Abstract
Background and objective
The bacterial profile of saliva is composed of bacteria from different oral surfaces. The objective of this study was to determine whether different diet intake, lifestyle, or socioeconomic status is associated with characteristic bacterial saliva profiles.
Design
Stimulated saliva samples from 292 participants with low levels of dental caries and periodontitis, enrolled in the Danish Health Examination Survey (DANHES), were analyzed for the presence of approximately 300 bacterial species by means of the Human Oral Microbe Identification Microarray (HOMIM). Using presence and levels (mean HOMIM-value) of bacterial probes as endpoints, the influence of diet intake, lifestyle, and socioeconomic status on the bacterial saliva profile was analyzed by Mann–Whitney tests with Benjamini–Hochberg's correction for multiple comparisons and principal component analysis.
Results
Targets for 131 different probes were identified in 292 samples, with Streptococcus and Veillonella being the most predominant genera identified. Two bacterial taxa (Streptococcus sobrinus and Eubacterium [11][G-3] brachy) were more associated with smokers than non-smokers (adjusted p-value<0.01). Stratification of the group based on extreme ends of the parameters age, gender, alcohol consumption, body mass index (BMI), and diet intake had no statistical influence on the composition of the bacterial profile of saliva. Conversely, differences in socioeconomic status were reflected by the bacterial profiles of saliva.
Conclusions
The bacterial profile of saliva seems independent of diet intake, but influenced by smoking and maybe socioeconomic status.
Keywords: saliva, HOMIM, bacteria, diet lifestyle
The oral cavity is an open environment harboring a complex community comprising >700 different oral bacterial species, of which only around 50% have been cultivated (1, 2). Over recent decades, molecular methods based on sequencing of the bacterial 16S ribosomal RNA (rRNA) gene have expanded the knowledge about oral bacteria considerably (3). Thus, studies have revealed a hitherto unknown diversity of the oral microbiota, with significant variation between different oral locations (4, 5). The resident oral microbiota is believed to play an essential role in maintaining oral homeostasis, protecting the oral cavity against the development of disease (3). However, dental caries and periodontitis, the two most prevalent oral diseases, have been found to be associated with site-specific changes in the oral microbiota (6, 7).
Differences in diet intake, lifestyle, and socioeconomic status are generally considered important factors in the multifactorial background of dental caries and periodontitis. Obesity often measured by a body mass index (BMI) above 30 kg/m2 is associated with high levels of dental caries (8) and periodontitis (9, 10), and a high frequency of sugar intake increases the risk of dental caries (11). Smoking increases the risk of acquiring periodontitis and of aggravating existing diseases (12). High intake of alcohol has been suggested to protect against periodontitis (13), as well as being a risk factor for periodontitis (14). In addition, low socioeconomic status is also known to be associated with periodontitis (15, 16) and dental caries (17, 18). At present, little is known about the influence of differences in diet, lifestyle, and socioeconomic status on composition of the oral microbiome, and whether differences in bacterial composition may be detected in saliva samples.
Being an easily collectable, non-invasive biological material, saliva is suitable for medical investigation, and several health and disease-associated factors are reflected in saliva (19). Thus, saliva is currently being thoroughly investigated in general and in dental medicine in the search for biomarkers of health and disease (20–22). A few studies have investigated the bacterial composition of saliva in oral health (5, 23–25) and verified that a salivary bacterial profile resembles that of the throat, tonsils, and dorsum of the tongue (26). In addition, the salivary microbiome has been shown to be highly diverse and is proposed to depend on lifestyle and diet intake (25, 27). Also, oral disease may influence the bacterial profile, and recently it has been shown that both dental caries and periodontitis are associated with characteristic bacterial profiles of saliva different from that of oral health (28, 29).
In this study, we investigated stimulated saliva samples collected from 292 participants of the Danish Health Examination Survey (DANHES). We used the Human Oral Microbe Identification Microarray (HOMIM), a 16S rRNA high-throughput based microarray technique able to identify around 300 different bacterial species simultaneously. The purpose of the study was to determine whether differences in diet intake, lifestyle, and socioeconomic status associate with the bacterial profile of saliva.
Materials and methods
Study population
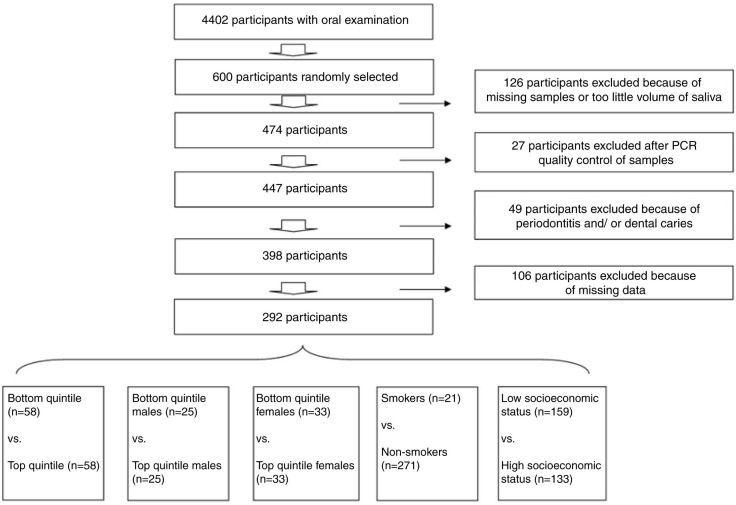
The cohort of the Danish population-based study DANHES (30) and the subpopulation participating in the oral part of the study (31) have been described in detail elsewhere. In brief, the DANHES study was carried out in 13 different municipalities in 2007–2008, and oral examinations were performed in 2008–2009. A total of 76,484 individuals participated in the questionnaire survey concerning general health, and 18,065 had a general medical examination performed. In addition to the general medical examination, 4,402 subjects had a dental examination performed, of which 4,337 delivered a stimulated saliva sample and 4,230 answered a questionnaire concerning self-perceived oral health. All participants received written information prior to participation and signed informed consent. The study was approved by the regional ethical committee (H-C-2007-0118) and reported to the Danish data authority (2007-41-1567). This study is part of a group of studies conducted on saliva samples collected from the DANHES cohort, including studies investigating how periodontitis and dental caries associate with the bacterial profile of saliva (28, 29). Population characteristics are presented in Table 1, and a flowchart depicting the sample selection strategy is shown in Fig. 1. In this study, dental caries was defined as: untreated caries lesions ≥3 surfaces, and manifest periodontitis was defined as bleeding on probing (BOP) on ≥25% of total sites+minimum 2 teeth with clinical attachment level ≥4 mm+minimum 2 teeth with probing depth ≥6 mm. Finally, a total of 292 participants with low levels of oral diseases from 12 different municipalities were included in this study.
Table 1.
Population characteristics
| Mean (SD) | Range | Lower quintile n=58, mean (SD) | Upper quintile n=58, mean (SD) | |
|---|---|---|---|---|
| Age (years) | 55 (13) | 20–81 | 35 (5) | 71 (4) |
| BMI | 25.1 (3.7) | 17.8–38.4 | 20.4 (1.2) | 30.6 (2.4) |
| Alcohol intake (units/week) | 7.7 (7.2) | 0–48 | 0.3 (0.6) | 19.5 (6.5) |
| Energy intake from fat (%) | 30.8 (9.8) | 3.8–86.5 | 19.3 (4.6) | 44.5 (10.6) |
| Energy intake from protein (%) | 17.0 (4.2) | 4.3–36.5 | 11.8 (1.8) | 23.0 (3.8) |
| Energy intake from carbohydrates (%) | 52.2 (9.1) | 9.1–79.1 | 39.0 (7.6) | 63.6 (4.1) |
| Proportions of carbohydrates originating from added sugar (%) | 10.8 (7.4) | 0–52 | 3.3 (1.3) | 22.3 (7.6) |
Fig. 1.
Flowchart of sample selection and group establishment.
Clinical examination and collection of saliva samples
The dental examination has been presented in detail previously (31). In brief, the oral examination was performed by 3 dental hygienists and 3 dental nurses working in pairs. The hygienists were calibrated intra- and interindividually prior and halfway through the project, with a perfect agreement for periodontal measurements between 60 and 80% and Kappa-coefficients between 0.85 and 0.93 for caries registration. The periodontal examination was performed in 2 randomly selected quadrants, and caries was registered full-mouth. As part of the periodontal examination, gingival margin position (GMP), probing pocket depth (PD), and BOP were recorded in six sites on each tooth. Caries was scored as a clinically manifested lesion present on surface and tooth levels. No information from dental radiographs was included in this study. Stimulated saliva samples were collected by a dental hygienist according to a standardized protocol: the participant was instructed to flush thoroughly and subsequently chew for 1 minute on 1 gram of paraffin gum until a solid bolus was formed. Finally, the participant was instructed to spit repeatedly for 4 minutes, and for the last 3 minutes saliva was collected in a plastic cup, transferred by pipette to a plastic tube, and stored at −80°C until further analysis.
DNA isolation and microarray analysis
DNA isolation from saliva samples was performed according to the manufacturer's guidelines (Roche, Mannheim, Germany). In brief, 200 µl of sample material was thoroughly vortexed, supplemented with 180 µl MagNA Pure Bacterial Lysis buffer (Roche) and 20 µl Proteinase K 20 mg/ml (Roche), and incubated for 1 hour at 37°C. Purification was performed in a Roche MagNA Pure 96 automated DNA extractor, using MagNA Pure 96 DNA and viral NA Small Volume kit (Roche) by specifications according to protocol Pathogen_Universal_200. The final elution volume was 50 µl. HOMIM analyses were performed as described (6). In brief, HOMIM is a molecular technique using 2 separate polymerase chain reaction (PCR) amplifications and subsequent DNA–DNA hybridization for identification of around 300 bacterial species, as single-stranded fluorescent labeled PCR products are captured by highly specific bacterial 16S rRNA probes (18–20 bases) printed on a customized aldehyde-coated glass slide. In conjunction with studies conducted on saliva samples from DANHES, all probes were thoroughly validated. Probes with cross-reactions and/or missing reactions were excluded from statistical analysis. Further analysis and generation of microbial profiles from scanned microarrays were performed with the HOMIM online tool for analysis (http://bioinformatics.forsyth.org/homim/).
Collection of data
The data used in this study were collected through a medical examination, performed by accredited medical personal, and a self-reported questionnaire. The medical examination and the questionnaire gathered information about the physical status and lifestyle-related factors of the participants. Age was registered in whole years at the time point of questionnaire answering. Height was recorded in centimeters (cm), and weight was recorded in kilograms (kg). BMI was calculated using the formula [height (cm)//weight (kg)*weight (kg)]. Smoking status was recorded by questionnaire, and the probands were grouped in current smokers and non-smokers. Information about alcohol consumption was gathered from the questionnaire and recorded as intake of units/week. Dietary data were collected using an Internet-based, 267 items food frequency questionnaire (FFQ), that included 32 photoseries of food portions to quantify portion sizes. The amount of daily energy originating from fat, protein, and carbohydrates was calculated using the Foodcalc program. In addition, the proportion of carbohydrates consisting of added sugar was determined. Information concerning the socioeconomic status of the 12 municipalities was gathered by information from Denmark's statistics using the parameters: average income per year (2007), percentage of unemployed inhabitants, and percentage of inhabitants with a non-western citizenship. In addition, average level of education (International Standard Classification of Education, levels 1–6) was calculated using individual educational information gathered from the questionnaire. In each of the four categories, the municipalities were scored from 1 to 12 (1 being the best ranked municipality) and the values were added (theoretical range 4–48).
Formation of subgroups for analysis
The influence of each of the following variables on the bacterial profile of saliva was tested separately: age, gender, BMI, smoking status, alcohol consumption, proportion of energy originating from fat, proportion of energy origination from protein, and proportion of energy originating from carbohydrates and proportion of carbohydrates originating from added sugar. Based on each of the variables, the total cohort of 292 was divided into quintiles of 58 individuals, and the top quintile was compared to the bottom quintile. In addition, the cohort was split into two subgroups according to gender, establishing a subgroup of males (n=126) and females (n=166). Each gender subgroup was split into quintiles (males: n=25, females: n=33), and based on the same parameters, the top quintile was compared to the bottom quintile separately by gender. The subgroup of smokers (n=21) was compared to the subgroup of non-smokers (n=271). The 12 municipalities were ranked socioeconomically from 1 to 12 based on each of the 4 socioeconomic parameters and the values added up. A total of 133 participants from 5 municipalities constituted a subgroup of high socioeconomic status (16–19 points) that was compared to a subgroup of 159 participants from 7 municipalities with low socioeconomic status (26–44 points) (Fig. 1).
Statistical analysis
Differences in hybridization patterns were analyzed using the Mann–Whitney test, and differences in data between groups were compared with unpaired t-test. For these analyses, a p-value <0.05 was considered statistically significant. Comparison of quintiles using frequency and levels (mean HOMIM-value) at taxon/cluster level as parameters of investigation was carried out using a Mann–Whitney test with Benjamini–Hochberg's correction for multiple comparisons (32). For these analyses, an adjusted p-value <0.01 was considered statistically significant. Principal component analysis was used for visualization through data reduction of patterns in this n-dimensional dataset. Graph Pad prism 5 (San Diego, CA) and MeV 4.8.1 (33) were used as statistical software.
Results
Population characteristics
Population characteristics are presented in Table 1. For each parameter presented in Table 1, the highest and the lowest quintile were highly significantly different (p<0.0001).
General findings
In 292 samples from participants with low levels of oral diseases, we observed positive identification for targets of 131 probes. The number of hits per sample was 24.61±9.3 (mean±SD). Sixty-six percent of the targets identified were present in >1% of the samples, while 42% were identified in >5% of the samples. A total of 6 bacterial phyla were identified (Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, Fusobacteria, and TM7). Firmicutes was the predominant phylum present, representing approximately 50% of the total probe signal, followed by Proteobacteria and Bacteroidetes, each of which accounted for approximately 17% of the targets present. No statistical differences were found in number of targets identified, mean probes per sample, or phylogenetic distribution between subgroups divided on the basis of the parameters investigated (data not shown). A complete list of targets identified is presented in the Supplementary file. The predominant bacterial profile (taxa/clusters identified in >50% of the samples) was dominated by taxa/clusters traditionally associated with oral health representing the genera Streptococcus and Veillonella. In general, the targets most frequently identified were also those present at the highest levels (mean HOMIM-value). Putative periodontal pathogens such as Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Prevotella intermedia and suggested cariogenic bacteria such as Streptococcus mutans and Lactobacillus species were all found in <4% of the samples.
Differences between subgroups at bacterial taxon/cluster level
By comparison of the parameters, that is, age, gender, BMI, alcohol consumption, proportion of energy originating from fat, proportion of energy origination from protein, and proportion of energy originating from carbohydrates and proportion of carbohydrates originating from added sugar separately, no statistical difference in presence or levels (mean HOMIM-value) of any taxon/cluster was observed (adjusted p-values>0.01) between the top and bottom quintile of the cohort. Based on the same parameters, no statistical difference in presence or levels of any taxon/cluster was observed when analyzing each gender separately by comparison of the top and bottom quintile of each gender subgroup (adjusted p-values>0.01).
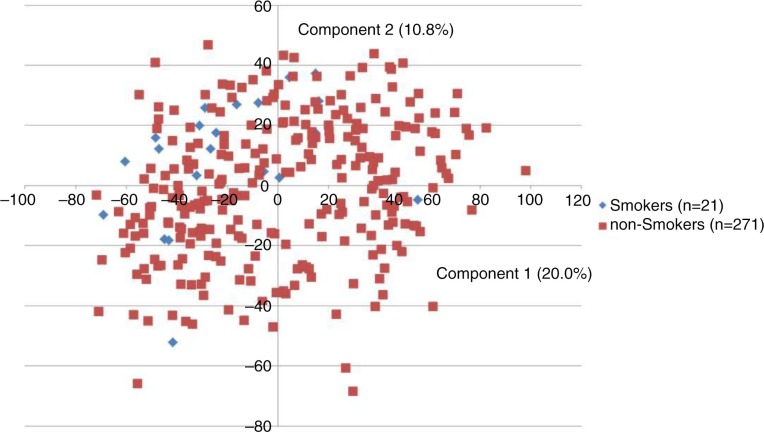
Comparison of smokers (n=21) with non-smokers (n=271) revealed that 2 bacterial taxa (Streptococcus sobrinus and Eubacterium[11][G-3] brachy) were identified at higher levels in smokers than in non-smokers (adjusted p-value<0.01). Principal component analysis displayed clustering of the 21 samples from smokers combined with a random distribution of the samples from non-smokers (Fig. 2). In addition current smokers (n=21) and never-smokers (n=148) were compared, and the same bacterial taxa (S. sobrinus and Eubacterium[11][G-3] brachy) were found to be associated with current smokers, when compared to never-smokers. Finally, former-smokers (n=123) were compared against never-smokers, and no statistical differences were observed between these groups (data not shown).
Fig. 2.
Principal component analysis of smoking status. Smokers (blue, n=21) and non-smokers (red, n=271) displayed 2-dimensionally with principal component analysis by the 2 principal components, accounting for 30.8% of the variation of the dataset using mean HOMIM-value (level), as parameter for investigation.
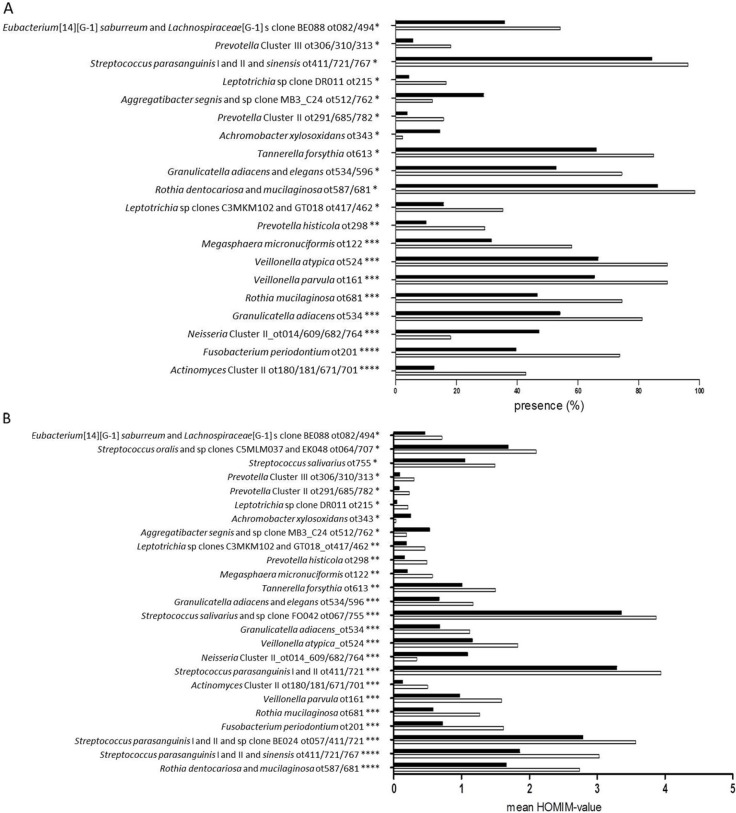
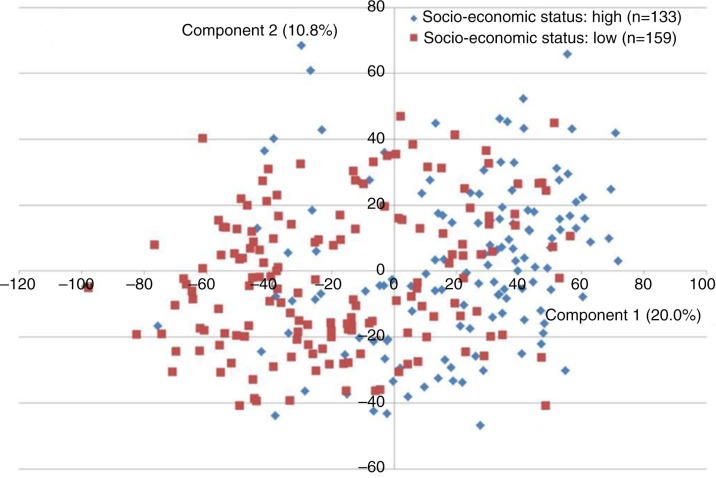
When comparing the high socioeconomic status subgroup to the low socioeconomic status subgroup, highly significant differences were observed, as 20 bacterial probes were present with a different frequency between subgroups. Seventeen probes (9 recognizing a bacterial taxon and 8 recognizing a bacterial cluster) were associated with the high socioeconomic status subgroup, and three (1 identifying a bacterial taxon and 2 identifying a bacterial cluster) were associated with the low socioeconomic subgroup. In addition, 25 bacterial probes were identified at different levels in the 2 subgroups. Twenty-two probes (10 recognizing a bacterial taxon and 12 recognizing a bacterial cluster) were more associated with the high socioeconomic status subgroup, and 3 (1 identifying a bacterial taxon and 2 identifying a bacterial cluster) were associated with the low socioeconomic status subgroup. A total of 19 out of 20 taxa/clusters identified with statistically different frequency between the high and low socioeconomic status subgroup were also identified at statistically different levels (Fig. 3A and 3B). As can be observed from Fig. 4, a marked difference in the distribution of samples from the high and low socioeconomic status group was identified using principal component analysis, based on the most decisive variable, representing 20% of the mathematical variation in the dataset.
Fig. 3.
Differences at taxon/cluster level by socioeconomic status. A. The 20 statistically significant probes are presented based on presence (%) of total amount of samples in the high and low socioeconomic status subgroup, respectively. Black bars: low socioeconomic status subgroup. White bars: high socioeconomic status subgroup. *Adjusted p-value < 0.01, *adjusted p-value < 0.001, ***adjusted p-value < 1.0×10−4, ****adjusted p-value < 1.0×10−6. B. Levels (mean HOMIM-value) of the 25 statistically significant probes are visualized based on means of HOMIM-value in samples from the high and low socioeconomic status subgroup. Black bars: low socioeconomic status subgroup. White bars: high socioeconomic status subgroup. *Adjusted p-value < 0.01, **adjusted p-value < 0.001, ***adjusted p-value < 1.0×10−5, ****adjusted p-value < 1.0×10−12.
Fig. 4.
Principal component analysis of socioeconomic status. The subgroup with high socioeconomic status (blue, n=133) and the subgroup with low socioeconomic status (red, n=159) visualized 2-dimensionally by principal component analysis, with axes expressing the 2 most crucial components accounting for 30.8% of the total variation of the dataset.
Discussion
The purpose of this study was to examine whether the bacterial saliva profiles in individuals with low levels of oral diseases were associated with differences in diet intake, lifestyle, or socioeconomic status. By comparison of high-throughput data collected using the HOMIM with available data from the DANHES, we were able to show that the bacterial saliva profile associated with low levels of oral diseases was independent of diet intake, but influenced by smoking and socioeconomic status. To the best of our knowledge, this is the first study to combine high-throughput microbiologic information with comprehensive data, characterizing the composition of the bacterial profile of saliva in individuals with low levels of oral diseases.
The bacterial profile described in this study was, in spite of high diversity, largely dominated by the phylum of Firmicutes, expressed by a massive probe signal representing Streptococcus and Veillonella species. These findings are in line with studies using 16S rRNA pyrosequencing of saliva samples showing that the microbial profile of saliva in oral health resembles the profiles found on the tongue, the tonsils, and the throat and is characterized by high numbers of Firmicutes (25–27). Using molecular methods, putative periodontal pathogens such as P. gingivalis and A. actinomycetemcomitans have been identified to be associated with saliva samples from patients with periodontitis (34) and S. mutans with samples from carious lesions (35). Interestingly, these bacteria were only found sporadically in this cohort, indicating that saliva is not a major reservoir of putative oral pathogens in individuals with low levels of dental caries and periodontitis.
One possible limitation of this study was that samples were not collected at the same time of the day. As studies have shown that the oral microbiome might vary considerably over time because of food consumption and mental and physical stress (36), it cannot be excluded that some of the differences observed might result from diurnal variation in sample collection. In addition, it is important to emphasize that the microarray technique used only provides information of bacterial taxa/clusters with a corresponding probe present on the microarray. Therefore, only parts of the complex saliva bacterial profile were visualized using this technique. Moreover, with a probe length of 18–20 bases, it is not possible to detect all bacteria at species level. As a consequence, some taxa are targeted using cluster probes recognizing 2 or more closely related bacterial taxa, which is why some detail may be lost.
It is highly interesting that the bacterial profile of saliva appears not to be influenced by diet. Based on different carbohydrate fermentation abilities, aciduric and acidogenic oral streptococci, for example, S. mutans, are believed to play an essential role in caries development (37). However, according to our data, the amount of carbohydrate intake had no influence on the presence of such bacteria in saliva. As frequency of sugar intake is an important risk factor for caries development (11), it is however possible that the bacterial profile of saliva might be influenced by frequency of sugar intake rather than the amount of sugar consumed, but no information about frequency of sugar intake was included in this study. High levels of BMI have been described to be associated with high occurrence of caries lesions (8), and intake of high amounts of alcohol has been suggested as a risk factor for periodontitis (14), as well as protective against periodontitis (13). In the population examined in this study, these parameters had no influence on the bacterial saliva profile, even when the upper and lower quintiles were compared. These findings suggest that the bacterial profile of saliva is, in general, not associated with the mechanism linking different lifestyle factors with major oral diseases. However, the value of these results might be limited by a problem often encountered in epidemiological research: individuals participating in medical studies tend to be healthier than the general population, as visualized by the small percentage of current smokers in this group (7.2%), compared to the background Danish population (22%). This may limit the variability, and hence the chance to find significant associations. This may also compromise the generalizability of the conclusions drawn to general population subsets. Furthermore, in this study quintiles were chosen for comparison of the most extreme ends of the cohort based on each parameter investigated. The advantage with this approach was accomplishment of major differences between the subgroups analyzed. However, as this strategy resulted in relatively small subgroups, there was risk of loss of statistical power, theoretically masking some of the associations investigated.
In this study, the bacterial taxa S. sobrinus and Eubacterium[11][G-3] brachy were found to be associated with saliva samples from smokers when compared to a group of non-smokers and never-smokers, respectively. As discussed in a recent review of the literature (38), a possible association between smoking and alterations of the subgingival bacterial profile remains controversial. Thus, even though our data suggest that smoking is associated with a different bacterial profile of saliva than the one observed in non-smokers, this needs to be addressed in future studies.
Our results suggest that the bacterial profile of saliva is influenced by socioeconomic status. Thus, when dividing the cohort into subgroups of high and low socioeconomic status, 20 and 25 probes differed significantly between subgroups based on presence and levels (mean HOMIM-value), receptively (Fig. 3A and 3B). As can be observed from Fig. 4, the 2 subgroups clustered almost completely separately based on the principal component, expressing 20% of the total variation of the dataset. This finding is in accordance with other studies suggesting that differences of the salivary microbiome observed in various geographic locations might be explained by different lifestyle and socioeconomic profiles between populations (25, 27). It is noteworthy that the differences observed at taxon/cluster level affected probes targeting bacterial taxa as Veillonella parvula and Veillonella atypica and clusters as Streptococcus parasanguinis, all being part of the predominant commensal bacterial profile of saliva. In this study, we used general information about the municipalities from Denmark's statistics concerning the parameters: average income a year, percentage of unemployed inhabitants, and percentage of inhabitants with a non-western citizenship, in combination with average level of education (International Standard Classification of Education, levels 1–6), calculated from individually based information. As the socioeconomic variable was mostly constructed using general municipality based information, there is a risk that this variable was not fully representative of the actual individuals participating in the study, questioning extrapolation of the results obtained.
In conclusion, with the limitations mentioned above, this study suggests that the bacterial saliva profile in individuals with low levels of oral diseases is not influenced by diet. Conversely, smoking and possibly socioeconomic status seem to affect the bacterial profile of saliva. Prospective studies may reveal if bacterial profiles of saliva can be used as biomarkers of health and disease.
Acknowledgements
The authors wish to thank the National Institute of Public Health, University of Southern Denmark, Copenhagen, Denmark, for conducting the DANHES, Dr. Lisa B. Christensen for her assistance analyzing socioeconomic parameters, Dr. Mogens Kilian for his help interpreting microbiological data, and Sean Cotton, Alexis Kokaras, and Christina Murphy from the Forsyth Institute for laboratory assistance.
To access the supplementary material for this article, please see Supplementary Files under Article Tools online.
Conflict of interest and funding
The authors declare no conflicts of interest in this study. This study was supported by external financial support from the following: the Danish Dental Association, the Danish Foundation for Mutual Efforts in Dental Care, TrygFonden, the Health Insurance Foundation, and the Simon Spies Foundation.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–95. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner AC, Sonis AL, Lif Holgerson P, Starr JR, Nunez Y, Kressirer CA, et al. White-spot lesions and gingivitis microbiotas in orthodontic patients. J Dent Res. 2012;91:853–8. doi: 10.1177/0022034512455031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cinar AB, Christensen LB, Hede B. Clustering of obesity and dental caries with lifestyle factors among Danish adolescents. Oral Health Prev Dent. 2011;9:123–30. [PubMed] [Google Scholar]
- 9.Suvan J, Petrie A, Moles DR, Nibali L, Patel K, Darbar U, et al. Body mass index as a predictive factor of periodontal therapy outcomes. J Dent Res. 2014;93:49–54. doi: 10.1177/0022034513511084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxlin T, Ylöstalo P, Suominen-Taipale L, Männistö S, Knuuttila M. Association between periodontal infection and obesity: results of the Health 2000 Survey. J Clin Periodontol. 2011;38:236–42. doi: 10.1111/j.1600-051X.2010.01677.x. [DOI] [PubMed] [Google Scholar]
- 11.Anderson CA, Curzon ME, Van LC, Tatsi C, Duggal MS. Sucrose and dental caries: a review of the evidence. Obes Rev. 2009;10(Suppl 1):41–54. doi: 10.1111/j.1467-789X.2008.00564.x. [DOI] [PubMed] [Google Scholar]
- 12.Van Dyke TE, Sheilesh D. Risk factors for periodontitis. J Int Acad Periodontol. 2005;7:3–7. [PMC free article] [PubMed] [Google Scholar]
- 13.Kongstad J, Hvidtfeldt UA, Gronbaek M, Jontell M, Stoltze K, Holmstrup P. Amount and type of alcohol and periodontitis in the Copenhagen City Heart Study. J Clin Periodontol. 2008;35:1032–9. doi: 10.1111/j.1600-051X.2008.01325.x. [DOI] [PubMed] [Google Scholar]
- 14.Tezal M, Grossi SG, Ho AW, Genco RJ. Alcohol consumption and periodontal disease. The Third National Health and Nutrition Examination Survey. J Clin Periodontol. 2004;31:484–8. doi: 10.1111/j.1600-051X.2004.00503.x. [DOI] [PubMed] [Google Scholar]
- 15.Borrell LN, Crawford ND. Socioeconomic position indicators and periodontitis: examining the evidence. Periodontol 2000. 2012;58:69–83. doi: 10.1111/j.1600-0757.2011.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchwald S, Kocher T, Biffar R, Harb A, Holtfreter B, Meisel P. Tooth loss and periodontitis by socio-economic status and inflammation in a longitudinal population-based study. J Clin Periodontol. 2013;40:203–11. doi: 10.1111/jcpe.12056. [DOI] [PubMed] [Google Scholar]
- 17.Ekback G, Persson C. Caries in five different socio-economic clusters in Orebro county. Community Dent Health. 2012;29:229–32. [PubMed] [Google Scholar]
- 18.Pieper K, Dressler S, Heinzel-Gutenbrunner M, Neuhäuser A, Krecker M, Wunderlich K, et al. The influence of social status on pre-school children's eating habits, caries experience and caries prevention behavior. Int J Public Health. 2012;57:207–15. doi: 10.1007/s00038-011-0291-3. [DOI] [PubMed] [Google Scholar]
- 19.Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent. 2009;22:241–8. [PMC free article] [PubMed] [Google Scholar]
- 20.Giannobile WV, McDevitt JT, Niedbala RS, Malamud D. Translational and clinical applications of salivary diagnostics. Adv Dent Res. 2011;23:375–80. doi: 10.1177/0022034511420434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathnayake N, Akerman S, Klinge B, Lundegren N, Jansson H, Tryselius Y, et al. Salivary biomarkers of oral health: a cross-sectional study. J Clin Periodontol. 2013;40:140–7. doi: 10.1111/jcpe.12038. [DOI] [PubMed] [Google Scholar]
- 22.Rathnayake N, Akerman S, Klinge B, Lundegren N, Jansson H, Tryselius Y, et al. Salivary biomarkers for detection of systemic diseases. PLoS One. 2013;8:e61356. doi: 10.1371/journal.pone.0061356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, Meline BS, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One. 2011;6:e23503. doi: 10.1371/journal.pone.0023503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–43. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasidze I, Li J, Schroeder R, Creasey JL, Li M, Stoneking M. High diversity of the saliva microbiome in Batwa Pygmies. PLoS One. 2011;6:e23352. doi: 10.1371/journal.pone.0023352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belstrøm D, Fiehn N-E, Nielsen CH, Kirkby N, Twetman S, Klepac-Ceraj V, et al. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41:104–12. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- 29.Belstrøm D, Fiehn N-E, Nielsen CH, Holmstrup P, Kirkby N, Klepac-Ceraj V, et al. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–75. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- 30.Eriksen L, Gronbaek M, Helge JW, Tolstrup JS, Curtis T. The Danish Health Examination Survey 2007–2008 (DANHES 2007–2008) Scand J Public Health. 2011;39:203–11. doi: 10.1177/1403494810393557. [DOI] [PubMed] [Google Scholar]
- 31.Kongstad J, Ekstrand K, Qvist V, Christensen LB, Cortsen B, Grønbæk M, et al. Findings from the oral health study of the Danish Health Examination Survey 2007–2008. Acta Odontol Scand. 2013;71:1560–9. doi: 10.3109/00016357.2013.776701. [DOI] [PubMed] [Google Scholar]
- 32.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 33.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 34.Paju S, Pussinen PJ, Suominen-Taipale L, Hyvönen M, Knuuttila M, Könönen E. Detection of multiple pathogenic species in saliva is associated with periodontal infection in adults. J Clin Microbiol. 2009;47:235–8. doi: 10.1128/JCM.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torlakovic L, Klepac-Ceraj V, Ogaard B, Cotton SL, Paster BJ, Olsen I. Microbial community succession on developing lesions on human enamel. J Oral Microbiol. 2012;4: 1–7 doi: 10.3402/jom.v4i0.16125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–43. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Tanzer JM, Thompson A, Sharma K, Vickerman MM, Haase EM, Scannapieco FA. Streptococcus mutans out-competes Streptococcus gordonii in vivo. J Dent Res. 2012;91:513–19. doi: 10.1177/0022034512442894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johannsen A, Susin C, Gustafsson A. Smoking and inflammation: evidence for a synergistic role in chronic disease. Periodontol 2000. 2014;64:111–26. doi: 10.1111/j.1600-0757.2012.00456.x. [DOI] [PubMed] [Google Scholar]