Summary
Background
Height is a critical variable for many biomedical analyses because it is an important component of Body Mass Index (BMI). Transforming EHR height measures into meaningful research-ready values is challenging and there is limited information available on methods for “cleaning” these data.
Objectives
We sought to develop an algorithm to clean adult height data extracted from EHR using only height values and associated ages.
Results
The algorithm we developed is sensitive to normal decreases in adult height associated with aging, is implemented using an open-source software tool and is thus easily modifiable, and is freely available. We checked the performance of our algorithm using data from the Northwestern biobank and a replication sample from the Marshfield Clinic biobank obtained through our participation in the eMERGE consortium. The algorithm identified 1262 erroneous values from a total of 33937 records in the Northwestern sample. Replacing erroneous height values with those identified as correct by the algorithm resulted in meaningful changes in height and BMI records; median change in recorded height after cleaning was 7.6 cm and median change in BMI was 2.9 kg/m2. Comparison of cleaned EHR height values to observer measured values showed that 94.5% (95% C.I 93.8-% – 95.2%) of cleaned values were within 3.5 cm of observer measured values.
Conclusions
Our freely available height algorithm cleans EHR height data with only height and age inputs. Use of this algorithm will benefit groups trying to perform research with height and BMI data extracted from EHR.
Key words: Height, dimensional measurement accuracy, electronic health record, body mass index, electronic medical record, phenotyping
Introduction
In recent years, there has been increased interest in extracting phenotypes from electronic health record (EHR) data for use in clinical, epidemiologic, and genetic research. However, much of the research to date has focused on dichotomous disease phenotypes, with less research focused on the extraction of labs and vitals observations, and demographic data. This is unfortunate given the importance of these variables both as outcomes and covariates in many research studies. More research is needed on the extraction of these variables to help investigators understand how to deal with frequent measurements at uneven intervals, temporal changes, and also measurement error.
Height is a critical variable for many biomedical analyses because it is a component of body mass index (BMI), the most commonly used measure of obesity in population studies. However, there has been little research into the proper extraction of height from electronic health records (EHRs). There is evidence of both data-entry errors in EHR height data [1] and errors in measuring height [2]. The cleaning of adult height data extracted from EHR is made somewhat complicated in older populations, given the documented decrease in adult height with aging [3].
This paper describes an algorithm we developed to transform EHR adult height data, using simply age and height values easily obtained from the EHR. The algorithm is portable between sites with different EHRs, and it is designed to account for the gradual decrease in adult height with aging, as height values are compared only to values obtained at similar ages. We describe the effects of cleaning adult height data using this algorithm on an example dataset derived from Northwestern’s EHR, in the Northwestern Medicine Enterprise Data Warehouse (NMEDW), and on another set obtained from Marshfield Clinic using a different EHR system.
Methods
IRB approval for this study was obtained at both study sites.
Algorithm development
The height cleaning algorithm is shown in ▶ Figure 1. Since height measurement can vary within a specific time frame due to factors such as lack of standard protocols, inaccurate measuring, and imprecise equipment set-up we created a cleaning algorithm. We tried various cutoffs values for allowable error in measurement in the algorithm and found good performance with 3.5 cm. This acceptable error threshold can be easily changed by others running the algorithm, if desirable. Because human height as a function of growth can be classified into three distinctive phases – subject grows and attains maximum height (~up to age 24 years), height remains constant (~age 25 to 50 years), and height might decrease (~age 51 and above) [3, 4] – the cleaning algorithm separates heights obtained in these three phases and runs the algorithm separately for each phase as appropriate. The algorithm is only designed for the cleaning of adult height values (height values obtained after age 18).
Fig. 1.
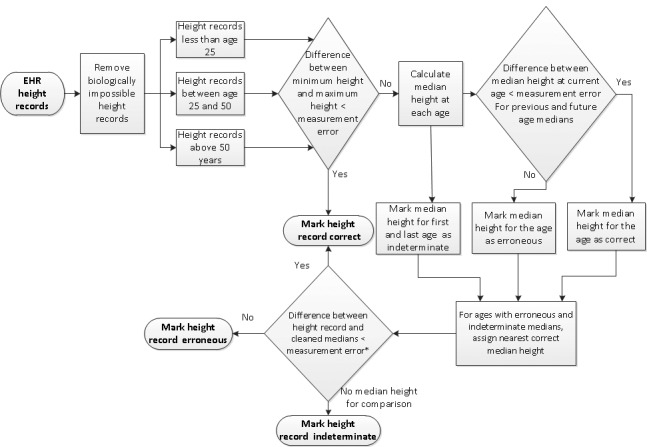
Height Cleaning Algorithm
If all height records for a single subject in a single phase differ by less than 3.5 cm, then all height records for that subject in that phase are marked correct by the algorithm. For all other subjects, median height at each age is calculated. The median height for each age is compared with that of the prior and next median; when the difference in medians is greater than 3.5 cm for both, the median height for that age is flagged as potentially erroneous. If there are only two valid medians and they differ by more than 3.5 cm, both medians are deemed indeterminate. For the ages that have an erroneous or indeterminate median, the algorithm assigns the nearest correct median height within a 3 year period. The algorithm then compares all other height measures to the corresponding cleaned median height at that age. If the recorded height for any age differs by more than 3.5 cm from the cleaned median height at that age (or 6 cm from an indeterminable median [5]), it is deemed erroneous.
We developed the algorithm in KNIME, an open-source data mining software tool (available from http://www.knime.org/), to facilitate portability of an executable algorithm and reduce implementation errors at other sites. Interested parties can clean their adult height data by downloading the workflow from the link http://tinyurl.com/EHR-Height-Cleaning and supplying a patient identifier, heights, and ages at which the heights were recorded.
Data
As part of the electronic Medical Records and Genomics (eMERGE) network [6, 7] we collected adult height data from several EHR systems. At our own site we used eMERGE participants selected from the NUgene biobank. The NUgene Project is a growing collection of DNA samples with associated health information collected from a questionnaire and the EHR. Participants give a broad consent allowing for the assessment of individual genetic variation, mining of the EHRs for phenotypes, and the use of these data for establishing correlations between phenotypes and genotypes (https://www.nugene.org/) [8]. eMERGE participants from the NUgene Project who had at least 1 non-zero height measure discretely recorded in the Northwestern Medicine (northwesternmedicine.org) Enterprise Data Warehouse (EDW) (edw.northwestern.org), (which comprises EHR data from the Cerner and Epic systems used at Northwestern for inpatient and outpatient care) were included.
In addition to the subject identifier, we used all available adult (over age 18) height values and age at height observation as inputs to the algorithm. The earliest available height record for Northwestern was in 1996 and the most recent was in 2013. All height measures were converted to centimeters. Using observation date and date of birth we calculated age at height observation and rounded it to closest age in years. For calculating BMI, we used weight in kilograms and age at weight measurements. We obtained height data from Marshfield Clinic’s EHR through our research partnership with eMERGE for use as a replication sample. The earliest height record at Marshfield was recorded in 1985 and the most recent was recorded in 2011.
As one assessment of the performance of the algorithm, we compared height measurements classified by the algorithm with available age-matched, cleaned, self-reported, height data gathered from individuals in our sample at the time of their enrollment into the NUgene biobank. We also compared height measurements classified by the algorithm with available age-matched, cleaned, height data gathered by rigorously trained research coordinators according to a standard protocol using a stadiometer from individuals at the time of their enrollment into Marshfield Clinic’s Personalized Medicine Research Project biobank. As a measure of concordance we determined whether the EHR height measure was within 3.5 cm of the self-reported or observer-measured height. 95% confidence interval estimates were calculated for positive predictive and negative predictive values of these comparisons using a freely available SAS macro made available by Erik Bergstralh on the Mayo clinic website http://www.mayo.edu/research/departments-divisions/department-health-sciences-research/division-biomedical-statistics-informatics/software/locally-written-sas-macros.
Results
▶ Table 1 describes the characteristics of the Northwestern study sample, information about height records in the EHR for this sample, and the preliminary results of cleaning this data using our algorithm. The sample in which we undertook this analysis was largely female and white. We do not anticipate that the utility of this algorithm will differ in populations with different racial or ethnic distributions. The majority (56.4 %) of our sample had 6 or more height records in the EHR. The median age where height measurements were obtained in our sample was 56. Therefore, the majority of height measurements in our sample occurred after age 50, an age when height might begin to decrease [3] within an individual. Before any cleaning, recorded height values ranged from 2.54 cm to 2,116 cm.
Table 1.
EHR Height Information in the Northwestern Study Sample
| Sample Characteristics | |
| Female (%) | 81.8 |
| Male (%) | 18.2 |
| White (%) | 87.4 |
| African American (%) | 12.6 |
| EHR Height information | |
| Number of individuals in sample | 4325 |
| Total number of height records in sample | 33,937 |
| Number of height records per subject | 6 [1–53] |
| % subjects with 1 height record | 9.9 |
| % subjects with 2–5 height records | 33.8 |
| % subjects with 6–10 height records | 29.3 |
| % subjects with 11–20 height records | 22.6 |
| % subjects with more than 20 height records | 4.5 |
| Age associated with height record (y) | 56 [18–99] |
| Maximum Height recorded (cm) | 165.1 [2.54–2116] |
| Height Cleaning Results | |
| Total records removed due to biologically implausible values* | 275 |
| Individuals with records removed due to biologically implausible values* | 232 |
| Total records removed with the algorithm | 1,248 |
| Total individuals with records removed due to the algorithm | 736 |
Mean [median]
*heights less than 100 cm or greater than 250 cm were considered to be biologically implausible
To clean the 33,937 recorded height values from the 4325 people in our sample, we first removed all height values we considered biologically implausible (based on Guinness records and literature review [9], we set the plausible thresholds greater than 100 cm and less than 250 cm, but the thresholds can easily be modified by other algorithm users if desirable). This removed 275 records total across 232 individuals in the data set. After removing biologically implausible data, we ran our height cleaning algorithm to remove additional erroneous values. Our algorithm identified 1248 additional erroneous records across 736 individuals in the dataset. The algorithm identified 31,152 records to be correct and was unable to classify 1262 records.
We describe the influence of removal of erroneous height records by our algorithm on BMI in ▶ Table 2. ▶ Table 2 summarizes information for the 1,189 height records, identified as erroneous by the algorithm, where we had access to 1) another EHR height measurement identified correct by the algorithm and 2) a weight recorded in the EHR. In cases where more than one “correct” height measurement was available in the EHR, the height measurement obtained closest to the date of the “erroneous” height record was used for comparison purposes. For these 1,189 records, the median change in height (between the “erroneous” record and the nearest “clean” record) was 7.6 cm. Fourteen records were incorrectly identified by the algorithm and therefore there was no change in height for these records. The median change in BMI resulting from the change in height was 2.9 kg/m2. In the most extreme example, an individual who was 180 cm tall had an erroneous height value of 119.38 cm identified by the algorithm. This represented a change of 50.3 units in BMI from 89.8 to 39.47.
Table 2.
Results of EHR height cleaning using the algorithm – Northwestern sample
| Erroneous height records with available correct height and weight for comparison | 1,189 |
| Mean height change (cm) | 11.8 |
| Median height change (cm) | 7.6 |
| Height range change (cm) | 0–80.1 |
| Mean BMI change (kg/m2) | 4.6 |
| Median BMI change (kg/m2) | 2.9 |
| BMI range change (kg/m2) | 0–50.3 |
Change calculated by comparing algorithm-identified “erroneous” EHR height measure to nearest algorithm-identified “correct” height EHR height measure, based on date measures obtained
We implemented the algorithm on data from a second site (Marshfield Clinic). The Marshfield dataset had more males (47% of the height records came from males) and more height records per person (median height records per person = 11). When we implemented the algorithm on the Marshfield dataset including 45,541 records, 1,357 records were found to be biologically implausible and then an additional 889 were identified as erroneous by the algorithm. In this sample, the median change in height based on the cleaning using the algorithm was 5.1 cm and the median change in BMI was 1.8 kg/m2.
We evaluated our algorithm by comparing the cleaned EMR height data to self-reported data collected at enrollment in the NUgene biobank. In this comparison, 94.5% (95% C.I 93.8-% – 95.2%) of EHR height records determined to be correct by the algorithm were within 3.5 cm of height collected at enrollment in (i.e. positive predictive value PPV), and 83.7% (95% C.I 77.2% – 89.0%) of the EHR height records determined to be erroneous were found to be more than 3.5 cm different from the height collected at enrollment in (i.e. negative predictive value NPV). We then compared cleaned Marshfield EMR height data to observer-measured data collected at biobank enrollment. In this comparison, 95.2% (95% C.I 94.7% – 95.6%) of EHR height records determined to be correct by the algorithm were within 3.5 cm of the observer-measured height collected at enrollment and 46.2% (95% C.I 38.8% – 53.7%) of the EHR height records determined to be erroneous were found to be more than 3.5 cm different from the height collected at enrollment. The smaller negative predictive value for Marshfield can be partially attributed to the smaller percentage of erroneous height measures present in this EHR system.
Conclusion
In this brief report, we described an algorithm developed to clean adult height data in EHR systems using only height records and age as inputs. We implemented the algorithm in KNIME and made the workflow publicly available so that it can be easily accessed by other groups. We believe this is the first freely disseminated program designed to clean EHR adult height data. This work employs several elements described in a previous report on using statistics to clean data from large databases [10]. Application of this algorithm using data extracted from two different EHR systems identified hundreds of errors at each site, and correction of erroneous errors resulted in meaningful changes in height measurements and associated BMI measurements.
Previous literature has demonstrated errors in height measures recorded in EHR. A study in the Veterans Health Administration (VHA) corporate data warehouse noted implausible variation in same-day and same-year heights that was attributed to measurement or data-entry error [1]. A comparison of an EHR and epidemiologic dataset in the UK found that the degree of random measurement error for height in the EHR data was greater than in the epidemiologic data [11]. It is critical to develop methods to clean EHR height data, so that height and BMI data from EHR records can be used with confidence as outcomes or covariates in observational studies, and so that clinical algorithms in the EHR designed to identify patients at lower or higher risk of disease can be improved [12].
There are limitations to our algorithm and analysis. The algorithm is only designed to be used to clean adult height (height measured after age 18). For the rare individual experiencing large (>3.5 cm/year) increases in height after age 18 due to normal growth, the algorithm may incorrectly flag some height values as erroneous. The algorithm will flag as erroneous dramatic changes in height due to serious illnesses and accidents, including potentially some severe cases of degenerative disc disease or osteoporosis. The algorithm also falsely identified some records as erroneous in middle aged individuals; it might be that a measurement error of 3.5 cm is not sufficiently large to account for normal measurement variability in the clinical setting. Both of these concerns could be partly addressed by changing the measurement error tolerance in the KNIME workflow. Additionally, one of our evaluation methods compared cleaned height to self-reported height. Self-reported height can be inaccurate, usually over reported rather than underreported [13–15]. The other used observer measured height obtained from a stadiometer, which produces higher quality data, but still suffers from a single measurement and cannot truly be considered a gold standard.
Strengths of this algorithm and analysis include that the algorithm performed well in an older population where one would expect a gradual decline in height [3] as well as that it is publicly available and easily modifiable. We encourage interested parties to obtain more information at http://tinyurl.com/EHR-Height-Cleaning.
Acknowledgements
All authors prepared and edited the manuscript, and gave final approval. AM conceived, planned, and implemented the presented work. JAP assisted with the design, analysis, and interpretation of the data, and cleaned the self-reported data. SA also cleaned the self-reported data and provided background research. PLP and JTF contributed to the design and conducted analysis, and interpretation on behalf of Marshfield. ANK and LRT supervised the work and provided feedback, with LRT leading the design, analysis, and interpretation. GT gave feedback on the algorithm and interpretation.
Footnotes
Clinical Relevance
This paper describes an algorithm we created to clean adult height values in EHR records. The algorithm can easily be downloaded, customized, and implemented by interested individuals or groups. Clean height values will be important for research using height or BMI as outcomes or covariates in analysis.
Conflict Of Interest
None.
Human Subject Protections
Northwestern University IRB #: STU00027497 and Marshfield Clinic Research Foundation IRB #: MCC21310PM-C
Funding
The eMERGE Network was initiated and funded by the National Human Genome Research Institute and includes the following grants: U01HG006389 (Essentia Institute of Rural Health); U01HG006388 (Northwestern University). This work was also funded in part by a grant from the National Center for Research Resources, UL1RR025741.
Data Sharing Statement
Northwestern University’s EHR Height Cleaning algorithm is freely available for non-commercial use and modification. Source code and instructions may be downloaded from: http://tinyurl.com/EHR-Height-Cleaning
References
- 1.Noel PH, Copeland LA, Perrin RA, Lancaster AE, Pugh MJ, Wang CP, Bollinger MJ, Hazuda HP. VHA Corporate Data Warehouse height and weight data: opportunities and challenges for health services research. Journal of rehabilitation research and development 2010; 47(8): 739–750. PubMed PMID: 21141302. Epub 2010/12/15. eng. [DOI] [PubMed] [Google Scholar]
- 2.Lipman TH, McGinley A, Hughes J, Minakami J, Layden VM, Ratcliffe S, Hench K. Evaluation of the accuracy of height assessment of premenopausal and menopausal women. J Obstet Gynecol Neonatal Nurs 2006; 35(4): 516–22. PubMed PMID: 16881996. Epub 2006/08/03. [DOI] [PubMed] [Google Scholar]
- 3.Sorkin JD, Muller DC, Andres R. Longitudinal change in the heights of men and women: consequential effects on body mass index. Epidemiologic reviews 1999; 21(2): 247–260. PubMed PMID: 10682261. Epub 2000/02/22. [DOI] [PubMed] [Google Scholar]
- 4.Joss EE, Temperli R, Mullis PE. Adult height in constitutionally tall stature: accuracy of five different height prediction methods. Arch Dis Child 1992; 67(11): 1357–1362. PubMed PMID: 1471886. Pubmed Central PMCID: 1793786. Epub 1992/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siminoski K, Warshawski RS, Jen H, Lee K. The accuracy of historical height loss for the detection of vertebral fractures in postmenopausal women. Osteoporosis international : a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA 2006; 17(2): 290–296. PubMed PMID: 16143833. Epub 2005/09/07. [DOI] [PubMed] [Google Scholar]
- 6.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011; 3(79): 79re1 PubMed PMID: 21508311. Epub 2011/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, Basford M, Chute CG, Kullo IJ, Li R, Pacheco JA, Rasmussen LV, Spangler L, Denny JC. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013. PubMed PMID: 23531748. Epub 2013/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, Denny JC, Peissig PL, Miller AW, Wei WQ, Bielinski SJ, Chute CG, Leibson CL, Jarvik GP, Crosslin DR, Carlson CS, Newton KM, Wolf WA, Chisholm RL, Lowe WL. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012; 19(2): 212–218. PubMed PMID: 22101970. Pubmed Central PMCID: 3277617. Epub 2011/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, den Heijer M, Franke B, Verbeek AL, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K. Many sequence variants affecting diversity of adult human height. Nat Genet 2008; 40(5): 609–615. PubMed PMID: 18391951. [DOI] [PubMed] [Google Scholar]
- 10.Hellerstein JM. Quantitative data cleaning for large databases. United Nations Economic Commission for Europe (UNECE) 2008. [Google Scholar]
- 11.Lyratzopoulos G, Heller RF, Hanily M, Lewis PS. Risk factor measurement quality in primary care routine data was variable but nondifferential between individuals. J Clin Epidemiol 2008; 61(3): 261–267. PubMed PMID: 18226749. Epub 2008/01/30. [DOI] [PubMed] [Google Scholar]
- 12.Green BB, Anderson ML, Cook AJ, Catz S, Fishman PA, McClure JB, Reid R. Using body mass index data in the electronic health record to calculate cardiovascular risk. Am J Prev Med 2012; 42(4): 342–347. PubMed PMID: 22424246. Pubmed Central PMCID: 3308122. Epub 2012/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoong SL, Carey ML, D’Este C, Sanson-Fisher RW. Agreement between self-reported and measured weight and height collected in general practice patients: a prospective study. BMC medical research methodology 2013; 13: 38 PubMed PMID: 23510189. Pubmed Central PMCID: 3599990. Epub 2013/03/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stommel M, Schoenborn CA. Accuracy and usefulness of BMI measures based on self-reported weight and height: findings from the NHANES & NHIS 2001-2006. BMC public health 2009; 9: 421 PubMed PMID: 19922675. Pubmed Central PMCID: 2784464. Epub 2009/11/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor Gorber S, Tremblay M, Moher D, Gorber B. A comparison of direct vs. self-report measures for assessing height, weight and body mass index: a systematic review. Obes Rev 2007; 8(4): 307–326. PubMed PMID: 17578381. Epub 2007/06/21. [DOI] [PubMed] [Google Scholar]


