Abstract
Angiopoietin-1 (Ang1) has potential therapeutic applications in inducing angiogenesis, enhancing endothelial cell survival, and preventing vascular leakage. However, production of Ang1 is hindered by aggregation and insolubility resulting from disulfide-linked higher-order structures. Here, by replacing the N-terminal portion of Ang1 with the short coiled-coil domain of cartilage oligomeric matrix protein (COMP), we have generated a soluble, stable, and potent Ang1 variant, COMP-Ang1. This variant is more potent than native Ang1 in phosphorylating the tyrosine kinase with Ig and epidermal growth factor homology domain 2 (Tie2) receptor and Akt in primary cultured endothelial cells, enhancing angiogenesis in vitro and increasing adult angiogenesis in vivo. Thus, COMP-Ang1 is an effective alternative to native Ang1 for therapeutic angiogenesis in vivo.
Angiopoietin-1 (Ang1) has been identified as a secreted protein ligand of tyrosine kinase with Ig and epidermal growth factor homology domain 2 (Tie2) (1, 2). Tie2 is a member of the receptor tyrosine kinase family and is expressed predominantly on vascular endothelial cells, early hematopoietic cells, and their embryonic precursors (3, 4). Ang1- and Tie2-deficient mice have similar phenotypes characterized by embryonic lethality with severe vascular remodeling defects, insufficient vessel stabilization, and perturbed vascular maturation (5, 6). Thus, Ang1 and Tie2 play critical roles in vascular development. Moreover, transgenic overexpression or gene transfer of Ang1 increases vessel formation (7–9). In addition, Ang1 can counteract vascular endothelial growth factor (VEGF)-induced side effects (10–12) while having an additive effect on vessel formation (9–11). Based on these observations, Ang1 is a candidate growth factor for therapeutic angiogenesis (13, 14). It has also been reported that overexpression or gene transfer of Ang1 protects the adult vasculature against vascular leakage (10, 11). This action seems related to the ability of Ang1 to maximize interactions between endothelial cells and surrounding support cells and matrix (2, 10, 11). Thus, Ang1 protein has potential therapeutic application in angiogenesis and prevention of vascular leakage (7–11). However, large-scale production of recombinant Ang1 is hindered by the aggregation and insolubility of the protein. The activity of the protein frequently varies after purification. These difficulties are due to its unique structural characteristics.
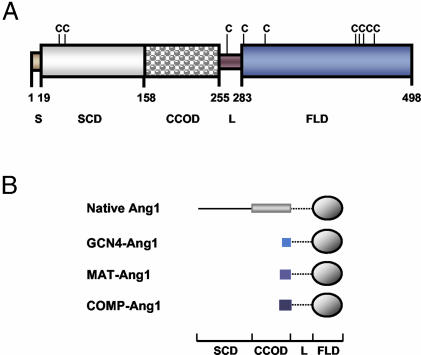
The Ang1 structure consists of a C-terminal fibrinogen-like domain that is responsible for receptor binding, a central coiled-coil domain that oligomerizes these fibrinogen-like domains, and a short N-terminal domain that superclusters these oligomers into variably sized multimers (Fig. 1A) (1, 15, 16). To achieve Tie2 receptor oligomerization and activation, Ang1 uses a modular and multimeric structure unlike that of any other known growth factor (15–17). However, the central coiled-coil domain and N-terminal superclustering domain responsible for the modular and multimeric structure of Ang1 result in protein aggregation and insolubility. Therefore, we sought to develop a modified Ang1 containing a minimal coiled-coil domain, long enough for oligomerization but short enough to avoid problems of aggregation and insolubility.
Fig. 1.
Schematic diagrams outlining the protein structure of native Ang1 and the Ang1 variants. (A) Amino acids 1–19 are the secretory signal sequence (S), amino acids 20–158 are the superclustering domain (SCD), amino acids 159–255 are the coiled-coil oligomeric domain (CCOD), amino acids 256–283 are the linker (L), and amino acids 284–498 are the fibrinogen-like domain (FLD). There are cysteines (C) at amino acids 41, 54, 265, 286, 315, 435, 437, 439, and 452. (B) Schematic diagram outlining the protein structure of native Ang1 and the Ang1 variants.
Methods
Gene Constructs. pCDNA vector (Invitrogen) contains a secretory signal sequence for hemagglutinin (MKTIIALSYIFCLVFA) and a FLAG tag (DYKDDDDK). This vector was used to express the full sequence of human Ang1 (native Ang1), the coiled-coil domain of yeast GCN4 (MKQLEDKVEELLSKNYHLENEVARLKKLVGE) fused to the fibrinogen-like domain of Ang1 (GCN4-Ang1), the coiled-coil domain of chicken matrilin 1 (EEDPCECKSIVKFQTKVEELINTLQQKLEAVAKAIEALENKII) fused to the fibrinogen-like domain of Ang1 (MAT-Ang1), and the coiled-coil domain of rat cartilage oligomeric matrix protein (COMP) (DLAPQMLRELQETNAALQDVRELLRQQVKEITFLKNTVMECDACG) fused to the fibrinogen-like domain of Ang1 (COMP-Ang1). All recombinant plasmids were constructed by using standard molecular cloning methods.
Transfection and Purification of Recombinant Proteins. Recombinant proteins were obtained by transient expression in COS-7 cells (American Type Culture Collection) using the liposomal transfection reagent Effectene according to the manufacturer's instructions (Qiagen, Hilden, Germany). Transfection efficiency of the genes was ≈30–40%. Some of the cells were selected with G418 and cloned as stable cell lines for producing the recombinant proteins for in vivo experiments. The supernatant was harvested from transfected cells after 48–96 h. The recombinant proteins containing the FLAG sequence were purified by column chromatography on an anti-FLAG M1 antibody agarose affinity gel (Sigma–Aldrich, St. Louis). After purification of COS-7 supernatants, recombinant proteins were quantitated by using the Bradford assay and confirmed with Coomassie blue staining of an SDS/PAGE gel. These analyses showed that the yield was 800–1,000 μg of each recombinant protein per liter of COS-7 cell supernatant. For comparison experiments, Ang1* was obtained from Regeneron Pharmaceuticals (Tarrytown, NY). Ang1* is a recombinant version of Ang1 with a modified NH2 terminus and mutated Cys-245.
Characterization of the Recombinant Proteins. SDS/PAGE analyses of proteins were performed under nonreducing and reducing (heating for 10 min in 0.435 M 2-mercaptoethanol) conditions. Binding of the recombinant proteins to the soluble extracellular domain of the Tie1 crystallizable fragment (Fc) (sTie1-Fc) or Tie2-Fc (sTie2-Fc) (Regeneron Pharmaceuticals or R & D Systems) was assayed by using an in vitro binding assay. Each recombinant protein (20 ng) was mixed with 100 ng of sTie1-Fc or sTie2-Fc and incubated in 500 μl of Tris buffer solution (50 mM Tris/100 mM NaCl, pH 7.4) containing 0.02% Triton X-100 at 4°C for 2 h. Then, 20 μl of protein A agarose beads (Oncogene Research Products, Boston) were added and incubated for 1 hat 4°C. The protein A-conjugated samples were washed twice with 1 ml of Tris buffer containing 0.02% Triton X-100. The samples were eluted with sample buffer, heat-denatured, further separated by 10% SDS/PAGE, electroblotted onto nitrocellulose membranes, and probed with anti-FLAG M1 antibody. Alternatively, the binding interaction between different concentrations of the recombinant proteins and immobilized sTie2-Fc protein was observed in real time by surface plasmon resonance using a Biacore 2000 biosensor (Piscataway, NJ). Briefly, 600 ng of sTie2-Fc protein was immobilized on a Sensor Chip CM5 (Biacore). As a control, 600 ng of Fc protein was immobilized on another portion of the same chip. Recombinant proteins were then injected over the sTie2-Fc surface, and the amount captured was recorded in sensorgrams as a resonance unit. All samples were in running buffer to minimize bulk effects. Kinetic parameters of the binding interactions were calculated from the sensorgrams by nonlinear curve fitting with biaevaluation software (Biacore). The binding affinity was obtained by subtracting the response value obtained with the Fc protein from that obtained with sTie2-Fc.
Glycerol Spraying/Low-Angle Rotary Metal-Shadowing and Transmission Electron Microscopy. For glycerol spraying/low-angle rotary metal-shadowing, 20 μl of protein samples (0.02–0.1 mg/ml in TBS plus 30% glycerol) was sprayed onto freshly cleaved mica at room temperature and rotary metal-shadowed in a BA 511 M freeze-etch apparatus (Balzers) with platinum/carbon at an elevation angle of 3–5° (18). Electron micrographs were taken with a Philips Morgagni transmission electron microscope operated at 80 kV and equipped with a MegaView III charge-coupled device camera.
Biochemical and Biological Assays. Tie2 and Akt (Ser-473) phosphorylation assays using human umbilical vein endothelial cells (HUVECs) and mice (FVB males, 12 weeks old) were performed as previously described (19, 20). Tie2 expression in mouse tissues was performed as previously described (19, 20). Apoptosis, migration, tube formation, and sprouting assays in endothelial cells were performed as previously described (15, 20–22). Animal care and experimental procedures were performed under approval from the Animal Care Committees of Korea Advanced Institute of Science and Technology and Chonbuk National University.
Mouse Corneal Angiogenesis Assay. Eight-week-old male C57BL/6J mice (The Jackson Laboratory) were used. After systemic and local eye anesthesia, a central, intrastromal linear keratotomy ≈0.6 mm in length was performed with a surgical blade, and a micropocket was dissected toward the temporal limbus by using a modified von Graefe knife (23). A sucrose aluminum sulfate pellet was coated with Hydron polymer containing one of the following: control buffer, VEGF (100 ng per pellet), native Ang1 (600 ng per pellet), or COMP-Ang1 (300 ng per pellet). The pellet was positioned 0.6–0.8 mm from the corneal limbus. On postoperative day 6, the arc of corneal circumference occupied by angiogenesis (circumferential angiogenesis, in degrees) and vessel lengths and numbers were measured. The mice then received an i.v. injection of 25 mg of FITC-dextran (molecular weight of 43,200; Sigma–Aldrich). After 30 min, the eyes were enucleated and fixed in 4% paraformaldehyde. The corneas were placed on glass slides and examined by an AxioCam charge-coupled device camera attached to an Axioplan 2 imaging fluorescence microscope (Zeiss).
Statistics. Data are expressed as mean ± standard deviation. Statistical significance was tested by using one-way ANOVA followed by the Student–Newman–Keuls test. Statistical significance was set at P < 0.05.
Results
Design and Production of Ang1 Variants. To explore this possibility, we generated designed oligomeric Ang1 variants in which the N-terminal portion of Ang1 was replaced with the short coiled-coil domains of yeast transcriptional activator GCN4 (24), matrilin 1 (25), and COMP (26). These three proteins form parallel two-, three-, and five-stranded structures, respectively. The corresponding chimeras were termed GCN4-Ang1, MAT-Ang1, and COMP-Ang1 (Fig. 1B). The recombinant proteins were efficiently secreted from COS-7 cells with >98% of the total proteins present in the culture medium. All recombinant proteins bound with high efficiency (>95%) to their respective purification matrices. Approximately 80% of the GCN4-Ang1, MAT-Ang1, and COMP-Ang1 variants was recovered from the purification matrix, whereas the recovery of recombinant native Ang1 was only ≈40–50% (Table 1). A solubility assay with acid precipitation indicated that GCN4-Ang1, MAT-Ang1, and COMP-Ang1 are highly soluble, but native Ang1 has a low solubility (Table 1). Furthermore, GCN4-Ang1, MAT-Ang1, and COMP-Ang1 are very stable and do not aggregate over time, whereas native Ang1 progressively lost its activity and aggregated over time (Table 1).
Table 1. Biochemical characteristics of chimeric Ang1 variants.
| Elution, % | Solubility, % | Stability, % | |
|---|---|---|---|
| Native Ang1 | 40-50 | 60-70 | 60-70 |
| GCN4-Ang1 | >80 | >95 | >95 |
| MAT-Ang1 | >80 | >95 | >95 |
| COMP-Ang1 | >80 | >95 | >95 |
Elution is the percent of protein released from anti-FLAG antibody-conjugated agarose beads. Solubility is the percent of soluble amount at pH 4.0 in acetic acid buffer. Insoluble proteins were precipitated. Stability is the phosphorylation activity on Tie2 in HUVECs after proteins were stored at -70°C for 3 months.
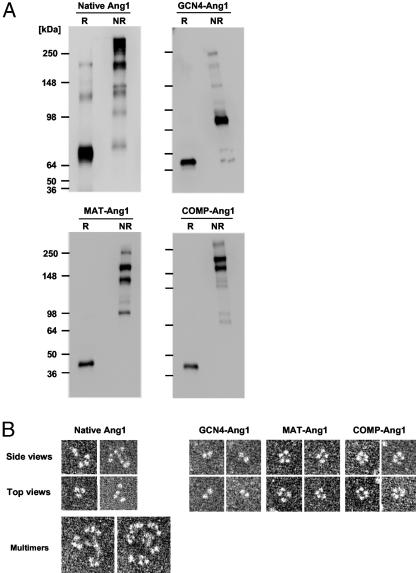
SDS/PAGE analysis of the purified proteins under denaturing conditions revealed predominantly single bands of the expected molecular masses (Fig. 2A). Under nonreducing conditions, full-length Ang1 formed characteristic disulfide-linked multimers of various sizes, whereas GCN4-Ang1 formed disulfide-linked dimers, MAT-Ang1 formed disulfide-linked trimers and tetramers, and COMP-Ang1 formed disulfide-linked tetramers and pentamers (Fig. 2 A). To confirm the oligomeric structure of native and chimeric Ang1 variants, we used rotary metal-shadowing transmission electron microscopy to directly image the molecules. Approximately 30–35% of the images obtained from native Ang1 showed the molecules in trimeric and tetrameric states yielding mushroom-shaped structures of variable size, consisting of an irregular bulbous “cap” structure (fibrinogen-like domain) at the end of a “stalk” structure (coiled-coil domain) with a small globular structure (superclustering domain) at the other end of the stalk (Fig. 2B). The majority of the images obtained from native Ang1 showed variations on the basic coiled-coil building block, with the trimers and tetramers coming together by clustering at a central point (Fig. 2B). These data indicate that recombinant native Ang1 is heterogeneous with respect to the degree of multimerization. They further indicate that variations in the activity of native Ang1 preparations could result from its degree of multimerization. These results are largely consistent with results were obtained by Davis et al. (16), although these authors concluded that the basic unit of Ang1 is a dimer. The most frequent images obtained for GCN4-Ang1 and COMP-Ang1 were dimers and pentamers, respectively. Interestingly, MAT-Ang1 seemed to form a tetramer (≈70%) rather than a trimer (Fig. 2).
Fig. 2.
Engineered Ang1 variants form distinct oligomers. (A) Reduced (R) and nonreduced (NR) proteins were separated by SDS/PAGE and immunoblotted with anti-FLAG antibody. Molecular masses (kDa) are indicated at left. (B) Glycerol spraying/low-angle rotary metal-shadowed specimens were imaged by transmission electron microscopy. Native Ang1 reveals variably sized multimers with a basic mushroom-shaped trimeric and tetrameric structure. GCN4-Ang1, MAT-Ang1, and COMP-Ang1 form dimeric, tetrameric, and pentameric structures, respectively.
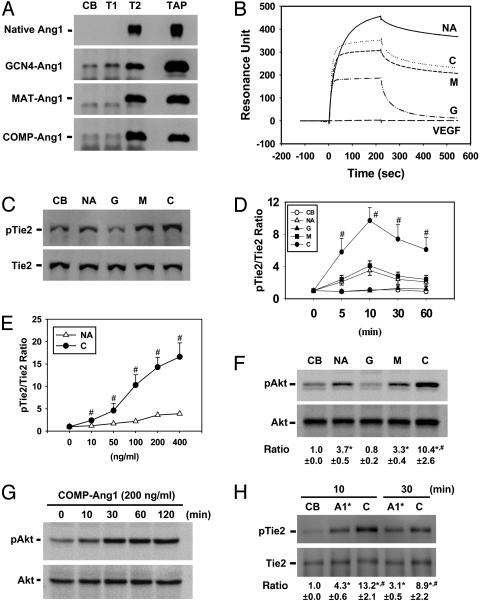
Binding Properties of Ang1 Variants to Tie2. In vitro binding assays revealed that native Ang1 and the three chimeric Ang1 variants all bound to soluble Tie2, but, as expected from its dimeric structure, the GCN4-Ang1 variant bound only weakly to the receptor (Fig. 3A). Approximately 75%, 20%, 95%, and 100% of the total protein amount of native Ang1, GCN4-Ang1, MAT-Ang1, and COMP-Ang1, respectively, bound to sTie2-Fc (Fig. 3A). The interaction between the recombinant proteins and the Tie2 receptor was further examined by biosensor surface plasmon resonance. The resulting affinity values [dissociation constant (Kd)] of native Ang1, GCN4-Ang1, MAT-Ang1, and COMP-Ang1 were 7.5, 158.5, 67, and 20.5 nM, respectively. These results indicated that the higher-order oligomerized forms of Ang1 had a higher affinity for Tie2. Interestingly, the association and dissociation rates of native Ang1 were slower than those of any of the three Ang1 variants (Fig. 3B). In addition, native Ang1 was not completely saturated with immobilized Tie2-Fc protein even at the higher concentration (250 nM), which is probably the result of self-aggregation. Virtually identical results were obtained by immobilizing the recombinant Ang1 variants and then measuring association and dissociation rates of sTie2-Fc (data not shown).
Fig. 3.
Ang1 variants have different receptor-binding and activation properties. (A) In vitro binding analysis with control buffer (CB), sTie1-Fc (T1), sTie2-Fc (T2), and total amount of protein input for binding assay (TAP). Approximately 75%, 20%, 95%, and 100% of the total protein amount of native Ang1, GCN4-Ang1, MAT-Ang1, and COMP-Ang1, respectively, bound to Tie2. (B) Biacore analysis of native Ang1 (NA), GCN4-Ang1 (G), MAT-Ang1 (M), and COMP-Ang1 (C) with sTie2-Fc immobilized on a chip. Typical sensorgram showing the association and dissociation profiles of 60 nM recombinant protein. An equal amount of VEGF was used as a control. (C–H) Serum-starved HUVECs were treated with 200 ng/ml of native Ang1, 100 ng/ml of Ang1 variants, or 200 ng/ml of Ang1* (A1*) for 10 (C) or 20 (F) min, for the indicated times (D, G, and H), and for 30 min (E) with the indicated amounts of native Ang1 and COMP-Ang1. The phosphorylations of Tie2 and Akt (Ser-473) were measured. The ratios indicate the densitometric analyses presented as the relative ratio of phospho-Tie2 (pTie2) to Tie2 or phospho-Akt (pAkt) to Akt at time 0 or with control buffer (CB) arbitrarily given as 1. (F and H) Ratio indicates the mean ± SD. from three experiments. *, P < 0.05 versus control buffer. #, P < 0.05 versus native Ang1 or Ang1*.
Biological Effects of Ang1 Variants in Primary Cultured Endothelial Cells. The activities of the recombinant Ang1 variants were tested by using primary cultured HUVECs. Similar levels of Tie2 phosphorylation were observed for MAT-Ang1 and native Ang1 (Fig. 3C). Interestingly, Tie2 phosphorylation induced by COMP-Ang1 was not only greater but was also significantly more persistent than that of native Ang1 (Fig. 3 D and E). As expected, the GCN4-Ang1 protein did not induce Tie2 phosphorylation (Fig. 3 C and D). Consistent results were obtained for the phosphorylation of Akt (Ser-473), which is known to be a component of the Tie2 signaling pathway in endothelial cells (27, 28). Likewise, Akt phosphorylation induced by COMP-Ang1 was higher and significantly more persistent than that induced by native Ang1 or MAT-Ang1 (Fig. 3 F and G). Akt phosphorylation by COMP-Ang1 persisted for 3–5 h (Fig. 3G), whereas that by native Ang1 lasted only 1–2 h (data not shown). In addition, we compared the Tie2 phosphorylation activity of COMP-Ang1 with Ang1*. Similarly, Tie2 phosphorylation induced by COMP-Ang1 was not only greater but was also significantly more persistent than that of Ang1* (Fig. 3H).
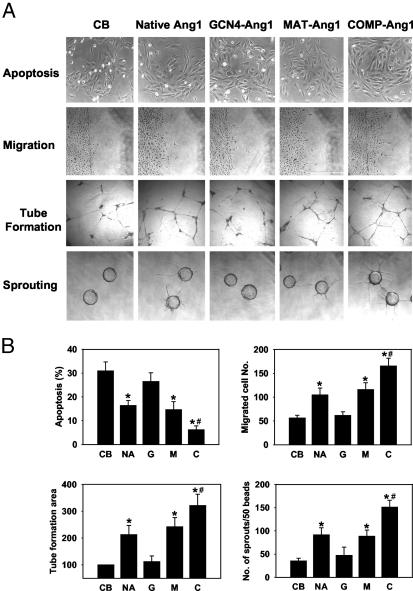
The Ang1–Tie2–Akt system is involved mainly in endothelial cell survival, migration, sprouting, and tube formation (20, 22, 27, 28). Consistent with these biochemical effects, COMP-Ang1 showed the strongest effect on these cellular activities, whereas GCN4-Ang1 was completely inactive (Fig. 4). The observation that GCN4-Ang1 did not provoke changes in Tie2/Akt phosphorylation or biological activities in endothelial cells suggested the possibility that this particular variant could act as an antagonist to native Ang1. To address this possibility, we examined the effect of COMP-Ang1 on endothelial cells in the presence of an ≈10-fold molar excess of GCN4-Ang1. Interestingly, pretreatment of endothelial cells with GCN4-Ang1 further enhanced the activity of COMP-Ang1. These results suggest that GCN4-Ang1 enhances the clustering of Tie2, which is likely to be an essential step in Tie2-mediated signal transduction.
Fig. 4.
COMP-Ang1 has a greater biological activity than native Ang1. (A) Representative photographs of assays of apoptosis, cell migration, tube formation, and sprouting in HUVECs. The HUVECs were treated with 200 ng/ml of native Ang1 or 100 ng/ml of Ang1 variants for 24 h (apoptosis, migration, and tube formation) or for 72 h (sprouting). CB, control buffer. (Magnification ×200.) (B) Quantification of biological activities. NA, native Ang1; G, GCN4-Ang1; M, MAT-Ang1; C, COMP-Ang1. *, P < 0.05 versus control buffer. #, P < 0.05 versus native Ang1 (200 ng/ml).
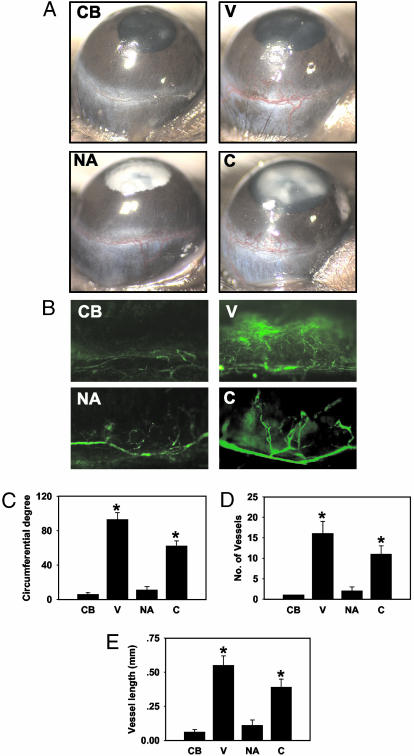
COMP-Ang1 Alone Induces Strong Angiogenesis in Vivo. To assess the effect of native Ang1 and COMP-Ang1 on in vivo angiogenesis, we performed a corneal micropocket assay with implantation of pellets containing the recombinant proteins. On day 6 after pellet implantation, corneal angiogenesis was evaluated by using a stereomicroscope. Consistent with a previous report (29), the pellet containing control buffer or native Ang1 did not induce significant angiogenesis, whereas VEGF induced strong angiogenesis (Fig. 5 A and C–E). However, the pellet containing COMP-Ang1 induced corneal angiogenesis extending from the limbus across the cornea, comparable with VEGF-induced angiogenesis (Fig. 5 A and C–E). Thus, COMP-Ang1 shows a potent angiogenic effect in vivo. Using in situ FITC-dextran fluorescent microscopy in the same animals (Fig. 5B), we also measured the stability of neovessels. Interestingly, COMP-Ang1-induced neovessels did not show any leakage, whereas VEGF-induced neovessels showed profound leakage. These data suggest that COMP-Ang1 alone can produce an angiogenic effect, with nonleaky neovessel formation.
Fig. 5.
COMP-Ang1 is sufficient to generate angiogenesis in the mouse corneal micropocket assay. (A) Representative macroscopic photographs of mouse corneas 6 days after pellet implantation. Corneal angiogenesis was not observed with control buffer (CB) or with native Ang1 (NA), whereas VEGF (V) or COMP-Ang1 (C) induced corneal angiogenesis extending from the limbus across the cornea. (B) In situ FITC-dextran fluorescent staining of corneal neovessels in the same animal. COMP-Ang1-induced neovessels show no leakage, whereas VEGF-induced neovessels show profound leakage. (C–E) Quantification of corneal angiogenesis presented as circumferential degree, number of microvessels, and vessel length, respectively. Amounts used of VEGF, native Ang1, and COMP-Ang1 were 100, 600, and 300 ng, respectively. *, P < 0.01 versus control buffer. For each group, n = 12.
Discussion
Detailed structural information on a protein ligand is indispensable for understanding how it activates its receptor and for finding ways to improve its activity by protein engineering. To achieve Tie2 receptor oligomerization and activation, Ang1 uses a modular and multimeric structure unlike that of any other known growth factor (15–17). Consistent with a recent report (16), our findings demonstrate that Ang1 tetramers can mediate receptor activation. Although Davis et al. (16) concluded that tetramers are the minimal unit required for receptor activation, the possibility that Tie2 could be activated by trimeric Ang1 variants cannot be excluded at present.
Davis et al. (16) found that engineered dimers of Ang1 lacking the superclustering domain were able to bind to Tie2 but were inactive in Tie2 phosphorylation. These findings are supported by our virtually identical results on GCN4-Ang1. Although these observations highlight the importance of the superclustering domain for native Ang1 function, interestingly, the engineered Ang1 variants MAT-Ang1 and Ang1-F1–Fc-F1 (16) could phosphorylate Tie2 with a similar efficiency to native Ang1, indicating that superclustering per se is not required for receptor activation. Although the details of how Tie2 clustering leads to receptor phosphorylation remain to be worked out, these findings suggest that tetramers and possibly even trimers of Tie2 could be the minimum units for autophosphorylation. Alternatively, the even greater activity of COMP-Ang1 as compared with native Ang1, Ang1*, or MAT-Ang1 may be explained by an even higher local concentration of clustered fibrinogen-like domains in the five-stranded Ang1 variant.
In this study, we have shown that a chimeric form of Ang1, COMP-Ang1, has many potential advantages over the native protein. The generation of native Ang1 is difficult, and the activity of the purified native Ang1 varies, possibly as a result of its tendency to form variably sized multimers. Furthermore, our results indicate that the superclustering domain of Ang1 is the major cause of the protein's insolubility. Multimerization of Ang1 by the superclustering domain results in large aggregates over time, and these aggregates show a strong tendency to precipitate and thereby lose the activity. The scientists at Regeneron Pharmaceutical (30) have generated a modified recombinant Ang1, Ang1*. However, Ang1* still has the same problems that have been observed in native Ang1. In contrast, the generation of COMP-Ang1 is relatively easy, its activity is potent and stable, and it does not aggregate over time. Thus, COMP-Ang1 can be used for therapeutic applications. Moreover, our present biotechnology could be applied to design and produce soluble and potent oligomeric ligands from other endogenous multimeric ligands such as other angiopoietins, angiopoietin-related proteins, adiponectin, ephrins, endostatin, or thrombospondins.
The ultimate goal of therapeutic angiogenesis using growth factors is to establish stable, healthy, and functional blood vessels in ischemic areas while avoiding side effects (13, 14). Among growth factors, VEGF and Ang1 are of particular interest because their receptors are specifically located on endothelial cells (2, 3, 13, 14). In fact, VEGF is already in clinical trials for therapeutic angiogenesis despite its deleterious effects, which include edema, inflammation, vascular leakage, and production of disorganized blood vessels in experimental systems (13, 14, 31). In a mouse corneal micropocket assay, Ang1 failed to stimulate an angiogenic response when administered alone (29). However, when administered with VEGF, Ang1 augmented postnatal angiogenesis (29). Consistent with these results, our study showed that native Ang1 failed to stimulate an angiogenic response. However, intriguingly, our study reveals that COMP-Ang1 alone stimulates angiogenesis with nonleaky neovessel formation, whereas VEGF stimulates angiogenesis with leaky neovessel formation. Thus, COMP-Ang1 can induce stable, healthy, and functional blood vessel formation in vivo. In addition, Ang1 can counteract deleterious VEGF-induced side effects, while having an additive effect on angiogenesis (9–12). Thus, COMP-Ang1 protein delivery could be useful for accurate and safe therapeutic angiogenesis. In conclusion, COMP-Ang1 is superior to native Ang1 in several ways, including efficiency of generation, potency, Tie2 activation, and angiogenesis in vivo. Thus, COMP-Ang1 can potentially be applied in therapeutic angiogenesis.
Acknowledgments
We thank T. Suda, S. Davis, N. Gale, and G. Thurston for critical comments and helpful discussion; Dr. B. Maco for excellent technical assistance in the metal-shadowing specimen preparation; J. Macke for help in preparing the manuscript; the Interdisciplinary Center for Microscopy of the University of Basel for access to the transmission electron microscope; and Regeneron Pharmaceuticals for providing Ang1*. R.A.K. is a Wellcome Trust Research Career Development Fellow. This work was supported by the Bio-Challenge Program (to G.Y.K.) and by National Research Laboratory Program Grant 2000-N-NL-01-C-228 (to G.M.L.) of the Korean Ministry of Science and Technology and the Brain Korea 21 Project.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ang1, angiopoietin-1; Tien, tyrosine kinase with Ig and epidermal growth factor homology domain n; VEGF, vascular endothelial growth factor; sTien, soluble extracellular domain of Tien; COMP, cartilage oligomeric matrix protein; Fc, crystallizable fragment; HUVEC, human umbilical vein endothelial cell.
References
- 1.Davis, S., Aldrich, T. H., Jones, P. F., Acheson, A., Compton, D. L., Jain, V., Ryan, T. E., Bruno, J., Radziejewski, C., Maisonpierre, P. C., et al. (1996) Cell 87, 1161–1169. [DOI] [PubMed] [Google Scholar]
- 2.Yancopoulos, G. D., Davis, S., Gale, N. W., Rudge, J. S., Wiegand, S. J. & Holash, J. (2000) Nature 407, 242–248. [DOI] [PubMed] [Google Scholar]
- 3.Jones, N., Iljin, K., Dumont, D. J. & Alitalo, K. (2001) Nat. Rev. Mol. Cell Biol. 2, 257–267. [DOI] [PubMed] [Google Scholar]
- 4.Takakura, N., Watanabe, T., Suenobu, S., Yamada, Y., Noda, T., Ito, Y., Satake, M. & Suda, T. (2000) Cell 102, 199–209. [DOI] [PubMed] [Google Scholar]
- 5.Suri, C., Jones, P. F., Patan, S., Bartunkova, S., Maisonpierre, P. C., Davis, S., Sato, T. N. & Yancopoulos, G. D. (1996) Cell 87, 1171–1180. [DOI] [PubMed] [Google Scholar]
- 6.Sato, T. N., Qin, Y., Kozak, C. A. & Audus, K. L. (1993) Proc. Natl. Acad. Sci. USA 90, 9355–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suri, C., McClain, J., Thurston, G., McDonald, D. M., Zhou, H., Oldmixon, E. H., Sato, T. N. & Yancopoulos, G. D. (1998) Science 282, 468–471. [DOI] [PubMed] [Google Scholar]
- 8.Shyu, K. G., Manor, O., Magner, M., Yancopoulos, G. D. & Isner, J. M. (1998) Circulation 98, 2081–2087. [DOI] [PubMed] [Google Scholar]
- 9.Chae, J. K., Kim, I., Lim, S. T., Chung, M. J., Kim, W. H., Kim, H. G., Ko, J. K. & Koh, G. Y. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 2573–2578. [DOI] [PubMed] [Google Scholar]
- 10.Thurston, G., Suri, C., Smith, K., McClain, J., Sato, T. N., Yancopoulos, G. D. & McDonald, D. M. (1999) Science 286, 2511–2514. [DOI] [PubMed] [Google Scholar]
- 11.Thurston, G., Rudge, J. S., Ioffe, E., Zhou, H., Ross, L., Croll, S. D., Glazer, N., Holash, J., McDonald, D. M. & Yancopoulos, G. D. (2000) Nat. Med. 6, 460–463. [DOI] [PubMed] [Google Scholar]
- 12.Kim, I., Moon, S. O., Park, S. K., Chae, S. W. & Koh, G. Y. (2001) Circ. Res. 89, 477–479. [DOI] [PubMed] [Google Scholar]
- 13.Carmeliet, P. (2000) Nat. Med. 6, 389–395. [DOI] [PubMed] [Google Scholar]
- 14.Yla-Herttuala, S. & Alitalo, K. (2003) Nat. Med. 6, 694–701. [DOI] [PubMed] [Google Scholar]
- 15.Procopio, W. N., Pelavin, P. I., Lee, W. M. & Yeilding, N. M. (1999) J. Biol. Chem. 274, 30196–30201. [DOI] [PubMed] [Google Scholar]
- 16.Davis, S., Papadopoulos, N., Aldrich, T. H., Maisonpierre, P. C., Huang, T., Kovac, L., Xu, A., Leidich, R., Radziejewska, E., Rafique, A., et al. (2003) Nat. Struct. Biol. 10, 38–44. [DOI] [PubMed] [Google Scholar]
- 17.Schlessinger, J. (2000) Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 18.Fowler, W. E. & Aebi, U. (1983) J. Ultrastruct. Res. 83, 319–334. [DOI] [PubMed] [Google Scholar]
- 19.Kim, I., Moon, S. O., Han, C. Y., Pak, Y. K., Moon, S. K., Kim, J. J. & Koh, G. Y. (2001) Cardiovasc. Res. 49, 872–881. [DOI] [PubMed] [Google Scholar]
- 20.Kim, I., Kim, H. G., So, J. N., Kim, J. H., Kwak, H. J. & Koh, G. Y. (2000) Circ. Res. 86, 24–29. [DOI] [PubMed] [Google Scholar]
- 21.Kwak, H. J., Lee, S. J., Lee, Y. H., Ryu, C. H., Koh, K. N., Choi, H. Y. & Koh, G. Y. (2000) Circulation 101, 2317–2324. [DOI] [PubMed] [Google Scholar]
- 22.Kim, I., Kim, H. G., Moon, S. O., Chae, S. W., So, J. N., Koh, K. N., Ahn, B. C. & Koh, G. Y. (2000) Circ. Res. 86, 952–959. [DOI] [PubMed] [Google Scholar]
- 23.Muthukkaruppan, V. & Auerbach, R. (1979) Science 205, 1416–1418. [DOI] [PubMed] [Google Scholar]
- 24.O'Shea, E. K., Klemm, J. D., Kim, P. S. & Alber, T. (1991) Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- 25.Dames, S. A., Kammerer, R. A., Wiltscheck, R., Engel, J. & Alexandrescu, A. T. (1998) Nat. Struct. Biol. 5, 687–691. [DOI] [PubMed] [Google Scholar]
- 26.Malashkevich, V. N., Kammerer, R. A., Efimov, V. P., Schulthess, T. & Engel, J. (1996) Science 274, 761–765. [DOI] [PubMed] [Google Scholar]
- 27.Kontos, C. D., Stauffer, T. P., Yang, W. P., York, J. D., Huang, L., Blanar, M. A., Meyer, T. & Peters, K. G. (1998) Mol. Cell. Biol. 18, 4131–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, N., Master, Z., Jones, J., Bouchard, D., Gunji, Y., Sasaki, H., Daly, R., Alitalo, K. & Dumont, D. J. (1999) J. Biol. Chem. 274, 30896–30905. [DOI] [PubMed] [Google Scholar]
- 29.Asahara, T., Chen, D., Takahashi, T., Fujikawa, K., Kearney, M., Magner, M., Yancopoulos, G. D. & Isner, J. M. (1998) Circ. Res. 83, 233–240. [DOI] [PubMed] [Google Scholar]
- 30.Maisonpierre, P. C., Suri, C., Jones, P. F., Bartunkova, S., Wiegand, S. J., Radziejewski, C., Compton, D., McClain, J., Aldrich, T. H., Papadopoulos, N., et al. (1997) Science 277, 55–60. [DOI] [PubMed] [Google Scholar]
- 31.Isner, J. M. & Asahara, T. (1999) J. Clin. Invest. 103, 1231–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]