLWTy1-1) | 2.1 |
| YER138c (YERCTy1-1) | 2.2 | |
| YER160c (YERCTy1-2) | 2.3 | |
| YHR214c-B (YHRCTy1-1) | 2.2 | |
| YML045w (YMLWTy1-2) | 2.6 | |
| YBR012w-A/-B (YBRWTy1-2) | 2.2/2.2 | |
| YCL019w/20w (YCLWTy2-1) | 2.1/2.4 | |
| YJR026w/27w (YJRWTy1-1)* | 2.2/2.5 | |
| YJR028w/29w (YJRWTy1-2) | 2.2/2.2 | |
| YML039w/40w (YMLWTy1-1) | 2.2/2.6 | |
| YMR045c/46c (YMRCTy1-3)* | 2.0/2.6 | |
| YMR050c/51c (YMRCTy1-4) | 2.3/2.6 |
Genes in bold have promoters containing putative regulatory elements scoring >7.0 when using the LORE or FeRE matrices generated by rsa-tools.
LORE-containing.
FeRE-containing.
The IZH4 promoter contains a potential LORE between –189 and –197 bp but does not contain an FeRE, suggesting that it is a target of Mga2p instead of Aft1p. To address this, we tested the effects of mga2 and aft1 mutations for their effects on IZH4-lacZ activity. Fig. 3B shows that although the induction of IZH4-lacZ in response to zinc was still 2-fold in an aft1 mutant strain, the basal level of activity of the reporter construct was increased 5-fold. Fig. 3C shows that basal and zinc-inducible expression of IZH4-lacZ depends on Mga2p. In addition, other stimuli that are known to induce the hypoxic response in yeast, such as high Co2+ and Ni2+ (12), also induce IZH4-lacZ. These responses are not seen in an mga2 strain. Fig. 3D confirms that the LORE-lacZ hypoxia reporter is also induced by high zinc in an Mga2p-dependent fashion, as well as by aft1 deletion. The IZH2 promoter also contains a putative LORE sequence between –137 and –145 bp. IZH2-lacZ is weakly induced by Co2+ and Ni2+ in an Mga2p-dependent manner. Like the control plasmids in wild-type yeast exposed to high zinc, elevated IZH2-lacZ activity is not maintained in an mga2 mutant, suggesting that Mga2p is responsible for maintaining elevated expression in high zinc (Fig. 3C).
IZH Genes Encode Homologs of Vertebrate Membrane Steroid Receptors. The IZH genes encode related proteins that belong to a large family of membrane proteins. An alignment of the four yeast proteins, a highly similar human protein (HsAdipoR1, NP_057083), three human membrane progestin receptors (HsmPRβ, NP_588608; HsmPRγ, NP_060175; and HsmPRα, NM_178422), and hemolysin III (Hly3, AAM90670) from Bacillus cereus is shown in Fig. 4. Each polypeptide has at least seven putative transmembrane domains (TMs) and a similar predicted topology with cytoplasmic N termini and extracytoplasmic C termini, although known vertebrate membrane progestin receptors possess an eighth predicted TM at the C terminus. This predicted topology has been confirmed for Izh4p (23). Five motifs that cluster on the cytoplasmic side of the membrane are conserved in the Izh-like proteins. These conserved regions, which contain potential metal-binding residues, can be summarized as follows: (i) a long motif N-terminal to TM1 that generally resembles PxnGYRxnNEx2Nx2T/SH; (ii) an Sx2Hx5S motif at the C terminus of TM2; (iii) a Dx9GS motif at the beginning of TM3; (iv) a Px2H motif in TM5 where the H residue is generally only conserved in higher eukaryotes; and (v) the loop between TM6 and TM7 containing PER/KxnPG and Hx2F/WH motifs with a conserved histidine in the middle of TM7 being most common. Hly3-like members of this family share most of these motifs with the striking exceptions of truncated motifs 1 and 5 and a complete lack of motif 4.
Fig. 4.
Multiple sequence alignment. Izh1p-4p and similar proteins are aligned by using clustalx. Predicted transmembrane segments are indicated by solid bars. Highly conserved regions are boxed. Stars indicate putative metal-binding residues that are highly conserved in the PAQR superfamily. Backslashes show locations where sequence was deleted to aid alignment.
IZH Genes Affect Zinc Tolerance. A collection of strains in which all combinations of the IZH genes have been deleted was generated. All of these strains were viable, indicating that no single gene or combination of genes was essential for viability. When stationary-phase cells were inoculated into either zinc-replete or -limiting media, all strains grew with wild-type characteristics in terms of lag-phase duration, exponential growth rate, and final yield of cell number (data not shown).
When these strains were inoculated into a high-zinc medium (6 mM), mutant phenotypes were observed. First, the izh3 strain showed slight but reproducible improvement in growth in high zinc when compared with the wild-type strain (Fig. 5A). This phenotype could be complemented by reintroduction of the IZH3 gene on a high-copy plasmid and was found to be attributed to a decrease in lag-phase duration in the mutant rather than a change in exponential-phase growth rate because the doubling time (DT) of zinc-treated izh3 mutant cells (3.25 ± 0.06 h) was identical to that of wild-type cells (3.22 ± 0.06 h). Mutation of izh2 also altered zinc sensitivity. In contrast to izh3, izh2 did not alter lag-phase duration but rather reduced the growth rate of mutant cells in exponential phase (DT = 4.28 ± 0.24 h).
Fig. 5.
Zn2+-dependent phenotypes. (A) Growth of wild type and izh3 mutants in liquid culture containing 6 mM zinc. pIZH3 is a 2-μ plasmid bearing the IZH3 gene under the control of its own promoter. (B) Growth of izh mutant strains on SD plates with or without 12 mM zinc added. (C) Complementation of zinc-dependent phenotypes by CEN (C) plasmids containing nativepromoter-driven IZH2 (pIZH2), IZH4 (pIZH4), and IZH1 (pIZH1) genes. (D) Complementation of the izh2 mutant phenotype by CEN plasmids harboring the GAL1-IZH2.1 and GAL1-IZH2.2 constructs on plates containing 2% glucose (Upper) or 2% galactose (Lower) with or without 12 mM zinc. Error bars represent ±1 SD.
Effects on zinc sensitivity were also apparent on agar plates containing high levels of zinc (12 mM). The izh2 mutant strain was sensitive under these conditions (Fig. 5B), a phenotype that could be complemented by a CEN plasmid containing the IZH2 ORF driven by its own promoter (Fig. 5C) or the GAL1 promoter (Fig. 5D). Both GAL1-IZH2.1 and GAL1-IZH2.2 (first and second in-frame ATG constructs) complement the izh2 phenotype when induced by galactose. As in liquid medium, strains bearing the izh3 mutation grew slightly better in high zinc than the corresponding isogenic IZH3 controls. For example, compare growth of izh1izh2izh4 with the quadruple mutant (Fig. 5B). A slight zinc sensitivity was also apparent in the izh1 single mutant (Fig. 5B). The contribution of IZH1 to zinc tolerance was clearly shown when comparing an izh2izh4 mutant with an izh1izh2izh4 strain. Moreover, the izh2izh4 mutant grew better than the izh2 strain indicating that mutation of IZH4 makes cells zinc tolerant. These latter two phenotypes could be complemented by CEN plasmids harboring the IZH1 or IZH4 genes (Fig. 5C) driven by their native promoters.
Because the IZH2 gene is adjacent to the PHO80 gene, it is possible that the izh2 phenotypes are due to PHO80 inactivation. pho80 mutants, like izh2 mutants, have been shown to be zinc sensitive (24). However, complementation of izh2 strains by plasmids bearing the IZH2 gene demonstrates that the zinc-sensitivity phenotype is not due to PHO80 inactivation.
Effects of IZH Overexpression on Zinc Homeostasis. By using the ZRE-lacZ reporter as a bioassay for Zap1p activity (indirect assay for labile zinc levels), the effect of IZH gene overexpression on cellular zinc homeostasis was measured. ZRE-lacZ activity was monitored over a range of zinc concentrations for wild-type strains in which the IZH genes were overexpressed from the GAL1 promoter by using the GEV system (5 μM β-estradiol induction) (Fig. 6A). Although there was no effect of IZH gene overexpression on the control HIS4-lacZ reporter, overexpression of any IZH gene significantly decreased ZRE-lacZ activity. Similar results were seen when the IZH genes were induced with 2% galactose (Fig. 6B), indicating that the observed effects were not an artifact of β-estradiol treatment. No significant effect on zinc accumulation was seen in strains overproducing these proteins (data not shown). GAL1-driven overexpression of either hZip1, a human zinc uptake transporter that does not function in yeast (25), or ZRT1Δ2, a nonfunctional mutant of yeast ZRT1 (26), has no significant effect on a ZRE-lacZ reporter, suggesting that the effect of IZH-gene dosage is not a generalized effect of aberrant protein folding or trafficking due to membrane protein overexpression (Fig. 6B).
Fig. 6.
IZH gene overexpression. (A) Overexpression of IZH1-4 using 5 μM β-estradiol in cells grown in low-zinc media medium results in a significant decrease in Zap1p activity measured with a ZRE-lacZ reporter construct (open symbols). No effect was seen when the same experiment was performed by using the HIS4-lacZ control reporter construct (filled symbols). IZH1-4 overexpression plasmids are LEU2-selectable and reporter plasmids are URA3-selectable. (B) Overexpression of IZH1-4 by using 2% galactose gives similar results (open symbols), whereas overexpression of control membrane proteins hZip1 and ZRT1Δ2 does not inhibit ZRE-lacZ activity (filled symbols). For the filled symbols in B the overexpression plasmids are URA3-selectable and the reporter plasmids are LEU2-selectable. Ctrl., control.
Discussion
IZH1, IZH2, IZH3, and IZH4 encode a family of paralogous proteins in yeast. The Izh proteins belong to a large and nearly ubiquitous family of proteins found in both prokaryotes and eukaryotes. This family has been named the PAQR (Progestin, AdipoQ-Receptor) family of proteins because several of its constituent members are steroid or adiponectin (AdipoQ) receptors. Structurally, PAQR proteins can be divided into two general subgroups: Hly3-like and Izh-like. Both subgroups are characterized by at least seven TMs and four highly conserved motifs rich in metal-binding amino acids. All of the conserved motifs are predicted to cluster on the cytoplasmic face of the membrane.
Little is known about the function of any member of this family. In the α-proteobacterium, Azospirillum brasilense, the Hly3-like gene encodes a chimeric protein with an Hly3-like N terminus fused to the CheA chemotaxis histidine kinase (27). Because A. brasilense is chemotactic toward hypoxic environments, Hly3-CheA may encode a sensor for hypoxia as well as the downstream kinase. Evidence that these proteins are receptors is more convincing for the vertebrate proteins. Expression of a sea trout mSR in cell culture resulted in progesterone-dependent inhibition of adenylate cyclase activity. This inhibition could be relieved by the addition of Pertussis toxin, suggesting that mSRs are inhibitory G protein-coupled receptors (4). Recombinant mSRs expressed in E. coli also bound progesterone with high affinity (28). Hly3 was originally discovered because of its hemolytic activity when overexpressed in E. coli, and evidence suggests that it is a pore-forming membrane protein with a pore diameter of ≈35 Å (29). These findings raise the possibility that PAQR proteins are receptors, channels, or both.
In yeast, gene regulation has provided the first functional clues. IZH1 and IZH2 are confirmed Zap1p-target genes, whereas IZH4 is induced by excess zinc. We demonstrated that deletion of either IZH1 or IZH2 results in increased sensitivity to elevated zinc, whereas deletion of IZH3 or IZH4 has the opposite effect. Zinc sensitivity is variable and requires long preculture times before re-inoculation in zinc-containing medium. The izh2 mutation increases the length of the cell cycle in zinc-treated cells, whereas izh3 mutation decreases the lag phase under the same conditions. Overexpression of any of these four genes results in decreased activity of the Zap1p transcription factor when cells are grown in zinc-limiting medium. Based on their metalloregulation and their impact on zinc tolerance and homeostasis, we propose a role for these genes in zinc metabolism.
Other lines of evidence suggested that the function of the IZH genes goes beyond a role in zinc metabolism. IZH4 is also induced by exposure to Zn2+, Co2+, Ni2+, and AFT1 deletion. Metalloregulation of IZH4 depends on the Mga2p hypoxia-responsive transcription factor. All three metal treatments, as well as aft1 deletion, are proven inducers of the hypoxic response (12). IZH2 has also been shown to be a weak target of Mga2p. The involvement of Hly3-CheA from A. brasilense in hypoxia sensing emphasizes the importance of this finding and suggests a conservation of function across species.
Furthermore, we confirmed earlier reports suggesting that IZH2 responds to exogenous myristate. Regulation of the IZH genes by fatty acids and hypoxia strongly suggests an alternative role in lipid metabolism. Preliminary phenotypic analysis shows that izh2 mutants are resistant to the sterol-binding antibiotic, nystatin (3), suggesting that izh2 mutants have altered membrane sterol composition. Furthermore, IZH3 transcription is induced and IZH4 transcription is repressed by defects in the ergosterol biosynthetic pathway (20). These observations, combined with the fact that some vertebrate orthologs function as receptors for structurally related steroids, identify ergosterol metabolism as a likely biochemical pathway in which to place the Izh proteins.
A role for the IZH genes in ergosterol metabolism helps explain the need for long precultures to see strong zinc-dependent izh mutant phenotypes. Even when cultures are vigorously shaken, yeast rapidly deplete the growth medium of O2 and cultures become increasingly anaerobic over time (30). Anaerobically grown yeast are auxotrophic for ergosterol (31); therefore, prolonged culturing may result in decreased sterol biosynthesis. If izh mutations exacerbate or ameliorate this situation they can have significant effects on sterol composition in preculture.
An anecdotal relationship between membrane sterol content and zinc metabolism has already been established. Depletion of cholesterol content in MCF-10A breast epithelial cells by using mevastatin significantly diminished the uptake of zinc but not 2-deoxyglucose (32). Nystatin treatment also inhibited 65Zn uptake in human fibroblasts (33) and induced the Zap1p regulon in S. cerevisiae (34). These studies suggest that changes in membrane sterols can specifically affect the permeability of certain ions. Therefore, it is possible that the sole role of the IZH genes is in ergosterol metabolism and that their effects on zinc tolerance are an indirect consequence of this role.
However, the induction of IZH1 and IZH2 by Zap1p under zinc deficiency, as well as the specific decrease in Zap1p activity in cells overexpressing Izh proteins, suggests a deeper connection between these genes, sterols, and zinc metabolism. A possible clue to the nature of this connection comes from the mSRs. Both transcriptional and posttranslational steroid signaling can be linked to zinc metabolism. The first obvious connection is that classical nuclear hormone receptors are zinc-binding proteins (35). That the most highly conserved residues in the mSR superfamily are potential metal-binding residues suggests that mSRs may indeed be metalloproteins as well. In addition, corticosteroids are known inducers of both zinc uptake and the expression of metallothionein (MT), a zinc-buffering protein (36). Sex hormones have been shown to induce zinc uptake as well as to regulate the expression of ZIP-family zinc transporters (37, 38). Lastly, zinc has been shown to regulate the aldosterone-mediated intracellular acidification of cultured kidney cells involving PKC (39). These studies suggest an intimate relationship between steroid signaling and zinc metabolism.
It is important to note here that posttranslational estrogen and glucocorticoid receptor signaling has been shown to involve a kinase cascade that includes PDK, a 3-phosphoinositide-dependent kinase (40) that phosphorylates a variety of substrates, including PKC (41). Yeast possess two orthologs of PDK, PKH1 and PKH2, which also act upstream of PKC (42). Intriguingly, IZH1 and IZH4 share promoter regions with the PKH1 and PKH2 genes, respectively. In yeast, such divergently transcribed genes often encode proteins in the same biochemical pathway (43). This finding suggests that there is a functional relationship between the IZH genes and the PDK cascade. This hypothesis is bolstered by the involvement of the PDK cascade in steroidal signaling in mammalian cells. It is also important to note here that signaling via both PDK (44) and PKC (45) is thought to be regulated directly by alterations in cellular zinc, a fact that connects this pathway to zinc homeostasis.
In conclusion, we propose three possible functions for the Izh proteins. First, these proteins may function solely in sterol metabolism. In this capacity they may influence the permeability of the plasma membrane and, consequently, the homeostasis of cations such as zinc. It is also possible that the Izh proteins function as transporters for zinc used in a signaling capacity, a possibility that may explain their regulation by Zap1p and their effect on Zap1p activity. The role of zinc in signal transduction is controversial. However, evidence for such a role is mounting. Perhaps the most compelling evidence is the finding that the CDF-1 zinc transporter in C. elegans positively regulates Ras-mediated signaling (46), another pathway that can transmit signals to PKC (47). It is possible that zinc, perhaps through Izh-like proteins, plays a widespread role in cellular kinase cascades. A third possibility is that the Izh proteins are involved in a signal transduction cascade that is independent of zinc, and that Zap1p is a downstream target of this pathway.
Supplementary Material
Acknowledgments
We thank C. E. Martin for generously providing hypoxia-responsive plasmids and A. Dancis for providing the FET3-lacZ plasmid. This work was supported by National Institutes of Health Grants GM20545 (to T.J.L.) and GM56285 (to D.J.E.), the University of Missouri Molecular Biology Program, and the University of Florida Department of Chemistry.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FeRE, iron response element; LORE, low-oxygen response element; mSR, membrane steroid receptor; TM, transmembrane domain.
References
- 1.Gaither, L. A. & Eide, D. J. (2001) Biometals 14, 251–270. [DOI] [PubMed] [Google Scholar]
- 2.Lyons, T. J., Gasch, A. P., Gaither, L. A., Botstein, D., Brown, P. O. & Eide, D. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karpichev, I. V., Cornivelli, L. & Small, G. M. (2002) J. Biol. Chem. 277, 19609–19617. [DOI] [PubMed] [Google Scholar]
- 4.Zhu, Y., Rice, C. D., Pang, Y., Pace, M. & Thomas, P. (2003) Proc. Natl. Acad. Sci. USA 100, 2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein, A. L., Pan, X. & McCusker, J. H. (1999) Yeast 15, 507–511. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein, A. L. & McCusker, J. H. (1999) Yeast 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- 8.Muhlrad, D., Hunter, R. & Parker, R. (1992) Yeast 8, 79–82. [DOI] [PubMed] [Google Scholar]
- 9.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch, A. G., Lucchini, G. & Fink, G. R. (1985) Proc. Natl. Acad. Sci. USA 82, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer, K., Prezant, T. & Guarente, L. (1987) Cell 49, 19–27. [DOI] [PubMed] [Google Scholar]
- 12.Vasconcelles, M. J., Jiang, Y., McDaid, K., Gilooly, L., Wretzel, S., Porter, D. L., Martin, C. E. & Goldberg, M. A. (2001) J. Biol. Chem. 276, 14374–14384. [DOI] [PubMed] [Google Scholar]
- 13.Bachhawat, N., Ouyang, Q. & Henry, S. A. (1995) J. Biol. Chem. 270, 25087–25095. [DOI] [PubMed] [Google Scholar]
- 14.Zhao, H., Butler, E., Rodgers, J., Spizzo, T., Duesterhoeft, S. & Eide, D. (1998) J. Biol. Chem. 273, 28713–28720. [DOI] [PubMed] [Google Scholar]
- 15.Hertz, G. Z. & Stormo, G. D. (1999) Bioinformatics 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 16.Liu, H., Krizek, J. & Bretscher, A. (1992) Genetics 132, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, C. Y. & Pinkham, J. L. (2000) BioTechniques 29, 1226–1231. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-del Valle, N. (1989) Mycopathologia 106, 23–29. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, S., Burkett, T. J., Yamashita, I. & Garfinkel, D. J. (1997) Mol. Cell. Biol. 17, 4718–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherry, J. M., Adler, C., Ball, C., Chervitz, S. A., Dwight, S. S., Hester, E. T., Jia, Y., Juvik, G., Roe, T., Schroeder, M., et al. (1998) Nucleic Acids Res. 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kastaniotis, A. J. & Zitomer, R. S. (2000) Adv. Exp. Med. Biol. 475, 185–195. [PubMed] [Google Scholar]
- 22.Rutherford, J. C., Jaron, S. & Winge, D. R. (2003) J. Biol. Chem. 278, 27636–27643. [DOI] [PubMed] [Google Scholar]
- 23.Kim, H., Melen, K. & von Heijne, G. (2003) J. Biol. Chem. 278, 10208–10213. [DOI] [PubMed] [Google Scholar]
- 24.Mao, X. C., Xia, Y. L., Hu, Y. F. & Lu, C. D. (2003) Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35, 86–91. [PubMed] [Google Scholar]
- 25.Gaither, L. A. & Eide, D. J. (2001) J. Biol. Chem. 276, 22258–22264. [DOI] [PubMed] [Google Scholar]
- 26.Gitan, R. S., Shababi, M., Kramer, M. & Eide, D. J. (2003) J. Biol. Chem. 278, 39558–39564. [DOI] [PubMed] [Google Scholar]
- 27.Hauwaerts, D., Alexandre, G., Das, S. K., Vanderleyden, J. & Zhulin, I. B. (2002) FEMS Microbiol. Lett. 208, 61–67. [DOI] [PubMed] [Google Scholar]
- 28.Zhu, Y., Bond, J. & Thomas, P. (2003) Proc. Natl. Acad. Sci. USA 100, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baida, G. E. & Kuzmin, N. P. (1996) Biochim. Biophys. Acta 1284, 122–124. [DOI] [PubMed] [Google Scholar]
- 30.Tolosa, L., Kostov, Y., Harms, P. & Rao, G. (2002) Biotechnol. Bioeng. 80, 594–597. [DOI] [PubMed] [Google Scholar]
- 31.Parks, L. W. & Casey, W. M. (1995) Annu. Rev. Microbiol. 49, 95–116. [DOI] [PubMed] [Google Scholar]
- 32.Mouat, M. F., Greenspan, P., Byerley, L. O. & Grider, A. (2003) J. Nutr. Biochem. 14, 74–80. [DOI] [PubMed] [Google Scholar]
- 33.Grider, A. & Vazquez, F. (1996) Biol. Trace Elem. Res. 54, 97–104. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, L., Zhang, Y., Zhou, Y., An, S. & Cheng, J. (2002) J. Antimicrob. Chemother. 49, 905–915. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, R. & Thompson, E. B. (1999) Steroids 64, 310–319. [DOI] [PubMed] [Google Scholar]
- 36.Cousins, R. J. & Coppen, D. E. (1987) Experientia Suppl. 52, 545–553. [DOI] [PubMed] [Google Scholar]
- 37.El-Tanani, M. K. & Green, C. D. (1997) J. Steroid Biochem. Mol. Biol. 60, 269–276. [DOI] [PubMed] [Google Scholar]
- 38.Costello, L. C., Liu, Y., Zou, J. & Franklin, R. B. (1999) J. Biol. Chem. 274, 17499–17504. [DOI] [PubMed] [Google Scholar]
- 39.Gekle, M., Silbernagl, S. & Wunsch, S. (1998) J. Physiol. 511, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simoncini, T., Fornari, L., Mannella, P., Varone, G., Caruso, A., Liao, J. K. & Genazzani, A. R. (2002) Steroids 67, 935–939. [DOI] [PubMed] [Google Scholar]
- 41.Sonnenburg, E. D., Gao, T. & Newton, A. C. (2001) J. Biol. Chem. 276, 45289–45297. [DOI] [PubMed] [Google Scholar]
- 42.Inagaki, M., Schmelzle, T., Yamaguchi, K., Irie, K., Hall, M. N. & Matsumoto, K. (1999) Mol. Cell. Biol. 19, 8344–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kruglyak, S. & Tang, H. (2000) Trends Genet. 16, 109–111. [DOI] [PubMed] [Google Scholar]
- 44.Min, Y. K., Park, J. H., Chong, S. A., Kim, Y. S., Ahn, Y. S., Seo, J. T., Bae, Y. S. & Chung, K. C. (2003) J. Neurosci. Res. 71, 689–700. [DOI] [PubMed] [Google Scholar]
- 45.Korichneva, I., Hoyos, B., Chua, R., Levi, E. & Hammerling, U. (2002) J. Biol. Chem. 277, 44327–44331. [DOI] [PubMed] [Google Scholar]
- 46.Bruinsma, J. J., Jirakulaporn, T., Muslin, A. J. & Kornfeld, K. (2002) Dev. Cell 2, 567–578. [DOI] [PubMed] [Google Scholar]
- 47.Price, B. D., Morris, J. D., Marshall, C. J. & Hall, A. (1989) Biochem. J. 260, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.