Abstract
Half the world's population is chronically infected with Helicobacter pylori1, causing gastritis, ulcers and increased incidence of gastric adenocarcinoma2. Its proton-gated inner-membrane urea channel, HpUreI, is essential for survival in the acidic environment of the stomach3. The channel is closed at neutral pH and opens at acidic pH to allow rapid urea access to cytoplasmic urease4. Urease produces NH3 and CO2 that neutralize entering protons and thus buffer the periplasm to pH ∼6.1 even in gastric juice at pH <2.0. Here we report the structure of HpUreI, revealing six protomers assembled in a hexameric ring surrounding a central bilayer plug of ordered lipids. Each protomer encloses a channel formed by a twisted bundle of six transmembrane helices. The bundle defines a novel fold comprising a two-helix hairpin motif repeated three times around the central axis of the channel, without the inverted repeat of mammalian urea transporters. Both the channel and the protomer interface contain residues conserved in the AmiS/UreI superfamily, suggesting preservation of channel architecture and oligomeric state in this superfamily. Predominantly aromatic or aliphatic side chains line the entire channel and define two consecutive constriction sites in the middle of the channel. Mutation of Trp153 in the cytoplasmic constriction site to Ala or Phe reduces the selectivity for urea compared to thiourea, suggesting that solute interaction with Trp153 contributes specificity. The novel hexameric channel structure described here provides a new paradigm for permeation of urea and other small amide solutes in prokaryotes and archaea.
Treatment of Helicobacter pylori infection is becoming less effective because of antibiotic resistance, suggesting that a specifically targeted approach to eradicate this organism would be beneficial5. Colonization of the acidic mammalian stomach by H. pylori depends on the presence of the inner membrane protein HpUreI, making it a viable clinical target3. HpUreI is a proton-gated urea channel closed at pH 7.0 and fully open at pH 5.0, enabling rapid entry of urea into the bacterium4. Urease activity (Supplementary Fig. 1) buffers the periplasm to pH 6.1, essential for survival of H. pylori at acidic pH6,7 due to the neutralizing capacity of NH3 and generation of HCO3- by periplasmic α-carbonic anhydrase. Conversely, the closure of HpUreI at neutral pH prevents over-alkalization in the presence of millimolar urea and consequent cell death8.
The structure of HpUreI was determined using multi-wavelength anomalous dispersion (Methods). The structure shows an arrangement of six protomers that form a compact hexameric ring (Fig. 1) about 95 Å in diameter and 45 Å in height. The center of the hexamer is filled with an ordered lipid plug that forms an asymmetric bilayer with electron density for six lipid tails in the periplasmic leaflet and for 18 tails in the cytoplasmic leaflet. This central lipid plug is reminiscent of those reported for other membrane protein oligomers such as bacteriorhodopsin9. The main inter-protomer contacts are between TMH1 and TMH2 of one protomer and TMH3 and TMH4 of a neighboring protomer, a region with conserved residues that are likely important for assembly (Fig. 2a). Native gel electrophoresis of HpUreI confirms that it is a hexamer (Supplementary Fig. 4) in contrast to the previously postulated trimer10. An electron cryomicroscopy 9 Å projection map of a Bacillus cereus amide channel, also a member of the AmiS/UreI family, revealed a similar hexameric arrangement11. Thus, the hexamer likely represents the physiological state rather than a crystallization artifact.
Figure 1. The Hp UreI urea channel hexamer.
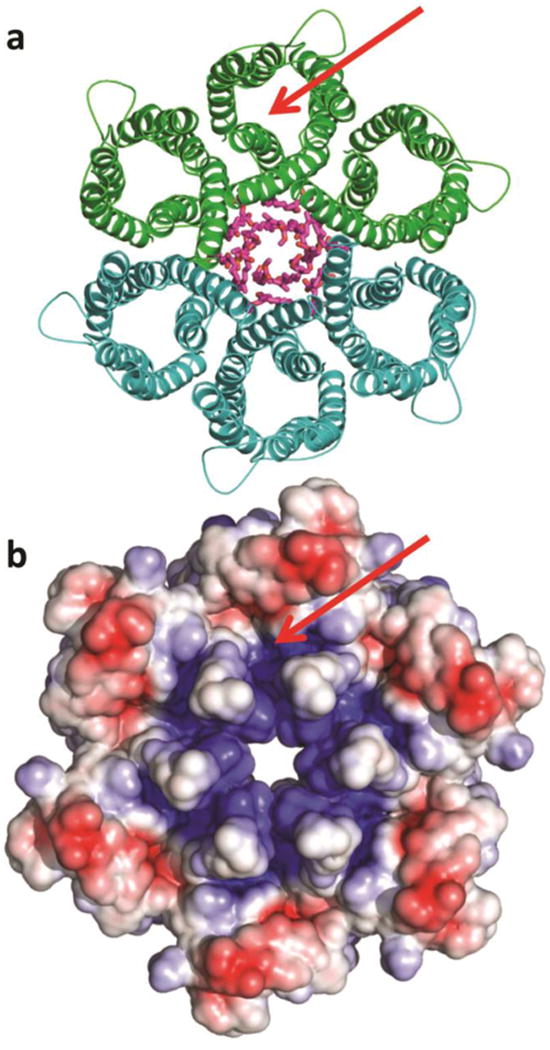
a, Ribbon diagram of hexamer surrounding the lipids of the central bilayer (purple sticks). The C6 hexamer is generated from the three protomers of one asymmetric unit (green) by the crystallographic two-fold axis (second asymmetric unit in teal). b, Electrostatic potential at the periplasmic hexamer surface computed at pH 5.3, the pH at which the crystals were grown (red: -4 kT/e, blue: +4 kT/e). The electrostatic potential was calculated with the program APBSmem22. The green arrow pinpoints the entrance to one of six urea channels.
Figure 2. Residues lining the channel.
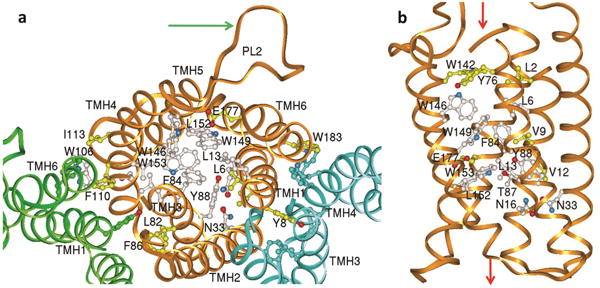
Residues conserved in the AmiS/UreI superfamily (aligned in Supplementary Fig. 11) are clustered in the channel and at the protomer interface. a, View from periplasm showing the conserved residues of protomer A (gold ribbon), and open channel of protomers B & C (green and blue ribbons) when only the protein backbone is shown. For clarity only selected side chains are labeled. Residues conserved in all members of the AmiS/UreI superfamily are white and additional residues conserved in the subset of known urea channels (including HpUreI) are yellow. The arrow points to the proton-sensing periplasmic loop 2 (PL2) of protomer A. b, View parallel to membrane with fully conserved residues (white) clustering in the middle of the channel pore and residues additionally conserved in the urea channels in yellow. The green arrows show the regions of urea entry (top, periplasmic side) and exit (bottom, cytoplasmic side).
Sequence analysis had predicted six transmembrane segments with periplasmic location of both N and C termini, generating two periplasmic loops (PL), one between transmembrane helix (TMH) 2 and TMH3 (PL1) and another between TMH4/TMH5 (PL2), as well as a short periplasmic C-terminal segment4. The crystal structure shows that each HpUreI protomer is a twisted bundle of six slightly tilted transmembrane helices whose inward-facing side chains define a central channel with a unique arrangement of residues (Fig. 2). Each protomer exhibits three-fold pseudosymmetry as evidenced by the high structural similarity after 120° and 240° rotations around an axis through the center of the channel (Fig. 3). The repeating motif is a helical hairpin composed of a pair of antiparallel TMHs connected by a short cytoplasmic loop. However, closer helix-helix packing is observed between helices from adjacent repeated motifs, namely, TMH2/TMH3, TMH4/TMH5, and TMH1/TMH6 (Fig. 2a). The mitochondrial ADP/ATP exchanger has a clear internal 3-fold tandem repeat of a sequence motif in its amino acid sequence12, whereas the 3-fold pseudosymmetry of HpUreI became apparent only after the crystal structure had been determined. Sequence alignments suggest that this motif is present in eubacteria and archaea (Supplementary Fig. 11), implying an ancient evolutionary origin.
Figure 3. Structural conservation of a 2-helical hairpin motif.

The internal 3-fold axis of pseudosymmetry is oriented perpendicular to the bilayer (black line) and is the result of a three-fold tandem non-inverted repeat of a helical hairpin motif (TMH pairs 1&2, 3&4 and 5&6). In this side view one HpUreI protomer (gold ribbon) has been overlaid on itself by rotations of 120° (green ribbon) and 240° (white ribbon) around the 3-fold axis through the center of the channel pore. The root mean square deviations for pairwise overlays of the backbone atoms are 1.52 Å for TMH1&2 onto TMH3&4; 1.13 Å for TMH3&4 onto TMH5&6; 1.76 Å for TMH5&6 onto TMH1&2, and 1.98 Å when all are superimposed simultaneously. Structural conservation is stronger for the cytoplasmic half of the channel. Analyzed with the program SymD23.
The channel is lined by conserved residues that identify the solute pathway in the AmiS/UreI channel superfamily and specifically for urea in HpUreI. The beginning of TMH1 is recessed within the helix bundle and starts with Met1 whose ordered side chain was crucial for selenomethionine phasing. TMH1 contains three of the eleven channel residues that are conserved in the AmiS/UreI superfamily (Fig. 2 and Supplementary Fig. 11), all facing inwards where they define one side of the channel. The longest helix, TMH2, is near the lipid-filled center, set back from the channel of the hexamer outside the urea pathway, with no conserved residues. This helix starts more than two helix turns earlier than predicted4. Upon reaching the periplasmic side, TMH2 continues for several helical turns until the disordered section of PL1, which contains the engineered 6-His tag. At this point, TMH2 reaches a height above the bilayer that is similar to the height of PL2. TMH3 is situated inwards of TMH2 and is a major contributor to the urea pathway with four conserved residues in the channel, including Phe84 and Tyr88, which form part of two constriction sites in the channel (Figs. 2 and 4). TMH3 contacts TMH2 of the same protomer as well as the cytoplasmic half of TMH2 from a neighboring protomer. TMH4, although part of the repeating structural hairpin motif with TMH3, barely contacts TMH3 and instead is in close contact with TMH5. TMH4 together with PL2, TMH5, CL3 and TMH6 constitute the outer, bilayer-facing edge of the HpUreI hexamer. TMH5 contains five tryptophans, three of which are highly conserved (Trp146, Trp149 and Trp153). These line the urea path, with the aromatic side chains of Trp149 and Trp153 being major components of the two constriction sites (Figs. 2b and 4). In the middle of the membrane, the side chain of Glu177 of TMH6 is in a hydrophobic environment and is thus predicted to have an elevated pKa of 6.7 and to be protonated at pH 5.3, the pH of crystallization. This residue is conserved only in the UreI family of AmiS/UreI members (Supplementary Fig. 12), implying a role in urea transport. It is located to the side of the channel and in position to hydrogen bond to urea. Beyond the C-terminal end of TMH6, the last five residues of HpUreI are not helical but bend back towards PL2, where the C-terminal segment is tucked under PL2.
Figure 4. Views of the HpUreI channel traversed by urea.

a,Side view with periplasm on top, showing the shape of channel in yellow and residues conserved in the UreI family (Supplementary Fig. 12). Constriction site 1 is largely due to the side chains of Leu6, Phe84 and Trp149. On the other side of Glu177, constriction site 2 is defined by the side chains of Leu13, Thr87, Tyr88, Leu152 and Trp153. TMH1 and TMH2 have been removed for clarity. Blue arrows show the positions of cross sections for panel b. b, Close-ups of constriction sites 1 and 2 viewed from the periplasm. c, Channel activity of mutants: mutating Trp153 to Ala or Phe (constriction site 2) changes selectivity from 4.2:1 (urea:thiourea) for the wild type to 1.5:1 and 1.2:1 for the Ala or Phe mutants, respectively. Mutations at Tyr88 and Trp149 inactivate or severely impair the channel whereas mutation Phe64Leu retains discrimination (n=10-20, error bars ± s.e.m.). d, Western analysis showing nearly equivalent expression for the different mutants.
There is likely no high-affinity urea-binding site in the channel since there is no saturation up to 100 mM urea when expressed in oocytes4 and the half-saturation concentration in proteoliposomes was reported to be 163 mM10. Although neither structurally nor mechanistically related, DvUT, a homolog of the mammalian urea transporters, also shows half-saturation concentrations above 100 mM urea13. However, it is constructed entirely differently, featuring a long (16 Å), narrow hydrophobic channel with inverted (point) symmetry also found in the mammalian urea transporter, UT-B14. Both of these contain phenylalanine residues at the entry and exit points of the channel.
HpUreI has much larger periplasmic loops than other members of the AmiS/UreI superfamily and proton-gated closure of the channel, affected by mutations of histidines or carboxylates in the periplasmic domain16, could be due to loop occlusion of the periplasmic vestibule. A similar effect has been shown for the FocA formate channel15, but here histidine protonation closes, rather than opens the channel. For HpUreI at pH 5.3, PL1 is disordered due to its engineered 6-His insertion and PL2 is oriented away from the channel (Fig. 1a), thus suggesting an open conformation for unimpeded periplasmic entry of urea into the periplasmic vestibule. Urea enters through this irregularly-shaped vestibule, which begins roughly at the height of the bilayer edge, defined by a set of largely hydrophobic side chains including Leu2, Tyr76, Trp142, and Trp146 (Fig. 2b). Next on its way to the cytoplasm, urea must pass through two constrictions on either side of Glu177. Constriction site 1 on the periplasmic side of Glu177 is defined by the side chains of conserved Leu6, Phe84 and Trp149 (Figs. 2b & 4b). Just beyond Glu177 is constriction site 2, defined by the side chains of conserved Leu13, Thr87, Tyr88, Leu152, and Trp153 (Figs. 2b & 4b). Trp153 has its aromatic plane oriented perpendicular to the channel axis, restricting the passage of solutes and is therefore a candidate for a functional mutation (Supplementary Fig. 8). Its indole side chain can reorient about χ2 due to minimal steric restraints, enabling passage of urea and other amides. Similar flexibility of other aromatic side chains in the channel would allow transient π–π stacking of their side-chain surfaces or NH2-π electrostatic interaction with planar urea to permit selective passage, while at the same time preventing acidification of the cytoplasm by blocking the passage of protons/hydronium. The constrictions in the middle third of the protomer likely represent the selectivity filter which discriminates tenfold between urea and thiourea in oocyte studies4. On the cytoplasmic side of constriction site 2, the channel widens again, with Tyr104 and the hydrogen-bonded pair of Asn16 and Asn33 at the beginning of a funnel-shaped vestibule, allowing urea exit into the cytoplasm.
Previously, mutations of protonatable residues in the periplasmic domain identified residues important for proton sensing or gating16. Here, we tested the effect of mutations Phe84Leu and Trp149Phe in the periplasmic constriction site 1 (Fig. 4b) and Tyr88Phe and Trp153Ala/Phe in the cytoplasmic constriction site 2 (Fig. 4b) on the discrimination between urea and thiourea (Fig. 4c), a property common to HpUreI, DvUT and UT-B13,14,16. The rate of urea or thiourea uptake was measured in Xenopus oocytes. Western analysis confirmed equal expression levels of mutant and wild-type proteins (Fig. 4d). Phe84Leu retained the discrimination but with reduced uptake. In the cytoplasmic constriction site, Trp153Ala and Trp153Phe retained urea transport but lost much of the discrimination between urea and thiourea (Fig. 4c). The selectivity for urea over thiourea is therefore largely determined by Trp153 in the cytoplasmic constriction site 2 of the channel. The buffering capacity generated by proton activation of HpUreI coupled with urea hydrolysis was estimated based on purified HpUreI (Supplemental Information).
Channel opening also correlates with acid-dependent recruitment of urease to the cytoplasmic side of the membrane, dependent on HpUreI and essential for survival at pH 3.020. About two thirds of the urease is recruited to the plasma membrane based on post-sectioning immuno-electron microscopy at pH 5.019, confirmed by SDS gel analysis of the membrane fraction of H. pylori that also demonstrates activation of the membrane-recruited urease20. Measurement of cytoplasmic pH changes in wild type and ureI deletion mutants also showed that HpUreI increases bilayer permeability of CO2 and NH3/NH4+, providing a physiological role for this association by allowing rapid backflow of these molecules to the periplasm20, thus increasing the rate of periplasmic buffering. These data suggest that a conformational change in the membrane domain of HpUreI may occur with acidification that is transmitted to its cytoplasmic surface.
In contrast to other neutral solute membrane channels, such as aquaporins21 and mammalian urea transporters14, which are constructed of two homologous antiparallel halves that generate a long, narrow channel, HpUreI's parallel three-fold pseudo-symmetric architecture defines a novel shorter hour-glass shaped channel with a central urea filter and no inverted symmetry. Thus, although both HpUreI and mammalian urea transporters homologous to DvUT selectively allow passage of urea, the three-dimensional structures and mechanisms of selectivity are clearly distinct.
This first three-dimensional structure of a channel from the AmiS/UreI superfamily reveals a novel fold with a unique channel architecture able to selectively filter polar organic solutes. As the structure of a validated target for H. pylori eradication it may guide discovery of small molecule inhibitors, providing the possibility of monotherapy without the use of conventional antibiotics.
Supplementary Methods
Engineering a 6His tag into HpUreI
DNA encoding HpUreI was isolated by PCR from Helicobacter pylori strain J99. A 6His tag was introduced into the protein at various locations to facilitate purification. The engineered proteins with a 6His tag at the N-terminus, in the first periplasmic loop (PL1), the second periplasmic loop (PL2) or at the C terminus were expressed in Xenopus oocytes and tested for channel activity. Wild-type HpUreI showed urea uptake of 11.55 +/-0.33 pmol/oocyte/6min (n=3) and the PL1 6His tag (after Gly61) showed an uptake of 3.0 +/-0.5 pmol/oocyte/6min (n=3). Mutants with the 6His insertion at the N terminus, in PL2 or at the C terminus were inactive. HpUreI with the 6His tag inserted in PL1 (HpUreI6HisPL1) was subsequently used for expression, purification, and crystallization.
HpUreI expression and membrane isolation
Small-scale expression
pET101UreI6HisPL1 was transformed into E. coli C43 (Avidis S.A.). For small-scale expression and crude membrane isolation bacterial cultures were grown to OD600 0.8 and then induced by addition of 1 mM IPTG. After 3 hours of induction, cells were harvested by centrifugation at 3000 rpm for 10 min. The pellet was resuspended in a solution of 50 mM Na2HPO4 pH 7.4, 1 mM EDTA, 30 μg/ml DNase I and sonicated to lyse the cells. Cell debris was removed by centrifugation (10,000 g, 10 min) and membranes were collected (100,000 g 45 min) and resuspended in the same buffer without DNase I (40-50 μl). Samples of membrane protein (25 μg, determined by BCA assay, Pierce Biotech, Inc.) were dissolved in gel loading buffer containing 1% mercaptoethanol and run without boiling on 4-12% SDS-polyacrylamide gels. After transfer to nitrocellulose, Western blot analysis was performed with either an anti-UreI or anti-His-tag antibody (GE).
Large-scale membrane expression for crystallization trials
A bioreactor (BioFlo 110, New Brunswick) containing 10 l Luria Bertani broth supplemented with 50 mM K2HPO4 pH7.8 and 1.5% w/v glycerol was inoculated with 0.2 l overnight culture of the E. coli harboring a pET101HpUreI6HisPL1 plasmid. When the OD600 reached 0.8-1.0 (∼2 hr), HpUreI expression was induced with 1 mM IPTG. Growth was maintained at 37 °C, with a continuous air supply at 5 liter/minute and 500 rpm stirring for 5 hours until the cell density reached 5-7 OD600. Cells were then collected by centrifugation and resuspended at 4 °C in 400 ml of buffer containing 50 mM Na2HPO4 pH 7.4, 30 μg/ml DNase and 10 mM mercaptoethanol. All subsequent steps were carried out at 4 °C. The bacterial suspension was passed twice through a French Press cell at 20,000 psi. The cell debris was removed (10,000 g, 20 min) and the remaining membranes were collected (100,000 g, 2 hr) to yield ∼4 grams of total membrane protein. The membrane pellet was resuspended in 350 ml of storage buffer (10 mM imidazole, 150 mM NaCl, 50 mM Na2HPO4 pH 7.4, 65 g/l glycerol and 2 mM mercaptoethanol) to yield 10-12 mg/ml of membrane protein which was then flash frozen in liquid nitrogen and stored at -80 °C.
Selenomethionine labeling
The pET101UreI6HisPL1 construct was transformed into the methionine auxotroph strain BL21-CodonPlus-RP-X strain (Stratagene) for optimal labeling of HpUreI with selenomethionine in minimal medium. A 10 L culture of M9 minimal media (60 g/L Na2HPO4, 30 g/L KH2PO4, 10 g/L NH4Cl, 5 g/L NaCl, 0.4% w/v glucose, 0.1 mM CaCl2, 2 mM MgSO4, and 0.1 L of MEM vitamin solution (Mediatech) supplemented with 60 μg/ml selenomethionine, was inoculated with 200 ml overnight culture grown in the same minimal medium with 0.1 mg/ml methionine in place of selenomethionine. Protein expression was induced with 1 mM IPTG at OD600 ∼0.8 and growth was continued for 40 h at 20 °C.
HpUreI purification
Solubilization and Metal Affinity Chromatography
Membranes in storage buffer were collected at 100,000 g for 90 min, resuspended at 10 mg/ml in a buffer of 10 mM imidazole, 150 mM NaCl, 50 mM sodium phosphate, pH 7.4, and partially dissolved in 2% decylmaltoside (DM, Anatrace) by adding a 20% detergent solution drop wise and stirring on ice for 30 min. The insoluble material was pelleted (160,000 g, 30 min) and the supernate was gently agitated overnight with 5 ml of cobalt-agarose resin (TALON resin, Clontech) at 4 °C to bind the 6His-UreI. All the solutions used in subsequent rinsing and elution of the resin contained 150 mM NaCl, 50 mM sodium phosphate, pH 7.4, 0.2% DM and 0.1 mg/ml E. coli polar lipids (Avanti) with increasing concentrations of imidazole. The resin was rinsed with 10 volumes of buffer with 10 mM imidazole, then 30 volumes with 20 mM imidazole. HpUreI was then eluted with 30 ml of the same buffer with 250 mM imidazole. The eluted HpUreI was concentrated to ∼10 mg/ml by using Amicon 50 kDa filters prior to gel filtration.
Size exclusion
HpUreI was purified on a Superose12 column (10/300, GE) with buffer containing 10 mM MES pH 6.5, 150 mM NaCl; 0.2% DM; 0.1 mg/ml E. coli polar lipid extract. The peak fractions were pooled and concentrated to 10 mg/ml (50 kDa filters, Amicon) for use in crystallization trials.
HpUreI crystallization
HpUreI protein at 10 mg/ml in Superose12 column buffer was diluted to give a solution comprised of 1.57 mg/ml HpUreI protein, 40 mM NaCl, 1 mM TlCl, 10 mM CaCl2, 7% PEG 400, 0.05% decylmaltoside, 2.25% octylglucoside, 0.8 mg/ml E. coli polar lipids (Avanti), 35 mM MES pH 5.3. This mixture (3.5 μl) was used for hanging drop diffusion over a reservoir (0.5 ml) of 20% PEG 400 in 0.1 M MES, pH 5.3. Crystals grew in 3-4 months at 11 °C and were dehydrated by raising the PEG 400 concentration in the reservoir in increments of 3% at two-day intervals until the final concentration in the well solution was 33% PEG 400. The crystal used for collection of the native dataset is shown in Supplementary Fig. 1.
X-ray data collection & data reduction
Single crystals were mounted in nylon loops and flash cooled in liquid nitrogen. X-ray diffraction data collection was carried out at 100 K by collecting 180 diffraction images 1° in width. Data were integrated, scaled & merged with the program XDS24 (Supplementary Tab. 1). Due to the large variability of the c-axis length (135 to 152 Å), data could not be merged between multiple crystals, making experimental phasing more challenging. After diffraction data from hundreds of heavy metal-soaked crystals had been collected without yielding interpretable maps, we resorted to selenomethionine (SeMet) phasing, despite the low abundance of methionine in HpUreI J99 (three residues out of 195, counting the N-terminal methionine). While fluorescence scans of the initially very small SeMet crystals indicated significant selenium incorporation, selenium sites could not be located in over 50 multi-wavelength anomalous dispersion (MAD) and single-wavelength anomalous dispersion (SAD) datasets collected with 30° wedges.
The successful MAD data set was collected from a rare larger crystal that took over five months to grow. Three-wavelength MAD diffraction data were collected in 10° wedges at beamline 12-2 at SSRL (Supplementary Tab. 1).
SeMet multi-wavelength anomalous dispersion (MAD) phasing
The program SHELXC25 was used to determine the approximate resolution cutoff for the anomalous signal (when the correlation coefficient between the anomalous differences at the peak and remote wavelengths decreased to below 30%). SeMet sites were initially obtained using the program SHELXD25. Of the nine possible sites (three protomers in the asymmetric unit with three SeMet sites per protomer, including all three N-terminal SeMet residues), eight sites were located. From the symmetry of these eight sites, a ninth site could be located, leading to the localization of all selenium anomalous scatterers. The boundary of the HpUreI hexamer and individual helices were readily apparent in the resulting low-resolution maps, but not surprisingly, detail of the maps and phasing power was poor. To improve the phases, the program autoSHARP26 was used. The figure of merit (FOM) from autoSHARP phasing was used to determine the high-resolution cutoff for the experimental phases. A cutoff of 0.3 for the FOM of acentric reflections was used for this estimation, suggesting useful phases to about 4.7 Å resolution (Supplementary Tab. 2).
Phase improvement & extension through solvent flattening, NCS and multi-crystal averaging
In order to improve & extend the initial experimental MAD phases, solvent flattening, 3-fold non-crystallographic symmetry (NCS) and multi-crystal averaging of data from crystals with different c-axis lengths were carried out. To obtain initial matrices for NCS averaging, 18 ideal helices were modeled into the experimental electron density representing the three molecules in the asymmetric unit. Subsequently the NCS matrices from molecule A to molecule B and from molecule A to molecule C were obtained by the SSM superpose function of the program COOT26. A mask was placed around the electron density of molecule A and 3-fold NCS averaging and solvent flattening (68.9% solvent) were performed with the program DM27, while NCS matrices were refined. Phases were extended in an iterative fashion from 4.5 to 3.5 Å using small resolution increments with the programs DM and DMMulti28. At this point the right-handed twist of the helices and density for some of the larger side chains could be seen.
Model building & refinement
Density representing each of the six transmembrane helices was cut out using the program PHENIX29. An ideal helix was built into this density, slightly curved if required, using the sequence representing each helix. Helix 6 contains a pi-bulge and was built by hand using the program COOT. Cytoplasmic loops 1 & 2 (CL1 and CL2) and periplasmic loop 2 (PL2) were initially modeled into density using the program RAPPER30. In places where the correct sequence could not be built initially, alanine was used temporarily. In order to generate the other two molecules of the asymmetric unit, the previously identified NCS matrices were used followed by rigid body refinement with the program PHENIX. The model was refined iteratively by cycles of manual adjustments using the program COOT and refinement with the program PHENIX, using TLS refinement with NCS and secondary structure restraints.
Upon analyzing the data with the UCLA anisotropy server31, the data were found to be severely anisotropic with an effective resolution of 3.1 Å in the a* and b* directions, but only 3.5 Å in the c* direction. The data were ellipsoidally truncated and rescaled to minimize inclusion of poor diffraction data. The model was refined with the newly truncated data using jelly body refinement with the program REFMAC32, leading to significantly improved electron density maps which allowed further model improvement. Because of disorder, no model was built for the majority of periplasmic loop 1 (PL1, residues 59 to 73), which contains the engineered 6His insertion.
A final round of refinement was carried out with the program REFMAC against the non-truncated data with two TLS groups in each protomer (residues 1-146 and 147-195), tight NCS restraints, and a jelly body value of 0.01. The final structure was refined to 3.26 Å with few Ramachandran outliers, small deviations from ideal geometry and predominantly preferred side chain rotamers (Supplementary Tab. 1).
SeMet anomalous difference maps
To validate the sequence assignment of the HpUreI crystal structure, additional methionine residues were engineered into various sites in the HpUreI sequence and labeled with SeMet. These proteins were crystallized and their anomalous differences Fourier maps inspected. For all five engineered sites, the location of the anomalous difference peak was less than 1 Å from the site of the methionine sulfur atom of the final model. The sites were Ile14, Ala148, Thr155, Leu173 and Ile191, in addition to the endogenous sites Met1, Met14 and Met127, the latter one being situated in periplasmic loop 2 (PL2).
Estimation of the number of HpUreI molecules per bacterial cell by Western blotting
Bacterial proteins were size-fractionated by sodium dodecyl sulfate (SDS)-tricine polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane followed by immunodetection by enhanced chemiluminescence (Amersham). The HpUreI antibody used for detection and quantitation was generated in rabbit against periplasmic loop 1 between TMH2 and TMH3 ( CEGAEDIAQVSHHLTSFYGPATG)33. Immunoblots were digitized (Micrcotek Scanmaker i800) at a resolution of 600 dpi. The scanned images were analyzed using Kodak 1D software and the amount of HpUreI was quantified using purified HpUreI as the reference. There are 5.3 ng of HpUreI per μg of total protein (Supplementary Fig. 2) in 5.4×106 cells (measured by colony forming units). The molecular weight of HpUreI is 21.7 kDa, thus there are 0.24 pmoles of HpUreI per 5.4×106 cells. 0.24 pmoles of HpUreI × 6.0×1023 / 5.4×106 cells is equivalent to approximately 27,000 channels per cell.
Urease activity assay
Bacteria grown on trypticase soy agar (TSA) plates were suspended in 1 ml of 1 mM phosphate buffer to a final concentration of OD 0.01 at 600 nm. Urease activity was measured radiometrically. Bacteria were added to 100 mM sodium phosphate buffer containing 5 mM KCl, 138 mM NaCl, 0.5 mM MgCl2, 1 mM CaCl2, 10 mM glucose and 5 mM 14C-urea with a specific activity of 10 μCi/μmol. To measure activity in the closed and open state, the pH of the buffer was set to pH 4.5 and 7.4 by mixing varying amounts of 100 mM monobasic sodium phosphate and 100 mM dibasic sodium phosphate to the desired pH. The pH of the buffer during the course of the experiment did not change by more than 0.1 pH units. Plastic wells containing 500 mM KOH-soaked filter paper hung from rubber stoppers were used to collect the total 14CO2 from the hydrolysis of urea. Urease activity was measured for 30 minutes at 37 °C with constant agitation. The reaction was terminated with 5 N H2SO4 to release all 14C as CO2 and incubated for 30 minutes at 37 °C. The wells were placed in scintillation cocktail (HiIonicFluor; Packard Instruments), and the radioactivity was measured by scintillation counting (1216 RackBeta; LKB Instruments). Protein concentration was determined by the BCA method (Pierce). Urease activity was ∼5 μmoles/min/mg, sufficient to hydrolyze all entering urea33.
Urea flux estimation
To estimate the urea flux per channel per second, the total urease activity of H. pylori cells was measured under HpUreI-mediated flux conditions. Assuming that all urea entering the cell is hydrolyzed by the large amount of cytoplasmic urease (∼8-10% of total protein) and knowing the number of HpUreI molecules per cell (27,000, see above), the lower bound of the flux was estimated to be ∼200 urea molecules per HpUreI channel per second at 5 mM medium urea calculated from the rate of urea hydrolysis and the number of channels per organism.
This is a tenfold higher influx of urea than that due to unfacilitated diffusion across phospholipid bilayers (4×10-6 sec-1)18 and agrees with the tenfold increase in urease activity observed when HpUreI is open. This calculation does not take into account the ∼fourfold increase in urease activity when incubated in acidic pH20 hence the rate could approximate 1,000 sec-1.
NH3 production due to urea influx through HpUreI
Using the calculated flux rate and the number of channels per cell from above it was estimated that at least 5.4×106 urea molecules are transported through HpUreI / cell / sec at gastric urea concentrations. At pH 5.0, with HpUreI fully open, this can generate about 30 mM NH3 per second neutralizing capacity in a bacterium with a volume of 0.6 fl.
Blue native gel electrophoresis of HpUreI
E. coli membranes were treated with 2% DDM and the soluble fraction was bound to TALON resin. After washes with buffer containing 20 mM imidazole, HpUreI was eluted with a buffer containing 150 mM imidazole, 50 mM sodium phosphate pH 7.4, 150 mM NaCl and 0.01% DDM. The eluate was concentrated on Amicon filters with 10 kDa cutoff to ∼5 mg/ml. 5 μl of the protein sample was mixed with 2× sample buffer (100 mM Tricine, 30 mM Bis-Tris pH 7.0, 30% glycerol and 0.02% DDM). The cathode buffer was 50 mM Tricine, 15 mM Bis-Tris pH 7.0 and the anode buffer was 50 mM Bis-Tris pH 7.0. The electrophoretic shift of proteins was achieved by including Coomassie Blue G250 dye in the cathode buffer. The running conditions were as following: initial concentration of Coomassie Blue G250 was 0.02% in the cathode buffer and the gel was run for 30 min at 100 V and an additional 2 hours at 200 V. The concentration of dye was then decreased to 0.002% and the gel was run for an additional 2 hours at 300 V. High molecular weight markers (Amersham, catalog number 17-0445-01), bovine serum albumin and trypsin inhibitor (Sigma) were mixed with the same sample buffer (Supplementary Fig. 3).
Solute uptake experiments in Xenopus laevis oocytes
Genes encoding wild-type HpUreI (H. pylori strain 43504) and 6His-tagged HpUreI (H. pylori strain J99 with the 6His-tag in PL1) were cloned into the pcDNA3.1 (Invitrogen) and pET101 (Invitrogen) vectors, respectively. For the experiments on site-specific mutants, substitutions were introduced into gene encoding 6His-tagged HpUreI using the QuickChange method (Stratagene). Capped and polyA-tailed RNA (cRNA) was prepared using the mMessage mMachine T7 Ultra Kit (Ambion). 50 nl of cRNA (1 μg μl-1) were injected into each oocyte. Oocytes were incubated for 2 days at 18 °C in Barth's solution (88 mM NaCl, 2.4 mM NaHCO3, 1 mM KCl, 0.4 mM CaCl2, 0.3 mM Ca(NO3)2, 0.8 mM MgSO4 and 10 mM HEPES-Tris pH 7.5). Oocytes were transferred to new vials containing 0.5 ml of Ringer's solution (100 mM NaCl, 2 mM KCl, 1 mM CaCl2 and MgCl2) buffered by 20 mM MES at pH 5.0 for comparison between urea and thiourea uptake through the open HpUreI channel. The transport reaction was started by adding 100 μM [14C]-urea or 100 μM [14C]-thiourea. After 10 minutes of incubation at room temperature the solution was aspirated to terminate transport and the oocytes were washed twice with 10 volumes of ice-cold radioisotope-free Barth's solution. Oocytes were transferred to individual scintillation vials and dissolved in 10% SDS prior to adding scintillation cocktail.
Western analysis of mutant expression levels
Ten oocytes were solubilized by pipetting into buffer containing 0.5 ml of 0.1 M NaCl, 0.02 M Tris HCl pH 7.6 and 1% Triton X100. The yolk was spun down by centrifugation at 14,000 rpm for 5 min and the supernatant was aspirated with care in order to minimize contamination with floating lipid. Twenty microliters of supernatant were size-fractionated by sodium dodecyl sulfate (SDS)-tricine polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane followed by immunodetection by alkaline phosphatase (Promega). The HpUreI antibody used for detection was generated in rabbit against periplasmic loop 1 between TMH2 and TMH3 (CEGAEDIAQVSHHLTSFYGPATG). All mutants were expressed at similar levels (Fig. 4d).
Supplementary Material
Acknowledgments
We are grateful for assistance with x-ray data collection at the following synchrotrons: the Stanford Synchrotron Radiation Lightsource (Tzanko Doukov), the Advanced Photon Source (Duilio Cascio & Raj Rajashankar), the Advanced Light Source (Jay Nix), and the Swiss Light Source (Clemens Schulze-Briese). We are thankful to Julian Whitelegge for mass spectrometry characterization of potential heavy atom derivatives and to David Scott for the urease assays and to Bruce Hirayama for advice on the oocyte experiments. We also acknowledge kind assistance with the development version of the program Phenix (Nathan Echols & Tom Terwilliger), fold characterization (Alexey Murzin), pseudosymmetry analysis (Tonya Silkov) and general suggestions (David Scott, Francesco Tombola, Venita De Souza, Kiran Luecke, Suraiya Luecke & Janos Lanyi).
This work was supported by NIH grants R01AI78000, P30CA062203, an NCI institutional training grant 5 T32 CA9054-34, the UC Irvine Center for Biomembrane Systems (H.L.), NIH grants R01DK53462 and R01DK58333 (G.S.) and the US Veterans Administration (G.S).
Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences.
Footnotes
Author Contributions: G.S. and H.L. contributed to the design of the project. D.S. and K.M. expressed, purified and crystallized HpUreI, D.S. carried out the oocyte uptake measurements, C.-K.C. helped with expression and native gels. R.M. and H.L. carried out the crystallographic experiments and analysis of data, S.M.S. assisted with aspects of phasing. G.S. and H.L. were responsible for overall project management and wrote the manuscript with K.M.
Competing financial interests: The authors declare no competing financial interests.
Data Deposition: The coordinates and structure factors have been deposited at the protein data bank with PDB code 3UX4.
Supplementary Information: The Supplementary Information file contains details on Supplementary Tables 1-2, Supplementary Figures 1-13 with legends and additional references.
References
- 1.Pounder RE, Ng D. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol Ther. 1995;9:33. [PubMed] [Google Scholar]
- 2.Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature Rev Cancer. 2002;228 doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 3.Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 5.Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;591143 doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- 6.Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. 2002;123:187. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 7.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nature Rev Microbiol. 2011;9:330. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer-Rosberg K, Scott DR, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996;111:886. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 9.Luecke H, et al. Structure of Bacteriorhodopsin at 1.55 Angstrom Resolution. J Mol Biol. 1999;291:899. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 10.Gray LR, Gu SX, Quick M, Khademi S. Transport kinetics and selectivity of HpUreI, the urea channel from Helicobacter pylori. Biochemistry. 2011;50:8656. doi: 10.1021/bi200887a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huysmans GHM, et al. A urea channel from Bacillus cereus reveals a novel hexameric structure. Biochem J. 2012;445:157. doi: 10.1042/BJ20120169. [DOI] [PubMed] [Google Scholar]
- 12.Pebay-Peyroula E, et al. Structure of mitochondrial ATP/ADP carrier in complex with carboxyatractyloside. Nature. 2003;426:39. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 13.Levin EJ, Quick M, Zhou M. Crystal structure of a bacterial homologue of the kidney urea transporter. Nature. 2009;462:757. doi: 10.1038/nature08558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin EJ, et al. Structure and permeation mechanism of a mammalian urea transporter. Proc Natl Acad Sci U S A. 2012;109:11194. doi: 10.1073/pnas.1207362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lü W et al. pH-dependent gating in a FocA formate channel. Science. 2011;332352 doi: 10.1126/science.1199098. [DOI] [PubMed] [Google Scholar]
- 16.Weeks DL, Sachs G. Sites of pH regulation of the urea channel of Helicobacter pylori. Mol Micro. 2001;40:1249. doi: 10.1046/j.1365-2958.2001.02466.x. [DOI] [PubMed] [Google Scholar]
- 17.Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orbach E, Finkelstein A. The nonelectrolyte permeability of planar lipid bilayer membranes. J Gen Phys. 1980;75427 doi: 10.1085/jgp.75.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong W, et al. Medium pH-dependent redistribution of the urease of Helicobacter pylori. J Med Microbiol. 2002;52:211. doi: 10.1099/jmm.0.05072-0. [DOI] [PubMed] [Google Scholar]
- 20.Scott DR, et al. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3 and NH4(+), is necessary for acid survival of Helicobacter pylori. J Bacteriol. 2010;192:94. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonen T, Sliz P, Kistler J, Chen Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429193 doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- 22.Callenberg KM, et al. APBSmem: a graphical interface for electrostatic calculations at the membrane. PLoS ONE. 2010;5:e12722. doi: 10.1371/journal.pone.0012722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Basner J, Lee B. Detecting internally symmetric protein structures. BMC Bioinformatics. 2010;11:303. doi: 10.1186/1471-2105-11-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W XDS. Acta Cryst. 2010;D66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheldrick GM. A short history of SHELX. Acta Cryst. 2008;D64:112. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Meth Mol Biol. 2007;364:215. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowtan K. Error estimation and bias correction in phase-improvement calculations. Acta Cryst. 1999;D55:1555. doi: 10.1107/s0907444999007416. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furnham N, et al. Knowledge-based real-space explorations for low-resolution structure determination. Structure. 2006;14:1313. doi: 10.1016/j.str.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Meth Enz. 2003;374:300. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 33.Scott DR, et al. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect Immun. 2000;68:470. doi: 10.1128/iai.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


