Abstract
micro RNAs (miRNAs) are small non-coding RNAs that act as posttranscriptional repressors by binding to the 3′-UTR of target mRNAs. On the other hand, mesenchymal-epithelial transition (EMT) and kidney fibrosis is a pathological process of chronic kidney disease (CKD), and its relationship to miRNAs is becoming recognized as a potential target for CKD therapies. To find new miRNAs involved in EMT, we examined miRNA expression in experimental models of EMT and renal epithelialization using microarray, and found that miR-34c attenuates EMT induced by TGF-β in a mouse tubular cell line. To confirm the effects of miR-34c in vivo, we administered the precursor of miR-34c to mice with unilateral ureteral obstruction, and miR-34c decreased kidney fibrosis area and the expression of connective tissue growth factor, α-SMA, collagen type 1, collagen type 3 and fibronectin. In conclusion, our study showed miR-34c attenuates EMT and kidney fibrosis of mice with ureteral obstruction.
micro RNAs (miRNAs) are small non-coding RNAs that act as posttranscriptional repressors by binding to the target mRNA, and are known to have important roles in controlling development and regulating cell proliferation and differentiation1. Moreover, many researchers have been elucidating involvement of miRNAs in different diseases, and have been becoming to recognize that miRNAs are potential targets for new therapeutic approaches2.
On the other hand, chronic kidney disease (CKD) is a major global health care problem, and causes enormous medical costs due to the therapies for end-stage renal disease (ESRD). Many clinicians are using the inhibitors of renin-angiotensin system (RAS) to slow the progression of CKD, however the inhibitors of RAS cannot fully stop the progression toward ESRD or regenerate the kidney function.
Epithelial-mesenchymal transition (EMT) is a pathological process which is characterized by dedifferentiation of epithelial cells, and results in tissue fibrosis with fibroblasts proliferation. This pathological process is thought to be a potential target for CKD therapies3. Actually, new therapies targeting kidney fibrosis in CKD have been being applied to clinical studies, and is thought to be a promising therapies for CKD with diabetic nephropathy4,5.
EMT is characterized by the up-regulation of mesenchymal genes such as Snail1 and Vimentin. Snail1 is reported to be expressed in tubular cells in experimental models of EMT such as TGF-β stimulation in vitro and unilateral ureteral obstruction (UUO)6,7. Previously, the origin of proliferated fibroblasts in kidney fibrosis was thought from tubular cells with EMT, however Lebleu recently reported that the origin of myofibroblasts in kidney fibrosis is mainly from local resident fibroblasts (50%) through proliferation, and the actual transition from tubular cells to myofibroblasts accounted for only 5%8. In contrast, it is also reported that tubular-specific induction of EMT causes kidney fibrosis9,10, thereby leading to speculation that tubular cells with EMT stimulate the proliferation of fibroblasts and result in kidney fibrosis.
Recent studies have been elucidating the role of miRNAs in kidney disease and EMT, and suggest miRNAs are potential targets for new therapies for CKD11,12,13,14,15,16,17. Especially regarding EMT, many researchers are trying to elucidate the role of miRNAs in EMT of kidneys. Chung et al. reported that miR-192 mediates TGF-β/Smad3-driven kidney fibrosis18. Similarly, Kriegel et al. also reported that miR-382 suppresses E-cadherin expression of human renal tubular cells via down-regulation of superoxide dismutase 219. In contrast to those reports, Krupa reported that loss of miR-192 promotes fibrogenesis in diabetic nephropathy20. Moreover, previous reports indicated that miR-200 family may have a critical role in the repression of E-cadherin by zinc finger E-box binding homeobox (ZEB)1 and ZEB2 during EMT21,22,23,24,25. In addition, miR-21 and miR-214 were shown to promote kidney fibrosis in animal models using UUO, and the previous studies suggested that the inhibition of those miRNAs might be a therapeutic approach to suppress kidney fibrosis26,27,28,29.
The aim of this study was to explore new miRNAs involved in EMT and to examine whether miRNA modification could ameliorate EMT. We have been elucidating the mechanisms of EMT and kidney fibrosis7,30, and have also been working on miRNA researches31. This time, we used EMT models using UUO and TGF-β, and also a renal epithelialization model using mouse embryonic stem (ES) cells which we previously shown32,33, and found a new miRNA which ameliorates EMT and kidney fibrosis.
Results
Ureteral obstruction induces epithelial-mesenchymal transition and alters the expression of miRNAs of kidneys
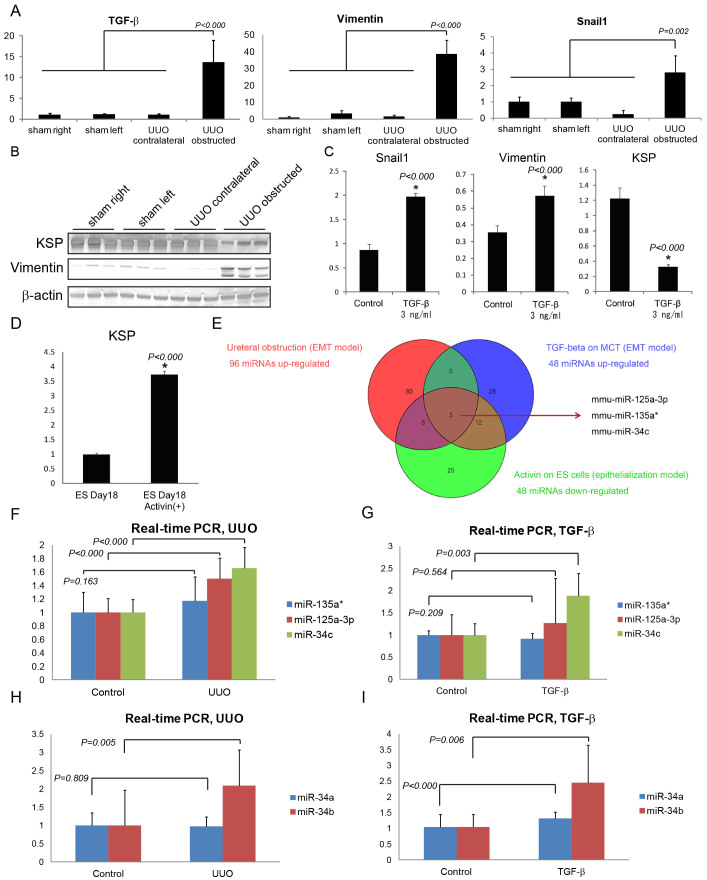
The unilateral ureter of 8 weeks ICR mice was ligated under anesthesia, and bilateral kidneys were harvested after one week of unilateral ureteral obstruction (UUO). Epithelial-mesenchymal transition (EMT) of kidneys was confirmed by PCR and Western blot. Snail1 and Vimentin, dedifferentiated markers of tubular cells, were significantly up-regulated compared with the contralateral kidney in UUO-operated mice and both sides of kidneys in sham-operated mice as well as TGF-β, an inductor of EMT. On the other hand, an epithelial marker, kidney specific protein (KSP) were down-regulated by ureteral obstruction (Fig. 1A, B).
Figure 1. Experimental models of EMT and epithelialization revealed miR-34c presumably involves in EMT.
(A, B) PCR and Western blot showing the expression of genes regarding EMT in kidneys of UUO and sham operated mice. Sham right; right kidneys of sham-operated mice. Sham left; left kidneys of sham-operated mice. UUO contralateral; contralateral kidneys of UUO-operated mice. UUO obstructed; UUO kidneys of UUO-operated mice. The original PCR and whole blot pictures are available in Supplementary Fig. S2 (n = 3). (A) PCR normalized to GAPDH. (B) Western blot. (C) Real-time PCR showing the expression of Snail1, Vimentin and KSP in MCT with or without TGF-β stimulation. (D) Real-time PCR showing the expression of KSP in mouse ES cells differentiated with or without Activin 10 ng/ml. (E) Venn diagram of microarray of three experimental models. (F) Real-time PCR of miRNAs in UUO mice. Control; contralateral kidneys of UUO-operated mice. UUO; UUO kidneys of UUO-operated mice. (G) Real-time PCR of miRNAs in MCT stimulated by TGF-β. (H, I) Real-time PCR of miR-34a and miR-34b in (H) UUO mice, and (I) MCT stimulated by TGF-β.
To elucidate miRNA involvement in EMT, the expressions of miRNAs in UUO kidneys were analyzed by miRNA microarray analysis, and were compared with contralateral kidneys (n = 4). The data was analyzed by GeneSpring GX (Agilent), and miRNAs up-regulated more than two-fold were sorted out (Supplementary Table S1). 96 miRNAs were up-regulated by UUO and were considered as possible candidate miRNAs for the induction of EMT (Fig. 1E).
TGF-β induces EMT of mouse tubule cell line and alters the expression of miRNAs
To find out miRNAs that were more likely to involve in EMT, an in vitro EMT model by TGF-β was also analyzed. A mouse proximal tubule cell line (MCT) was stimulated by TGF-β 3 ng/ml for 72 hours (n = 3), and the samples were harvested after TGF-β stimulation. Real-time PCR showed the up-regulation of Snail1 and Vimentin, and the down-regulation of KSP by TGF-β (Fig. 1C), and indicated that TGF-β induced EMT in MCT.
miRNA microarray was also performed to analyze the alterations of miRNA expression by TGF-β in MCT (n = 3), and 48 miRNAs were up-regulated by TGF-β more than two-fold (Supplementary Table S2). Among these 48 miRNAs, eight miRNAs were also up-regulated by ureteral obstruction (Fig. 1E).
Activin enhances renal epithelial induction of mouse embryonic stem cells and alters miRNAs
We previously reported that Activin 10 ng/ml enhances the differentiation of mouse embryonic stem (ES) cells and induced pluripotent stem (iPS) cells toward renal epithelial cells32. To narrow the results of microarray, we used this differentiation method of mouse ES cells with Activin as a counter model of EMT. Mouse ES cells were differentiated through embryoid body formation for three days, and were subsequently cultured by adhesion culture for additional 15 days. Activin 10 ng/ml was added to the differentiation medium for all differentiation period. The expression of KSP was analyzed by real-time PCR (Fig. 1D), and miRNA microarray analysis was carried out at day 18 of the differentiation. Activin enhanced the expression of KSP as we previously reported, and down-regulated 48 miRNAs less than half compared with the control samples without Activin (Supplementary Table S3).
To find out miRNAs involved in these three models; EMT by ureteral obstruction in mice, EMT by TGF-β in MCT and epithelialization by Activin in ES cells;, Venn diagram was made with GeneSpring GX (Fig. 1E). Only three miRNAs, mmu-miR-125a-3p, mmu-miR-135a* and mmu-miR-34c were involved in all three experimental models.
To confirm the expression change of these three miRNAs, real-time PCR of miRNAs was done in UUO mice and MCT stimulated by TGF-β (Fig. 1F, G). In these three miRNAs, only miR-34c showed statistically significant change, therefore we have decided to analyze miR-34c in detail.
The miR-34 family comprises three processed miRNAs in mammalians that are encoded by two different genes: miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript34. Therefore, we also performed real-time PCR of miR-34a and miR-34b in UUO mice and MCT stimulated by TGF-β (Fig. 1H, I). Interestingly, miR-34b was also up-regulated in those two experimental models while miR-34a showed only slight up-regulation in MCT with TGF-β. This results is compatible with previous reports showing homology of miR-34b and miR-34c in terms of the sequence and the function34,35,36.
Over-expression of miR-34c attenuates EMT induced by TGF-β in MCT
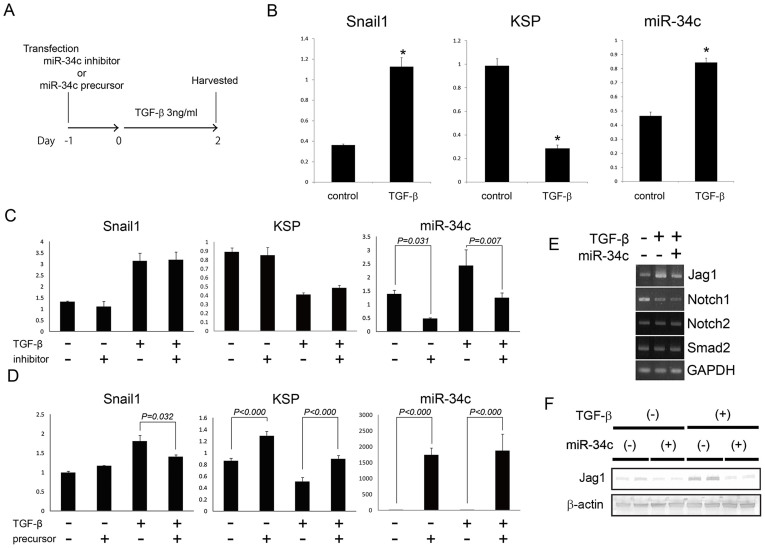
To examine the effects of miR-34c on EMT, knock-down of miR-34c were carried out using inhibitor of miR-34c in MCT. MCT was transfected with the inhibitor of miR-34c or the negative control of the inhibitor, and were subsequently stimulated with TGF-β 3 ng/ml from the next day (n = 3) (Figure 2A). Here, we changed the period of TGF-β stimulation from 3 days to 2 days since the effects of inhibitor of miR-34c was unlikely to last for a long time. Hence, the expression of Snail1, KSP and miR-34c was examined by real-time PCR after 2 days of TGF-β stimulation, and we have confirmed that EMT in MCT was induced by 2 days of TGF-β stimulation and miR-34c was up-regulated by TGF-β (Fig. 2B). Cells transfected with miR-34c inhibitor were harvested after two days of stimulation with TGF-β, and real-time PCR was carried out to analyze the expression of Snail1, KSP and miR-34c with triplicate. The expression of miR-34c was significantly up-regulated by TGF-β, and down-regulated by the inhibitor of miR-34c. However, the expression of Snail1 and KSP was not altered by knock-down of miR-34c (Fig. 2C).
Figure 2. miR-34c suppressed Jag1, and attenuated EMT induced by TGF-β in MCT.
(A) The protocol of transfection of miR-34c inhibitor or precursor. (B) Real-time PCR showing the expression of Snail1, KSP and miR-34c in MCT treated by TGF-β for 2 days. (C, D) Real-time PCR showing the expression of Snail1, KSP and miR-34c in MCT under indicated conditions. (C) Inhibitors of miR-34c. (D) Precursors of miR-34c. (E) PCR showing the expression of genes regarding Notch signal in MCT under indicated conditions. (F) Western blot of Jag1 and β-actin in MCT under indicated conditions. (n = 2) The original whole blot picture is available in Supplementary Fig. S3.
Therefore, we examined the over-expression of miR-34c using precursor of miR-34c in MCT. MCT was transfected with the precursor of miR-34c or the negative control, and were subsequently treated with TGF-β 3 ng/ml from the next day (n = 3). Cells were harvested and analyzed in the same way of the experiments of down-regulation of miR-34c. The expression of miR-34c was significantly up-regulated by the transfection of miR-34c precursors, and the over-expression of miR-34c significantly suppressed the up-regulation of Snail1 (Fig. 2D). The over-expression of miR-34c also significantly rescued the KSP expression.
To evaluate the target of miR-34c, we examined following gene expression based on target scan of miR-34c by GeneSpring GX and association with TGF-β pathway; Jag1, Notch1, Notch2 and Smad2. PCR showed only Jag1 was up-regulated by TGF-β and suppressed by over-expression of miR-34c (Fig. 2E). Western blot was also carried out to examine the expression of Jag1, and confirmed that miR-34c suppressed the expression of Jag1 (Fig. 2F). Bae et al. recently reported that miR-34c regulates Notch1, Notch2 and Jag1 in a direct manner37. Therefore, our results suggested that miR-34c attenuated EMT induced by TGF-β presumably via suppression of Notch/Jag1 pathway, though further experiments are needed to prove the direct relationship between miR-34c and Jag1.
miR-34c ameliorates EMT and kidney fibrosis induced by UUO
To confirm the effects of miR-34c on EMT, in vivo over-expression of miR-34c was carried out. The unilateral ureter of ICR mouse was ligated under anesthesia, and the mouse was subsequently injected with miR-34c precursor (n = 4) or sham (n = 5) from the tail vein on the next day. On day 4 of UUO, the injection of miR-34c precursor or sham was repeated in the same way. On day 7 of UUO, bilateral kidneys were harvested.
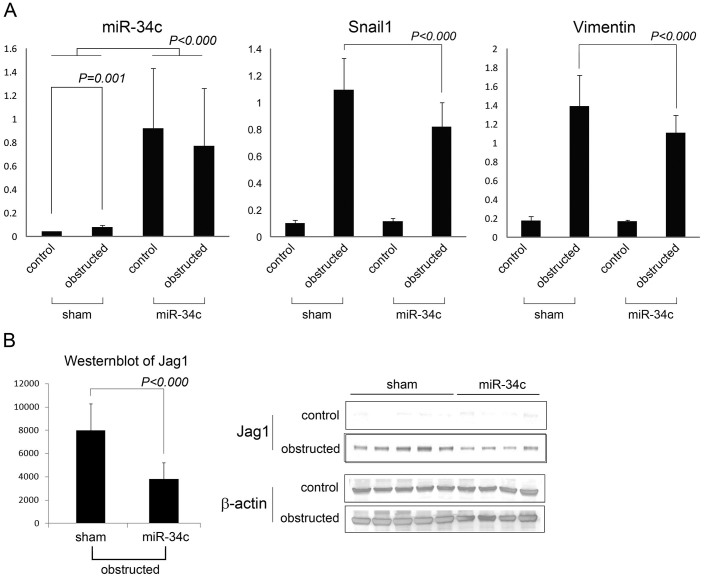
Real-time PCR of bilateral kidney was carried out to examine the expression of miR-34c, Snail1 and Vimentin (Figure 3A). The expression of miR-34c was significantly enhanced by the injection of miR-34c precursor while the up-regulation of miR-34c by UUO was very small. The over-expression of miR-34c suppressed the expression of Snail1 and Vimentin enhanced by UUO. Western blot was also carried out to examine the expression of Jag1. miR-34c significantly suppressed Jag1 expression while Jag1 was significantly up-regulated by UUO (Fig. 3B, Supplementary Fig. S4).
Figure 3. miR-34c attenuated EMT induced by UUO via down-regulation of Jag1.
(A) Real-time PCR of miR34c, Snail1 and Vimentin in mouse kidneys with or without ureteral obstruction under administration of miR-34c or sham. (B) Western blot of Jag1 and β-actin. The bands of Jag1 were quantified using ImageJ. Control; contralateral kidneys of UUO-operated mice. Obstructed; UUO kidneys of UUO-operated mice. Sham; sham-injected mice. miR-34c; precursor of miR-34 injected mice. The original whole blot pictures are available in Supplementary Fig. S4. The original whole blot pictures of control v.s. obstructed in a same membrane are also available in Supplementary Fig. S4. (sham n = 5, miR-34c n = 4).
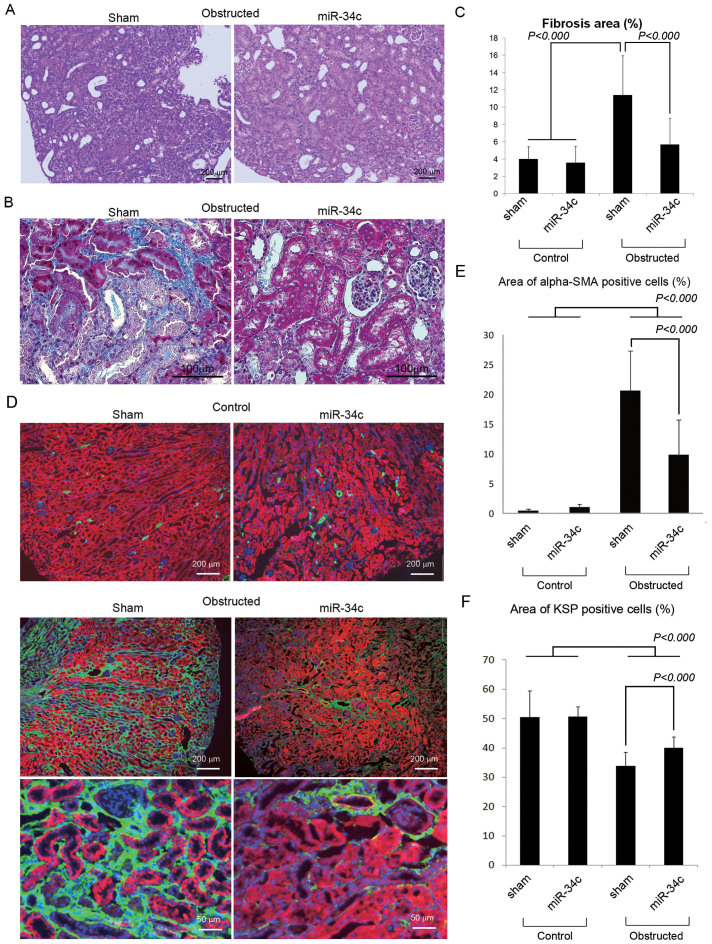
Hematoxylin-Eosin stain and Masson trichrome stain of kidneys revealed that miR-34c-injected mice had significantly less proliferation of interstitial cells and fibrosis than sham-injected mice (Fig. 4A, B, C, Supplementary Fig. S5). Moreover, immunohistochemistry of alpha-smooth muscle actin (α-SMA)38,39 revealed that the over-expression of miR-34c significantly suppressed the α-SMA positive area down to 9.9% while α-SMA positive area were increased up to 20.7% by ureteral obstruction in sham-injected mice (Fig. 4D, E). KSP positive area was also calculated to quantify the tubular cell area (Fig. 4F), and the result showed that miR-34c ameliorated the decrease of tubular cells.
Figure 4. miR-34c decreased fibrotic area in UUO kidneys.
(A) Hematoxylin-Eosin stain of kidneys with ureteral obstruction under administration of miR-34c or sham. Scale bars were indicated at the lower right corner of the pictures. (B) Masson trichrome stain of kidneys with ureteral obstruction under administration of miR-34c or sham. Scale bars were indicated at the lower right corner of the pictures. Additional pictures are available in Supplementary Fig. S5. (C) A graph showing the percent of fibrosis area in Masson trichrome stain samples. Quantification was done using ImageJ. (D) Immunohistochemistry showing α-SMA (green), KSP (red) and DAPI (blue) in kidneys with or without ureteral obstruction under administration of miR-34c or sham. Scale bars were indicated at the lower right corner of the pictures. (E) A graph showing the percent of α-SMA positive area. (F) A graph showing the percent of KSP positive area.
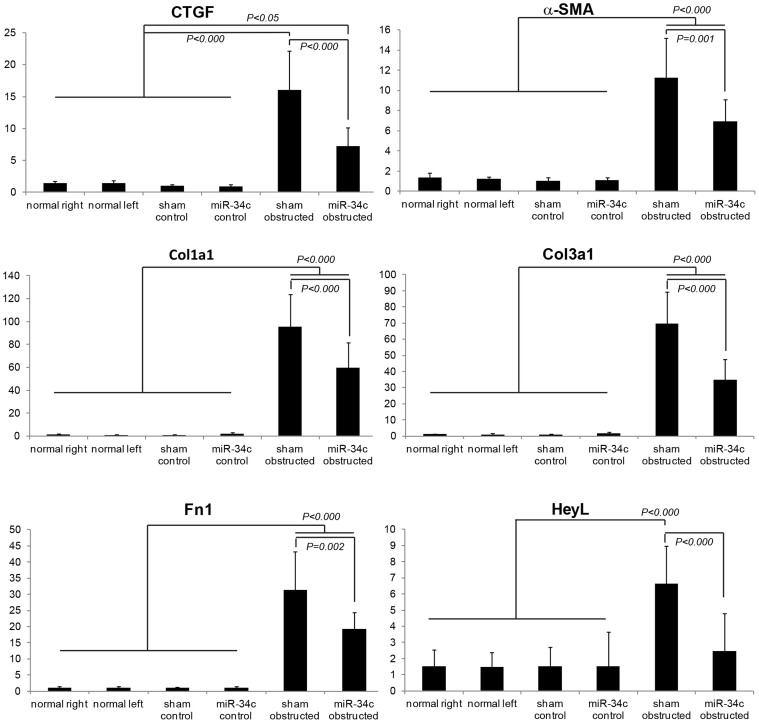
For further evaluation of kidney fibrosis, we performed real-time PCR of genes related to kidney fibrosis; connective tissue growth factor (CTGF), α-SMA, collagen type 1 (Col1a1), collagen type 3 (Col3a1), fibronectin (Fn1), and hairy/enhancer-of-split related with YRPW motif-like (HeyL)9. These kidney fibrosis markers were significantly up-regulated in UUO kidneys compared with contralateral kidneys and normal kidneys of sham operated mice, however administration of miR-34c significantly suppressed up-regulation of these genes (Fig. 5). Interestingly, a down-stream effecter of Notch/Jag1 pathway, HeyL was also significantly suppressed by miR-34c, suggesting that miR-34c ameliorated kidney fibrosis via suppressing Notch/Jag1 pathway.
Figure 5. miR-34c attenuated kidney fibrosis in UUO mice.
Real-time PCR of connective tissue growth factor (CTGF), alpha-smooth muscle actin (α-SMA), collagen type 1 (Col1a1), collagen type 3 (Col3a1), fibronectin (Fn1), and hairy/enhancer-of-split related with YRPW motif-like (HeyL). Normal right: right kidneys of sham-operated mice (n = 3), Normal left: left kidneys of sham-operated mice (n = 3), Sham control: contralateral kidneys of sham-injected mice with UUO (n = 5), miR-34c control: contralateral kidneys of miR-34c injected mice with UUO (n = 4), Sham obstructed: UUO kidneys of sham-injected mice (n = 5), miR-34c obstructed, UUO kidneys of miR-34c injected mice (n = 4).
Discussion
We have found a new miRNA, miR-34c which suppresses EMT and kidney fibrosis via suppression of Notch/Jag1 pathway. We showed miR-34c over-expression in renal tubular cells ameliorated EMT induced by TGF-β in vitro, and also demonstrated that administration of miR-34c precursors into mice ameliorated kidney fibrosis induced by UUO.
Our microarray results showed that miR-34c was up-regulated by ureteral obstruction and TGF-β, and was down-regulated through renal differentiation by Activin in ES cells. Based on these results, we hypothesized that miR-34c promoted EMT under TGF-β signaling, however inhibition of miR-34c under TGF-β stimulation did not show any changes of EMT markers. On the other hand, over-expression of miR-34c inhibited Snail1 up-regulation induced by TGF-β, and also restored KSP expression. We confirmed these effects of miR-34c using an in vivo mouse model with UUO. Taken together, these results suggest that TGF-β and UUO up-regulates miR-34c which competes with EMT, however the up-regulation of miR-34c is too small to oppose EMT under the experimental models.
Moreover, we tested the effects of miR-34c over-expression on the differentiation of mouse ES cells and mouse embryonic kidneys. Over-expression of miR-34c significantly up-regulated the expression of KSP (Supplementary Fig. S1), suggesting that miR-34c might involve in kidney differentiation. We also found the low level of miR-34c expression in E14.5 mouse embryonic kidneys which suggested that miR-34c might involve in kidney development, however, further experiments are required to examine the effects of miR-34c on the kidney differentiation.
miR-34c is now reported to have several functions against different biological processes. miR-34c was first reported to be a target of p53 by Corney35, and several reports also showed that miR-34c is induced by p53, and mediates apoptosis, cellular senescence and the cell cycle40,41,42. Recent reports indicated that down-regulation of miR-34c is associated with cancer growth and metastasis, and miR-34c acts as a tumor-suppressor43,44,45,46,47. Moreover, Cannell et al. reported that miR-34c is induced by p38 MAPK/MK2 and prevents Myc-dependent DNA replication via inhibition of translation of c-Myc through a highly conserved target-site within the 3′-UTR48,49. We have analyzed the expression of miR-34c in several organs of adult mice by real-time PCR, and confirmed that liver, lung, testis, spleen and kidney expressed miR-34c (data not shown). Especially lung and testis showed a high level of miR-34c expression. There are only few reports regarding the role of miR-34c in organ development50,51, and nobody has reported about the role of miR-34c in relationship kidney development. We showed that over-expression of miR-34c promoted the mesenchymal epithelial transition (MET) in mouse ES cells and mouse embryonic kidneys. These results suggested that miR-34c may have a role in kidney development, especially in MET, though further experiments are required to explore the functions of miR-34c on kidney development.
Moreover, there are a few reports suggesting the role of miR-34c in EMT. Li et al. reported that rno-miR-34 family is up-regulated in dimethylnitrosamine-induced hepatic fibrosis in rats52, and Yu et al. showed that down-regulation of miR34c promotes self-renewal and EMT in breast tumor-initiating cells53. Our results showed that UUO and TGF-β slightly up-regulated miR-34c, and the over-expression of miR-34c attenuated EMT induced by UUO and TGF-β, and are consistent with those previous reports.
The target genes of miR-34c are also important to understand the molecular pathway of miR-34c. Hermeking summarized previous reports about miR-34 family, and also described the targets of miR-34c34. miR-34c has some functions against different biological processes as we described above, and is also known to have some different targets. Previous reports indicated that E2F3, MET and c-Myc are direct targets of miR-34c for inhibition of proliferation, inhibition of invasion and migration, and G1-arrest respectively43,54,55. In addition to those reports, Bae et al. recently reported that miR-34c regulates Notch signaling during bone development, and showed that miR-34c targets Notch1, Notch2 and Jag1 in a direct manner, and influences osteoclast differentiation in a non-cell-autonomous fashion37.
Notch signaling in renal epithelial cells is now known to be an important pathway for kidney fibrosis and TGF-β in mouse and human kidneys9,56, and Smad signaling of TGF-β is also recognized to play a role in regulating EMT3. We have inferred the targets of miR-34c via target scan with GeneSpring GX and microRNA.org, and examined the expression of Jag1, Notch1, Notch2 Smad2 and HeyL which are potential targets of miR-34c. Our experiments showed that Jag1 and HeyL were up-regulated by UUO, and the up-regulation was attenuated by over-expression of miR-34c when TGF-β was present. In light of Bae's report37, our results suggest that miR-34c ameliorated EMT and kidney fibrosis induced by UUO presumably via suppression of Notch/Jag1 pathway, though further experiments are needed to prove the direct relationship between miR-34c and Jag1.
Recently, Lebleu reported that the origin of myofibroblasts in kidney fibrosis is mainly from local resident fibroblasts (50%) through proliferation, and the remaining cause of kidney fibrosis is from bone marrow (35%), endothelial-to-mesenchymal transition program (10%) and the epithelial-to-mesenchymal transition program (5%)8. On the other hand, Bielesz reported that Notch activation in tubular epithelial cells causes kidney fibrosis in the kidneys9. From these reports, we speculate that kidney fibrosis is mainly caused by myofibroblasts proliferation via Notch activation in tubular epithelial cells, and tubular epithelial cells influence fibroblasts resulting in kidney fibrosis. Hence, we infer that miR-34c ameliorated kidney fibrosis through suppression of Notch activation in tubular epithelial cells, though further experiments are required to know how tubular epithelial cells influenced fibroblasts.
In conclusion, we have demonstrated that administration of miR-34c into mice with UUO suppresses fibrosis of kidneys presumably through suppression of Notch/Jag1 pathway. Our study may provide new insight into developing therapies against CKD targeting kidney fibrosis and miRNAs.
Methods
Cell culture
A mouse renal tubular cell line, MCT was kindly provided by Dr. Neilson (University of Pennsylvania), and was maintained in Dulbecco's modified Eagles's medium (DMEM, GIBCO) and 10% fetal calf serum (FCS) as previously reported57. Prior to TGF-β stimulation, the medium was changed to DMEM supplemented with 0.5% FCS, and TGF-β (R&D Systems) was subsequently added to the medium at 3 ng/ml on the next day.
A mouse ES cell line, EB3 was kindly provided by Dr. Niwa (Riken Center for Developmental Biology, Japan), was maintained and differentiated with Activin (R&D Systems) as previously reported32,33.
RNA extraction and PCR
RNA was purified with RNeasy Mini Kit (QIAGEN), and potentially contaminating genomic DNA was digested by RNase-Free DNase set (QIAGEN) according to the manufacturer's protocol. cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems). GoTaq (Promega), TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and QuantiFast SYBR Green PCR Kit (QUIAGEN) were used for PCR and real-time PCR. All primers and probes were originally designed. GAPDH was used for internal control.
miRNA was purified with miRNeasy Mini Kit (QIAGEN). cDNA synthesis and real-time PCR were performed with TaqMan miRNA assays (Applied Biosystems). sno202 was used for internal control.
Western blot analysis
Cells were collected with RIPA buffer supplemented with Proteinase Inhibitor Cocktail (P8340, Sigma) after two times washing with PBS. Lysates were cleared by centrifugation at 4°C for 15 min at 12,000 g, and protein concentrations were determined using BCA Protein Assay Kit (Pierce). Samples were subjected to SDS-PAGE, and transferred to a membrane. Primay antibodies were anti-Vimentin (Cell Signaling, #5741)58, anti-KSP33, anti-Jag1 (Cell Signaling, #2620)59 and anti-β-actin (Sigma, A1978)60. Alkaline phosphatase conjugated antibody (Promega) was used as a second antibody. Detection was carried out with Western blue (Promega).
miRNA microarray
627 mouse miRNAs were scanned by Mouse miRNA Microarray (V2, G4471A) (Agilent Technologies) according to manufacturer's protocol. Microarray results were analyzed using GeneSpring GX (Agilent Technologies). The microarray data was deposited in the Gene Expression Omnibus (GSE42719, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42719).
Transfection of miRNA precursors and inhibitors
miRVana&trade miRNA mimic (Applied Biosystems) or miRVana&trade miRNA inhibitor (Applied Biosystem) of miR-34c was transfected into MCT at 30 nM of final concentration by Lipofectamine2000 (Invitrogen) according to manufacturer's protocol. mirVana&trade miRNA mimic negative control #1 (Applied Biosystem) or mirVana&trade miRNA inhibitor negative control #1 (Applied Biosystem) was used for negative control.
In vivo transfection was carried out by use of mirVana&trade miRNA Mimic of miR-34c (In Vivo Ready) (Applied Biosystems) and Invivofectamine (Invitrogen) according to the manufacturer's protocol, and 5% glucose was used for sham. The mimic of miR-34c was administered to mice on day 1 and day 4 after UUO. All mice experiments were approved by the Institutional Animal Center in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. mirVana miRNA mimic negative control was also administered to mice with UUO to examine the effects of miRNA transfection itself, however no significant difference between mirVana miRNA mimic negative control and 5% glucose (sham) was found (n = 3). (data not shown).
Immunhistochemistry
Kidneys were fixed with 4% paraformaldehyde, and were subsequently immersed in 30% (w/v) sucrose. Samples were subsequently blocked in PBS containing 3.0% (v/v) FBS after slides were made using a cryostat, and were incubated with anti-α-SMA antibody (Sigma, A5228) and anti-KSP antibody conjugated with biotin33. Alexa Fluor 488 (Invitrogen) and streptavidin-PE (Beckman Coulter) was used as a second antibody. Nuclei were stained with DAPI (Invitrogen). Pictures were taken with Olympus IX81, and α-SMA-positive cells were counted in 20 random high power fields (×100) by a blinded observer using ImageJ.
Statistical analysis
Results are given as mean ± SD. Statistical analysis of the data was performed with t-test or ANOVA followed by Tukey's post-hoc test. P < 0.05 was considered significant.
Author Contributions
R.M. designed and performed the experiments, and wrote the manuscript and figures. T.M. and H.I. designed the project and experiments. S.F., K.H., S.Y. and K.H. performed the experiments. R.M. and S.F. equally contributed to this work.
Supplementary Material
Supplemntary data
Acknowledgments
We thank Mari Fujiwara for technical assistance at the Core Instrumentation Facility, Keio University School of Medicine. This work was supported in part by Grant-in-Aid for Scientific Research (KAKENHI, 23890203, 21591038, 24591211) and grant from Daiwa Securities Health Foundation.
References
- Benfey P. N. Molecular biology: microRNA is here to stay. Nature 425, 244–245; 10.1038/425244a (2003). [DOI] [PubMed] [Google Scholar]
- Hurst D. R., Edmonds M. D. & Welch D. R. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res 69, 7495–7498; 10.1158/0008-5472.CAN-09-2111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. New insights into epithelial-mesenchymal transition in kidney fibrosis. J Am Soc Nephrol 21, 212–222; 10.1681/ASN.2008121226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K. et al. Pirfenidone for diabetic nephropathy. J Am Soc Nephrol 22, 1144–1151; 10.1681/ASN.2010101049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decleves A. E. & Sharma K. New pharmacological treatments for improving renal outcomes in diabetes. Nat Rev Nephrol 6, 371–380; 10.1038/nrneph.2010.57 (2010). [DOI] [PubMed] [Google Scholar]
- Sato M., Muragaki Y., Saika S., Roberts A. B. & Ooshima A. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest 112, 1486–1494; 10.1172/JCI19270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J. et al. Snail1 is involved in the renal epithelial-mesenchymal transition. Biochem Biophys Res Commun 362, 63–68; 10.1016/j.bbrc.2007.07.146 (2007). [DOI] [PubMed] [Google Scholar]
- Lebleu V. S. et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19, 1047–1053; 10.1038/nm.3218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielesz B. et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest 120, 4040–4054; 10.1172/JCI43025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet A. et al. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J 25, 5603–5613; 10.1038/sj.emboj.7601421 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen J. M., Haller H. & Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol 7, 286–294; 10.1038/nrneph.2011.26 (2011). [DOI] [PubMed] [Google Scholar]
- Ho J. et al. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol 19, 2069–2075; 10.1681/ASN.2008020162 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q. et al. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol 21, 756–761; 10.1681/ASN.2009070718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M. TGF-beta-induced signaling circuit loops mediated by microRNAs as new therapeutic targets for renal fibrosis? Kidney Int 84, 1067–1069; 10.1038/ki.2013.297 (2013). [DOI] [PubMed] [Google Scholar]
- Wu J. et al. Downregulation of MicroRNA-30 Facilitates Podocyte Injury and Is Prevented by Glucocorticoids. J Am Soc Nephrol 25, 92–104; 10.1681/ASN.2012111101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C. F. et al. MicroRNA-155 drives TH17 immune response and tissue injury in experimental crescentic GN. J Am Soc Nephrol 24, 1955–1965; 10.1681/ASN.2013020130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Koya D. & Kanasaki K. MicroRNAs in kidney fibrosis and diabetic nephropathy: roles on EMT and EndMT. Biomed Res Int 2013, 125469; 10.1155/2013/125469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A. C., Huang X. R., Meng X. & Lan H. Y. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21, 1317–1325; 10.1681/ASN.2010020134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel A. J. et al. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382. Nucleic Acids Res 38, 8338–8347; 10.1093/nar/gkq718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa A. et al. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 21, 438–447; 10.1681/ASN.2009050530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10, 593–601; 10.1038/ncb1722 (2008). [DOI] [PubMed] [Google Scholar]
- Korpal M., Lee E. S., Hu G. & Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283, 14910–14914; 10.1074/jbc.C800074200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M., Gaur A. B., Lengyel E. & Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22, 894–907; 10.1101/gad.1640608 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran K. et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int 81, 617–627; 10.1038/ki.2011.448 (2012). [DOI] [PubMed] [Google Scholar]
- Xiong M. et al. The miR-200 family regulates TGF-beta1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol 302, F369–379; 10.1152/ajprenal.00268.2011 (2012). [DOI] [PubMed] [Google Scholar]
- Denby L. et al. miR-21 and miR-214 are consistently modulated during renal injury in rodent models. Am J Pathol 179, 661–672; 10.1016/j.ajpath.2011.04.021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Chung A. C., Chen H. Y., Meng X. M. & Lan H. Y. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol 22, 1668–1681; 10.1681/ASN.2010111168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. et al. miR-21-containing microvesicles from injured tubular epithelial cells promote tubular phenotype transition by targeting PTEN protein. Am J Pathol 183, 1183–1196; 10.1016/j.ajpath.2013.06.032 (2013). [DOI] [PubMed] [Google Scholar]
- Denby L. et al. MicroRNA-214 antagonism protects against renal fibrosis. J Am Soc Nephrol 25, 65–80; 10.1681/ASN.2013010072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T. et al. Fibroblast expression of an IkappaB dominant-negative transgene attenuates renal fibrosis. J Am Soc Nephrol 21, 2047–2052; 10.1681/ASN.2010010003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. et al. The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells. Atherosclerosis 219, 468–474; 10.1016/j.atherosclerosis.2011.09.004 (2011). [DOI] [PubMed] [Google Scholar]
- Morizane R., Monkawa T. & Itoh H. Differentiation of murine embryonic stem and induced pluripotent stem cells to renal lineage in vitro. Biochem Biophys Res Commun 390, 1334–1339; 10.1016/j.bbrc.2009.10.148 (2009). [DOI] [PubMed] [Google Scholar]
- Morizane R. et al. Kidney specific protein-positive cells derived from embryonic stem cells reproduce tubular structures in vitro and differentiate into renal tubular cells. PLoS One 8, e64843; 10.1371/journal.pone.0064843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 17, 193–199; 10.1038/cdd.2009.56 (2010). [DOI] [PubMed] [Google Scholar]
- Corney D. C., Flesken-Nikitin A., Godwin A. K., Wang W. & Nikitin A. Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res 67, 8433–8438; 10.1158/0008-5472.CAN-07-1585 (2007). [DOI] [PubMed] [Google Scholar]
- Cardinaud B. et al. miR-34b/miR-34c: a regulator of TCL1 expression in 11q- chronic lymphocytic leukaemia? Leukemia 23, 2174–2177; 10.1038/leu.2009.125 (2009). [DOI] [PubMed] [Google Scholar]
- Bae Y. et al. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet 21, 2991–3000; 10.1093/hmg/dds129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J. & Bottinger E. P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 24, 5764–5774; 10.1038/sj.onc.1208927 (2005). [DOI] [PubMed] [Google Scholar]
- Humphreys B. D. et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176, 85–97; 10.2353/ajpath.2010.090517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., He L. & Hannon G. J. The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res 67, 11099–11101; 10.1158/0008-5472.CAN-07-2672 (2007). [DOI] [PubMed] [Google Scholar]
- Kumamoto K. et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res 68, 3193–3203; 10.1158/0008-5472.CAN-07-2780 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D. et al. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 130, 1249–1257; 10.1038/jid.2009.438 (2010). [DOI] [PubMed] [Google Scholar]
- Migliore C. et al. MicroRNAs impair MET-mediated invasive growth. Cancer Res 68, 10128–10136; 10.1158/0008-5472.CAN-08-2148 (2008). [DOI] [PubMed] [Google Scholar]
- Cai K. M. et al. Hsa-miR-34c suppresses growth and invasion of human laryngeal carcinoma cells via targeting c-Met. Int J Mol Med 25, 565–571; 10.3892/ijmm_00000378 (2010). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res 15, 1177–1183; 10.1158/1078-0432.CCR-08-1355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Liu X. & Dmitrovsky E. Tumor-suppressive microRNAs in Lung cancer: diagnostic and therapeutic opportunities. ScientificWorldJournal 9, 626–628; 10.1100/tsw.2009.88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman Z. et al. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer 127, 2768–2776; 10.1002/ijc.25269 (2010). [DOI] [PubMed] [Google Scholar]
- Cannell I. G. & Bushell M. Regulation of Myc by miR-34c: A mechanism to prevent genomic instability? Cell Cycle 9, 2726–2730; 10.4161/cc.9.14.12182 (2010). [PubMed] [Google Scholar]
- Cannell I. G. et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci U S A 107, 5375–5380; 10.1073/pnas.0910015107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhallier F. et al. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16, 720–731; 10.1261/rna.1963810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. et al. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One 7, e33861; 10.1371/journal.pone.0033861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Q. et al. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J 278, 1522–1532; 10.1111/j.1742-4658.2011.08075.x (2011). [DOI] [PubMed] [Google Scholar]
- Yu F. et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem 287, 465–473; 10.1074/jbc.M111.280768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 105, 13556–13561; 10.1073/pnas.0803055105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. W. et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A 105, 8866–8871; 10.1073/pnas.0800650105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhan K. C. et al. Jagged/Notch signalling is required for a subset of TGFbeta1 responses in human kidney epithelial cells. Biochim Biophys Acta 1803, 1386–1395; 10.1016/j.bbamcr.2010.09.001 (2010). [DOI] [PubMed] [Google Scholar]
- Okada H., Danoff T. M., Kalluri R. & Neilson E. G. Early role of Fsp1 in epithelial-mesenchymal transformation. Am J Physiol 273, F563–574 (1997). [DOI] [PubMed] [Google Scholar]
- Matsuyama M. et al. Defect of mitotic vimentin phosphorylation causes microophthalmia and cataract via aneuploidy and senescence in lens epithelial cells. J Biol Chem 288, 35626–35635; 10.1074/jbc.M113.514737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steg A. D. et al. Targeting the notch ligand JAGGED1 in both tumor cells and stroma in ovarian cancer. Clin Cancer Res 17, 5674–5685; 10.1158/1078-0432.CCR-11-0432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacor P. N. et al. Reelin secretion from glutamatergic neurons in culture is independent from neurotransmitter regulation. Proc Natl Acad Sci U S A 97, 3556–3561; 10.1073/pnas.050589597 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemntary data