Abstract
Multi-atlas segmentation has been widely applied in medical image analysis. This technique relies on image registration to transfer segmentation labels from pre-labeled atlases to a novel target image and applies label fusion to reduce errors produced by registration-based label transfer. To improve the performance of registration-based label transfer against registration errors, our first contribution is to propose a label transfer scheme that generates multiple warped versions of each atlas to one target image through registration paths obtained by composing inter-atlas registrations and atlas-target registrations. The problem of decreasing quality of warped atlases caused by accumulative errors in composing multiple registrations is properly addressed by an atlas selection method that is guided by atlas segmentations. To improve the performance of label fusion against registration errors, our second contribution is to integrate the probabilistic correspondence model employed by the non-local mean approach with the joint label fusion technique, both of which have shown excellent performance for label fusion. Experiments on mitral-valve segmentation in 3D transesophageal echocardiography (TEE) show the effectiveness of the proposed techniques.
1 Introduction
Label fusion based multi-atlas segmentation has been widely applied in medical image analysis. This technique applies deformable image registration to establish one-to-one correspondence between each pre-labeled training image, called an atlas, and a novel target image. Then, the segmentation label is transferred to the target image by warping each atlas based on the correspondence. Each warped atlas provides one candidate segmentation for the target image. To reduce segmentation errors produced by registration-based label transfer, label fusion is applied to combine all candidate segmentations into a consensus segmentation.
As empirical studies [1, 13] have shown, the performance of multi-atlas segmentation usually can be improved when the number of applied atlases increases. However, generating atlases with high quality segmentation is time consuming and labor intensive. Hence, atlases that can be applied in practice are often very limited. Under such circumstances, how to effectively apply limited atlases for optimal segmentation performance is an important problem. To address this problem, our main contribution is to propose techniques that allow more effective label transfer and more accurate label fusion.
Most existing multi-atlas segmentation techniques register and warp each atlas to a target image only once. If the registration fails, the information provided by the atlas for segmenting the target image may be completely wasted. Since any single registration may be unreliable, one natural solution to make the solution more robust to registration failures is to generate multiple registrations and warps for each atlas.
To address this problem, [7] proposed an atlas propagation approach that warps each atlas to a novel target image through other novel target images. To generate multiple warped versions of each atlas for a target image, multiple registration paths obtained by composing registrations between different novel target images are applied. Although this approach significantly improves single atlas segmentation performance [7], the requirement for inter-target registrations makes it computationally expensive.
To make the idea of atlas propagation through various registration paths more practical and more effective, our first contribution is to propose warping each atlas to a target image through registration paths obtained from composing inter-atlas registrations. This approach has two key advantages: 1) Since inter-atlas registrations can be computed off-line, this approach does not increase on-line registration burden. 2) Since the manual segmentation of each atlas is known, it allows an atlas segmentation guided atlas selection method, which can reliably detect and remove poor quality warps obtained through composing inter-atlas registrations.
To fuse the warped atlases, we apply local weighted voting with the joint label fusion technique [17]. Unlike most other label fusion techniques, which rely on the assumption of independence between atlases, joint label fusion explicitly incorporate correlations between atlases in label fusion. As shown in [17], joint label fusion performed better than label fusion with independent voting weight estimation. To further improve the performance of joint label fusion against registration errors, our second contribution is to integrate the probabilistic correspondence model employed by the non-local mean approach, which has proven to be highly effective for addressing image registration related uncertainties [5, 2], into the joint label fusion approach.
For validation, we apply our method to mitral valve segmentation in 3D transesophageal echocardiography (TEE) and compare with the original joint label fusion approach. We show that our atlas propagation approach and the enhanced joint label fusion approach produce significant improvements.
2 A Bayesian view for multi-atlas segmentation
First, we give a brief overview for multi-atlas segmentation. Let be n independently constructed atlases, where is an atlas image and is the atlas segmentation. For a target image TF, its segmentation, TS, can be estimated using the atlases as follows:
| (1) |
where D = (DF, DS) is one feasible warp of the atlases into the native space of TF. DF and DS denote the warped atlas image and the corresponding warped atlas segmentation, obtained by performing deformable image registration between one atlas to the target image. p(D|TF,
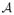 ) is the probability of observing the warped atlas D given the target image TF and the atlas set
) is the probability of observing the warped atlas D given the target image TF and the atlas set
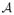 . One common way to estimate this probability is based on image similarity between the warped atlas image and the target image over local patches, under the assumption that high similarities indicate high probabilities of observing the warps.
. One common way to estimate this probability is based on image similarity between the warped atlas image and the target image over local patches, under the assumption that high similarities indicate high probabilities of observing the warps.
Due to the uncertainty in image registration, each atlas may generate multiple feasible warps for one target image. However, most work only produces one warp for each atlas, where the warped atlas is often generated by a maximum a posterior (MAP) registration result produced by some registration algorithm. Under the assumption that the posterior distribution of feasible warps produced by one atlas is narrowly peaked around its maximum, this approach usually can give reasonable solutions when the registration algorithms can reliably find the MAP solution. However, when the assumption is invalid or the registration algorithm works poorly, only using the MAP registration warp produced by one registration algorithm may be inadequate.
By contrast, since the Bayesian approach requires to marginalize over all feasible warped atlases, it is more robust against any single registration failures. This advantage can be crucial for applications such as mitral valve segmentation in ultrasound images addressed in this paper, where reliable image registrations are hard to generate due to poor image quality and large motion induced deformations. Although computing all feasible warped atlases is intractable, it is still beneficial if multiple feasible warps can be generated for each atlas. In the next section, we discuss some possible solutions to address this problem.
3 Sampling strategies for registration-based label transfer
3.1 Sampling with different registrations
One way to generate additional independently warped atlases using a fixed atlas set is by applying different registration algorithms with different parameter settings to compute the registrations for warping each atlas. The main limitation of this approach is that the computational cost for image registration increases linearly with the number of warped atlases. Given the fact that multi-atlas segmentation is already one of the most computationally expensive segmentation techniques due to the requirement for one registration between each atlas and a target image, further increasing registration costs will make this technique less practical.
3.2 Atlas propagation through various registration paths obtained from composing registrations
As discussed in the introduction, warping one atlas to a target image through various registration paths constructed by composing registrations is an efficient approach for generating multiple warps for each atlas because each registration can be utilized in multiple registration paths for generating new warps. [7] first applied this idea through composing inter-target registrations and showed its effectiveness. To make the idea more practical and more effective, we propose to generate multiple warps through composing inter-atlas registrations.
Let φi→T be the diffeomorphic map between atlas i and a target image computed by some registration algorithm. We call warped atlases obtained from such single registrations first-order warps. Let φi→j be the estimated diffeomorphic map from atlas i to atlas j. Then the composed diffeomorphic map φi→j→T = φi→j ∘ φj→T gives a correspondence map from atlas i to T as well. We call the warped atlases obtained from composing two independent registrations second-order warps. Similarly, higher-order warps can be produced from composing lower-order warps, e.g. φi→j→k→T = φi→j ∘ φj→k→T for i ≠ j ≠ k ≠ i.
Advantages
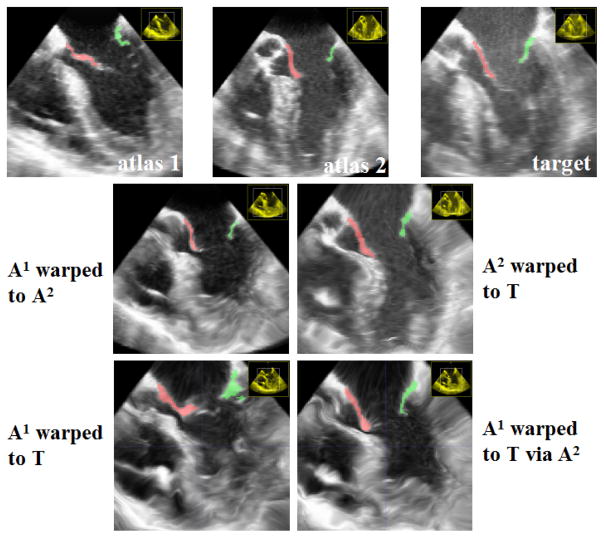
Since pairwise registrations among the atlases can be computed offline, one advantage is that it can significantly increase the number of warped atlases without substantially increasing the online computational burden, as only the registrations from each atlas to the target image are required. Furthermore, in some cases, high-order warps may improve the accuracy of the warped atlas. Typically, image registration can be more reliably done when the deformations between two images are small. When the deformations are large, registration is more likely to fail. One approach to address this problem is to decompose a large deformation into a series of small deformations [8]. As shown in [18, 4, 12], high-order atlas propagation is one way to achieve this goal. When each of the first-order registrations required for warping one atlas to a target image can be reliably estimated, high-order warps obtained by composing these first-order registrations may produce a more accurate warp than the one obtained from directly registering the atlas to the target image (see Fig. 1 for one example).
Fig. 1.
Atlas warping through inter-atlas registration for mitral valve segmentation in 3D transesophageal echocardiography, shown in the septolateral plane of the valve. The anterior and posterior leaflets are in red and green, respectively. Directly registering atlas A1 to the target image produces a low quality warp due to large deformations. However, since the registration between A1 and A2 and the registration between A2 and T can be done more accurately, warping A1 to T through A2 produces a more accurate warp than directly warping A1 to T.
Limitations
Since generating high-order warps requires composing multiple independently estimated registrations, errors produced in estimating these registrations will accumulate in the composed solution. Hence, it is reasonable to expect that high-order warps should overall produce less accurate candidate segmentations than low-order warps.
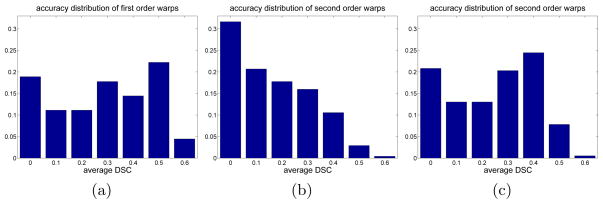
For demonstration, we conducted an empirical study to quantitatively measure the segmentation accuracy produced by first and second-order atlas warping. We used one set of 10 atlases used in our mitral valve segmentation experiments (data description in section 5). We computed pairwise deformable image registration between each pair of the atlases, from which each atlas was warped to each of the remaining 9 atlases through first-order and second-order warping. Fig. 2(a) shows the distribution of segmentation accuracy produced by first-order and second-order warping, respectively. It clearly shows that second-order warping produces less accurate segmentations than first-order warping.
Fig. 2.
Distribution of segmentation accuracy produced by first-order atlas warping (a) and second-order atlas warping (b) for the mitral valve segmentation problem. (c) shows the results of second-order warping with atlas segmentation guided atlas selection with a threshold of average Dice similarity coefficient (DSC)=0.5 (see text in section 3 for more detail). For this mitral valve segmentation problem, reliable image registrations are hard to generate. The average DSC produced by first-order warping is 0.328. The average DSC produced by second-order warping without/with atlas selection is 0.209/0.285, respectively. Atlas segmentation guided atlas selection significantly improves the overall quality of the warped atlases produced by second-order warping.
Atlas selection guided by atlas segmentations
To make label fusion more reliable with high-order warps, we propose an atlas selection technique that is capable of removing most of the low quality warped atlases. Since the ground truth segmentation of the target image is unknown, it is difficult to quantitatively estimate the registration accuracy in the final warped atlases. However, since the manual segmentation of each intermediate atlas is known, it is possible to quantitatively evaluate the quality of intermediately warped atlases. For instance, let atlas j be the last propagating atlas before warping atlas i to a target image. Let be the segmentation obtained from propagating atlas i’s segmentation to atlas j. Note that may or may not be obtained through high-order warping. Under the assumption that the segmentation of atlas i and atlas j are different only due to random effects in producing manual segmentation, the overlap between the manual segmentation of atlas Aj and is a good indicator of accumulative errors for propagating atlas i to atlas j. We measure the Dice similarity coefficient (DSC) [6] for each label between and and compute the average DSC over all labels. Then a threshold can be applied to remove those atlases that propagate poorly. Fig. 2 shows that this approach can significantly improve the overall quality of high-order atlas warps.
4 Label fusion
We apply local weighted voting with the joint label fusion algorithm [17] to fuse warped atlases. Comparing to other label fusion methods, the key advantage of joint label fusion is that it explicitly considers atlas correlations to reduce bias in the atlas set. To improve the performance of joint label fusion against registration errors, we adopt the probabilistic correspondence model employed by the non-local mean approach to improve the estimation accuracy of atlas correlations. For self completeness, we briefly summarize the joint label fusion approach below.
4.1 Joint label fusion
Joint label fusion models segmentation errors produced by each warped atlas as . TS,l(x), are the observed votes for label l produced by the target image and the ith warped atlas, respectively. Hence, δi(x) ∈ {−1, 0, 1} is the observed label difference. The correlation between the segmentation errors produced by any two atlases at location x are captured by a dependency matrix Mx, with measuring the probability that atlas i and j produce the same label error for the target image. The expected label difference between the consensus solution obtained from weighted voting and the target segmentation is:
| (2) |
where is the voting weight for atlas i at location x and t stands for transpose. To minimize the expected label difference, the optimal voting weights can be solved by , where n is the number of warped atlases and 1n = [1; 1; …; 1] is a vector of size n.
4.2 Robust estimation of pairwise atlas correlations
To estimate the pairwise atlas dependency matrix Mx, [17] applies an image similarity based model over local patches as follows:
| (3) |
where
is the vector of absolute intensity difference between a warped atlas and the target image over a local patch
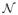 (x) centered at x and 〈·, ·〉 is the dot product. β is a model parameter.
(x) centered at x and 〈·, ·〉 is the dot product. β is a model parameter.
To make the estimation more robust against registration errors, [17] applies a local search algorithm to find the patch from each warped atlas within a small neighborhood
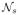 (x) that is the most similar to the target patch in the target image. Under the assumption that more similar patches are more likely to be correct correspondences, instead of the original corresponding patches in the warped atlases, the searched patches are applied for label fusion. However, since image similarities over local patches are not always reliable indicators of registration errors, the searched most similar patch may still give an incorrect correspondence. To address the unreliability in choosing any single candidate corresponding patch for label fusion, we propose to consider all feasible corresponding patches. We achieve this goal by integrating the non-local mean patch-based label fusion technique [5, 2], which is shown to be effective for handling the unreliability in determining the correct correspondence, into the above atlas correlation estimation method (3).
(x) that is the most similar to the target patch in the target image. Under the assumption that more similar patches are more likely to be correct correspondences, instead of the original corresponding patches in the warped atlases, the searched patches are applied for label fusion. However, since image similarities over local patches are not always reliable indicators of registration errors, the searched most similar patch may still give an incorrect correspondence. To address the unreliability in choosing any single candidate corresponding patch for label fusion, we propose to consider all feasible corresponding patches. We achieve this goal by integrating the non-local mean patch-based label fusion technique [5, 2], which is shown to be effective for handling the unreliability in determining the correct correspondence, into the above atlas correlation estimation method (3).
First, a probability model is applied to represent the correspondence between each warped atlas and a target image. For each voxel x in the target image, the probability that a voxel xi in the warped atlas i is the correct corresponding voxel is estimated by:
| (4) |
where a Gaussian model is applied to transfer image similarity into a probability measure. In our experiments, we normalize the intensity vector obtained from each local image intensity patch, such that the normalized vector has zero mean and a constant norm and σ is fixed to be 0.1. is a normalization factor. Combining this probability correspondence representation model and the patch-based atlas correlation model in (3), we estimate the probability that two atlases produce the same segmentation error for the target image by:
| (5) |
In comparison to the local search algorithm, the key difference in (5) is that each patch within the searching area are considered as a potential match, but weighted by the estimated probability of being the correct match. Hence, it is more robust to the errors produced by the hard decision made by selecting the correspondence based on local image similarities.
5 Experiments
We apply our approach to segment the mitral valve in 3D transesophageal echocardiography (3D TEE). The mitral valve supports physiologically normal cardiac function by maintaining unidirectional blood flow across the left heart. Common valve diseases, such as ischemic and degenerative mitral regurgitation, are associated with pathological alterations in mitral leaflet and annular morphology. 3D examination of these morphological abnormalities, which vary substantially between individuals, is critical to the diagnosis and personalized surgical treatment of mitral valve disease. 3D TEE is the most practical imaging modality for 3D mitral valve assessment in the operating room and has been effectively used in both research and clinical settings to visualize and quantify mitral leaflet and annular geometry in vivo [11, 15, 16]. Segmentation of the mitral valve from 3D TEE is crucial for quantitative assessment.
Three new methods are proposed in this paper, which include atlas propagation through composing inter-atlas registrations (AP), atlas segmentation guided atlas selection (AS) and non-local mean based robust atlas correlation estimation (NL). To evaluate the performance of each proposed method, in addition to the overall performance produced by combining the three components, we also evaluated the effectiveness of each component. The tested methods include LWJoint-NL, LWJoint-AP, LWJoint-AP-AS, LWJoint-AP-AS-NL, where the method’s name shows how different methods are combined. In our experiments, we only included first and second-order warps for AP. For atlas segmentation guided atlas selection, since the empirical study on atlases in Fig. 2.(c) shows that a threshold 0.5 average DSC can effectively remove most poor quality warped atlases, we fixed the threshold to be 0.5 average DSC.
Data acquisition and manual segmentation
Twenty patients undergoing cardiac surgery were imaged pre-operatively using real-time 3D TEE. This cohort included 6 subjects with normal mitral valve anatomy and function, and 14 subjects with mild to severe mitral valve disease. All studies were performed after induction of general anesthesia and before initiation of cardiopulmonary bypass. Electrocardiographically gated full-volume images were acquired with the iE33 platform (Philips Medical Systems, Andover, MA) using a 2 to 7 MHz transesophageal matrix-array transducer over four consecutive cardiac cycles. The frame rate was 17 to 30 Hz, and the imaging depth was 12 to 16 cm. From each subject’s data series, 3D TEE images of the mitral valve at mid-diastole were selected for analysis. These 3D TEE images were exported in Cartesian format (224×208×208 voxels), with an approximate isotropic resolution of 0.6 to 0.8 mm. The 20 images selected for analysis were uploaded to ITK-SNAP [19], an open source software package for medical image segmentation. An expert observer manually segmented the anterior and posterior leaflets in their entirety, associating the two leaflets with separate labels.
Experimental setup
For cross-validation evaluation, we randomly selected 10 images to be the atlases and the other 10 images for testing. Each atlas was registered to each test image, as well as to each other atlas. Global registration was performed using the FSL FLIRT tool [14] with 12 degrees of freedom and using the default parameters (normalized mutual information similarity metric; search range from −5 to 5 in x, y and z). Deformable registration was performed using the ANTS Symmetric Normalization (SyN) algorithm [3], with the cross-correlation similarity metric (with radius 2) and a Gaussian regularizer with σ = 3. The cross-validation experiment was repeated 10 times. In each experiment, a different set of atlases and test images were randomly selected. The results reported below are averaged over the 10 experiments.
The baseline performance is produced by joint label fusion (LWJoint) without high-order warps. As described in section 4, LWJoint has three free parameters: r, the radius of the local appearance window
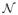 used in similarity-based Mx estimation; rs, the radius of the local searching window
used in similarity-based Mx estimation; rs, the radius of the local searching window
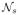 ; and β, the parameter used for estimating atlas correlation in (3). For each cross-validation experiment, the parameters are optimized by exhaustive search among a range of values in each parameter (r ∈ {1, 2, 3}; rs ∈ {0, 1, 2, 3}; β ∈ {0.5, 0.75, …, 2}) using the atlases in a leave-one-out strategy. We measure the average DSC between the automatic segmentation of each atlas obtained via the remaining atlases and the manual segmentation of that atlas, and find the optimal parameters that maximize this average DSC. For the 10 cross-validation experiments, the most frequently selected parameters are (r, rs, β)=(2,3,1). The same parameters selected for LWJoint were also applied for other LWJoint-based methods.
; and β, the parameter used for estimating atlas correlation in (3). For each cross-validation experiment, the parameters are optimized by exhaustive search among a range of values in each parameter (r ∈ {1, 2, 3}; rs ∈ {0, 1, 2, 3}; β ∈ {0.5, 0.75, …, 2}) using the atlases in a leave-one-out strategy. We measure the average DSC between the automatic segmentation of each atlas obtained via the remaining atlases and the manual segmentation of that atlas, and find the optimal parameters that maximize this average DSC. For the 10 cross-validation experiments, the most frequently selected parameters are (r, rs, β)=(2,3,1). The same parameters selected for LWJoint were also applied for other LWJoint-based methods.
For more comprehensive comparison, we also produced results by majority voting (MV), image similarity based local weighted voting with the inverse weighting model (LWInverse) [1, 10] and the Gaussian weighting model (LWGaussian) [13]. The weighting model parameters, σ and β, are optimized along with r and rs using the atlases through the leave-one-out strategy as well.
Results
Table 1 summarizes the performance of each automatic segmentation method. The results are given in DSC between manual segmentation and automatic segmentation. Due to the large mitral valve deformations across different subjects and high noises in the TEE images, most warped atlases produced by image registration are in low qualities. Hence, the results produced by MV have low accuracy. Image similarity based local weighted voting produced significantly better segmentation accuracy than MV. Among the three baseline local weighted voting methods, LWJoint produced the best results.
Table 1.
The performance of mitral valve segmentation in terms of Dice similarity coefficient produced by each method.
| method | anterior leaflets | posterior leaflets |
|---|---|---|
| MV | 0.348±0.229 | 0.250±0.187 |
| LWInverse | 0.573±0.144 | 0.422±0.172 |
| LWGaussian | 0.576±0.159 | 0.411±0.183 |
| LWJoint | 0.609±0.157 | 0.453±0.179 |
| LWJoint-NL | 0.616±0.150 | 0.464±0.174 |
| LWJoint-AP | 0.615±0.150 | 0.468±0.176 |
| LWJoint-AP-AS | 0.619±0.131 | 0.482±0.164 |
| LWJoint-AP-AS-NL | 0.623±0.126 | 0.490±0.158 |
Overall, each of the three methods proposed in this paper produced prominent improvements. The non-local mean based robust atlas correlation estimation technique consistently improved the accuracy of LWJoint with/without high-order atlas warps. The overall improvements produced by combining the three methods, i.e. LWJoint-AP-AS-NL, over LWJoint are statistically significant, with p<0.05 on the paired Students t-test. Fig. 3 shows some results produced by LWJoint and the full proposed method.
Fig. 3.
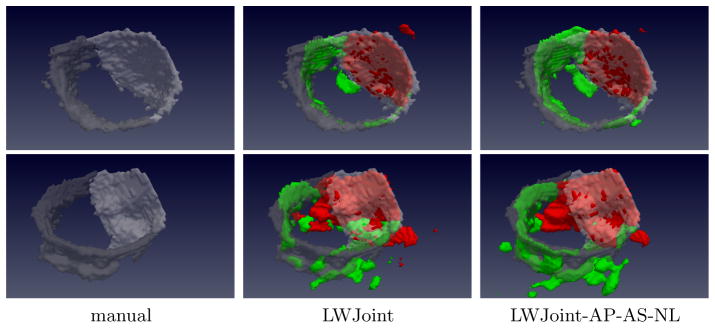
Mitral valve segmentation. The anterior and posterior leaflets are shown in red(bright) and green(gray) in automatic(manual) segmentation, respectively.
To report the mean surface distance between manual and automatic segmentation, for each voxel on one segmentation surface, we search for the closest voxel on the other segmentation surface and calculate the Euclidean distance between them. To make the measurement symmetric, the point-to-surface distance is calculated in two directions, from the automatic segmentation to manual segmentation surfaces and vice versa, and the average distance is taken as the final measurement for surface distance. When the mitral valve segmentation is evaluated as a whole, the average surface distances produced by MV, LWInverse, LGaussian, LWJoint and LWJoint-AP-AS-NL are 3.84 mm, 2.07 mm, 1.86 mm, 1.66 mm and 1.52 mm, respectively.
In the literature, [9] is the only existing work addressing fully automatic mitral valve segmentation. This work is based on discriminative learning and model fitting and was evaluated through a three-fold cross-validation on a set of 1516 TEE volumes. The mean point-to-surface distance between reference segmentation and automatic segmentation is 1.54 mm, which is comparable to that produced by our full method. However, we only used 10 training images, which is significantly fewer than those used in [9].
6 Conclusions and Discussion
To improve the performance of label fusion based multi-atlas segmentation, we proposed a robust label transfer technique that can efficiently generates multiple warps for each atlas through registration paths obtained from composing inter-atlas registrations. To address the potential risk caused by accumulative errors in composing multiple registrations, we also proposed an atlas selection method that is guided by atlas segmentations to remove poorly warped atlases. To improve the performance of joint label fusion, we proposed a new technique for robust estimation of atlas correlations. The proposed methods were validated in a mitral valve segmentation problem using 3D transesophageal echocardiography and produced significant improvements over the state of the art label fusion algorithm. Using significantly fewer manually labeled images for training, our mitral valve segmentation accuracy is also comparable to previous fully automatic mitral valve segmentation work.
In our current experiments, we only included second-order atlas warps for label fusion and we used a fixed threshold for atlas segmentation guided atlas selection. In future work, higher-order warps will be included. A study on the impact of the threshold choice on the performance will be studied as well.
References
- 1.Artaechevarria X, Munoz-Barrutia A, de Solorzano CO. Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE TMI. 2009;28(8):1266–1277. doi: 10.1109/TMI.2009.2014372. [DOI] [PubMed] [Google Scholar]
- 2.Asman A, Landman B. Non-local staple: An intensity-driven multi-atlas rater model. Medical Image Computing and Computer-Assisted Intervention–MICCAI; 2012; 2012. pp. 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avants B, Epstein C, Grossman M, Gee J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardoso MJ, Wolz R, Modat M, Fox NC, Rueckert D, Ourselin S. MICCAI. 2012. Geodesic information flows. [DOI] [PubMed] [Google Scholar]
- 5.Coupe P, Manjon J, Fonov V, Pruessner J, Robles N, Collins D. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 2011;54(2):940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Dice L. Measure of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 7.Gass T, Goksel O, Szkely G. Semi-supervised segmentation using multiple segmentation hypotheses from a single atlas. MICCAI Workshop on Medical Computer Vision; October 2012. [Google Scholar]
- 8.Hamm J, Ye D, Verma R, Davatzikos C. Gram: A framework for geodesic registration on anatomical manifolds. MedIA. 2010;14(5):633–642. doi: 10.1016/j.media.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ionasec R, Voigt I, Georgescu B, Wang Y, Houle H, Vega-Higuera F, Navab N, Comaniciu D. Patient-specific modeling and quantification of the aortic and mitral valves from 4-d cardiac ct and tee. Medical Imaging, IEEE Transactions on. 2010;29(9):1636–1651. doi: 10.1109/TMI.2010.2048756. [DOI] [PubMed] [Google Scholar]
- 10.Isgum I, Staring M, Rutten A, Prokop M, Viergever M, van Ginneken B. Multi-atlas-based segmentation with local decision fusion–application to cardiac and aortic segmentation in CT scans. IEEE Trans on MI. 2009;28(7):1000–1010. doi: 10.1109/TMI.2008.2011480. [DOI] [PubMed] [Google Scholar]
- 11.JJG, Mankad S, Freeman W, Click R, Suri R, Abel M, Oh J, Pellikka P, Nesbitt G, Syed I, Mulvagh S, Miller F. Real-time three-dimensional trans-esophageal echocardiography in the intraoperative assessment of mitral valve disease. J Am Soc Echocardiogr. 2009;22(1):34–41. doi: 10.1016/j.echo.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Jia H, Yap P, Shen D. Iterative multi-atlas-based multi-image segmentation with tree-based registration. Neuroimage. 2012;59(1):422–430. doi: 10.1016/j.neuroimage.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabuncu M, Yeo B, Leemput KV, Fischl B, Golland P. A generative model for image segmentation based on label fusion. IEEE TMI. 2010;29(10):1714–1720. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S, Jenkinson M, Woolrich M, Beckmann CaH, JohansenBerg TB, Bannister P, Luca M, Drobnjak I, Flitney D, Niazy R, Saunders J, Vickers J, Zhang Y, Stefano N, Brady J, Matthews P. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 15.Sugeng L, Shernan S, IS, Salgo, Weinert L, Shook D, Raman J, Jeevanandam V, Dupont F, Settlemier S, Savord B, Fox J, Mor-Avi V, Lang R. Live 3-dimensional transesophageal echocardiography initial experience using the fully-sampled matrix array probe. J Am Coll Cardiol. 2008;52(6):446–449. doi: 10.1016/j.jacc.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 16.Vergnat M, Jassar A, Jackson B, Ryan L, Eperjesi T, Pouch A, Weiss S, Cheung A, Acker M, Gorman J, Gorman R. Ischemic mitral regurgitation: a quantitative three-dimensional echocardiographic analysis. Ann Thorac Surg. 2011;91(1):157–164. doi: 10.1016/j.athoracsur.2010.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Suh JW, Das S, Pluta J, Craige C, Yushkevich P. Multi-atlas segmentation with joint label fusion. IEEE Trans on PAMI. 2013;35(3):611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolz R, Aljabar P, Hajnal J, Hammers A, Rueckert D. Leap: Learning embeddings for atlas propagation. NeuroImage. 2010;49(2):1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yushkevich P, Piven J, Hazlett H, Smith R, Ho S, Gee J, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]