Abstract
Fanconi Anemia (FA) is a rare disease characterized by congenital defects, progressive bone marrow failure and heightened cancer susceptibility. The FA proteins, BRCA1, and FANCD1/BRCA2, function cooperatively in the FA-BRCA pathway, to repair damaged DNA. Activation of the FA-BRCA pathway occurs via the monoubiquitination of the FANCD2 and FANCI proteins, targeting these proteins to discrete nuclear foci where they function in DNA repair. The cellular regulation of FANCD2/I monoubiquitination, however, remains poorly understood. In this study, we have examined the roles of the p53 tumor suppressor protein, as well as its downstream target the p21Cip1/Waf1 cyclin-dependent kinase inhibitor, in the regulation of the activation of the FA-BRCA pathway. We demonstrate that, in contrast to p53, p21 plays a major role in the regulation of the activation of the FA-BRCA pathway: p21 promotes S-phase and DNA damage-inducible FANCD2/I monoubiquitination and nuclear foci formation. Several lines of evidence establish that this effect is not a consequence of a defective G1-S checkpoint or altered cell cycle progression in the absence of p21. Instead, we demonstrate that p21 is required for the transcriptional repression of the USP1 deubiquitinating enzyme upon exposure to DNA damaging agents. In the absence of p21, persistent USP1 expression precludes the DNA damage-inducible accumulation of monoubiquitinated FANCD2 and FANCI. Consequently, p21−/− cells exhibit increased levels of mitomycin C-inducible complex chromosomal aberrations and elevated γ-H2AX nuclear foci formation. Our results demonstrate that p21 plays a critical role in the regulation of the activation of the FA-BRCA pathway and suggest a broader role for p21 in the orchestration of DNA repair processes following exposure to DNA crosslinking agents.
Keywords: Fanconi anemia, ubiquitination, deubiquitination, genome stability, DNA repair
Introduction
Fanconi anemia (FA) is a rare recessive disease characterized by developmental abnormalities, progressive bone marrow failure and elevated cancer susceptibility (Harney et al., 2008; Mathew, 2006). There are fourteen defined FA complementation groups, caused by biallelic mutations in the following genes: FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ/BRIP1, FANCL, FANCM, FANCN/PALB2, and FANCP/SLX4 (Kim et al., 2011; Moldovan and D’Andrea, 2009; Stoepker et al., 2011). In addition, biallelic mutations in the RAD51C gene have recently been uncovered in a FA-like disorder (Vaz et al., 2010). The protein products of these genes are hypothesized to function cooperatively in the FA-BRCA pathway, the primary function of which is to repair DNA damage thereby maintaining genome stability (Kee and D’Andrea, 2010; Rego et al., 2009). Indeed, the unifying trait of FA patient cells, as well as BRCA1/2-deficient cancer cells, is hypersensitivity to DNA crosslinking agents, e.g. mitomycin C (MMC) (Garcia-Higuera et al., 2001; Howlett et al., 2002).
Upon exposure to DNA damaging agents, and during S-phase of the cell cycle, the core FA complex, comprising FANCA, B, C, E, F, G, L, M, as well as FAAP100 and FAAP24, promotes the monoubiquitination of the FANCD2 and FANCI proteins (Garcia-Higuera et al., 2001; Smogorzewska et al., 2007). The FANCL and UBE2T proteins are the E3 ubiquitin ligase and E2 ubiquitin-conjugating enzymes, respectively (Machida et al., 2006; Meetei et al., 2003). Monoubiquitination of FANCD2 and FANCI targets these proteins to discrete nuclear foci, where they co-localize with DNA repair and replication proteins, including BRCA1, FANCD1/BRCA2, PCNA, and RAD51 (Garcia-Higuera et al., 2001; Howlett et al., 2005; Taniguchi et al., 2002; Wang et al., 2004). Monoubiquitinated FANCD2 and FANCI are deubiquitinated by the ubiquitin-specific protease USP1 (Nijman et al., 2005). Despite the clear importance of FANCD2/I monoubiquitination for cellular DNA crosslink repair and the molecular etiology of FA, its regulation remains poorly understood.
The p21Cip1/Waf1 protein plays a major role in the regulation of cell cycle progression, apoptosis, and cellular senescence (Abbas and Dutta, 2009). p21 is a member of a family of cyclin-dependent kinase (CDK) inhibitors and regulates cell cycle progression by binding CDKs via a CDK-binding domain and by binding PCNA via a PCNA-interaction motif (PIP-box) (Abukhdeir and Park, 2008; Prives and Gottifredi, 2008). p21 inhibits DNA replication by physically blocking the interaction between PCNA and essential replication factors, e.g. DNA polymerase δ (Podust et al., 1995) and FEN1 (Chen et al., 1996). p21 also forms inhibitory complexes with cyclin-CDKs thereby preventing G1-S and G2-M progression under adverse cellular conditions (Abukhdeir and Park, 2008). The role of p21 in DNA interstrand crosslink repair remains uncharacterized.
We have recently described an important functional interaction between FANCD2 and PCNA: disruption of a FANCD2 PIP-box abrogates spontaneous and DNA damage-inducible FANCD2 monoubiquitination (Howlett et al., 2009). In light of the strong connections between PCNA and p21, and to gain a greater understanding of the regulation of the FA-BRCA pathway, here we have analyzed the roles of the p53 and p21 proteins in the regulation of the monoubiquitination of FANCD2 and FANCI. Importantly, we demonstrate that DNA damage-inducible FANCD2 monoubiquitination is p53-independent. In contrast, the p21 protein plays a major role in the regulation of the activation of the FA-BRCA pathway: p21 is required for both S-phase and DNA damage-inducible FANCD2/I monoubiquitination and nuclear foci formation. Several lines of experimental evidence demonstrate that defective FANCD2/I monoubiquitination is not a consequence of an abrogated G1-S checkpoint or altered cell cycle progression in the absence of p21. Instead, we demonstrate that p21 is required for the transcriptional repression of the USP1 deubiquitinating enzyme upon exposure to DNA damaging agents. In the absence of p21, persistent USP1 expression precludes the DNA damage-inducible accumulation of monoubiquitinated FANCD2 and FANCI. Furthermore, we show that defective FANCD2/I monoubiquitination can be rescued by transient expression of a p21 transgene, siRNA-mediated USP1 knockdown, and transcription inhibition. Finally, we demonstrate that p21−/− cells display increased MMC-inducible complex chromosome aberrations and elevated γH2AX nuclear foci formation, similar to FA patient cells, establishing an important function for p21 in DNA crosslink repair. Our results indicate that p21 plays a central role in the regulation of the activation of a major cellular tumor suppressor network, and suggest that p21 may play a broader role in the promotion of conservative, error-free DNA repair.
Results
The p53 tumor suppressor protein does not play an overt role in the regulation of the monoubiquitination of FANCD2
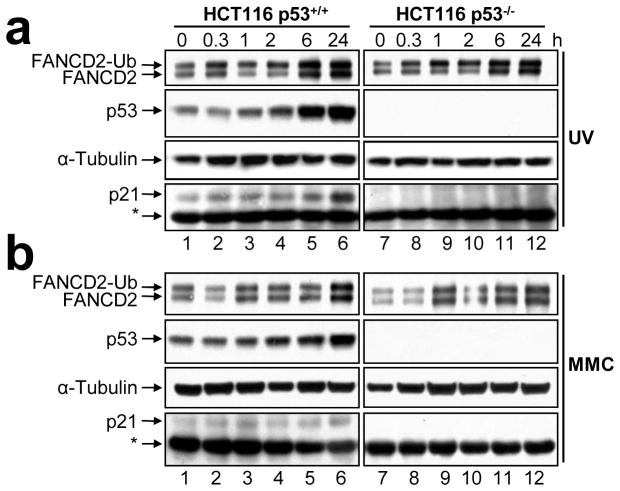
To examine the role of p53 in the activation of the FA-BRCA pathway, HCT116 p53+/+ and p53−/− cells (Bunz et al., 1998) were exposed to UV-C irradiation, MMC, and etoposide (VP-16), and FANCD2 monoubiquitination was assessed by immunoblotting. Treatment of both p53+/+ and p53−/− cells with all three types of DNA-damaging agents resulted in robust activation of FANCD2 monoubiquitination: no discernible differences in the kinetics or extent of FANCD2 monoubiquitination were observed (Figures 1a and b, Supplementary Figure 1). For example, an approximate 2-fold increase in the FANCD2-Ub:FANCD2 ratio was observed one hour following exposure to UV-C irradiation in both the p53+/+ and p53−/− cells (Figure 1a, lanes 3 and 9). Similar intact DNA damage-inducible FANCD2 monoubiquitination has previously been observed in several p53 defective cancer cell lines including HeLa, MDA-MB-231, NCI-H1703, SW900, and T47D (results not shown and (Garcia-Higuera et al., 2001)). Furthermore, the chicken B lymphocyte cell line DT40, which lacks functional p53, is fully competent for FANCD2 monoubiquitination (Yamamoto et al., 2005; Yamazoe et al., 2004). These results indicate that the p53 protein does not play an overt role in the regulation of the monoubiquitination of FANCD2.
Figure 1.
FANCD2 monoubiquitination is p53-independent. (a) HCT116 p53+/+ and p53−/− cells were treated with 20 J/m2 UV-C irradiation (UV) and 100 nM mitomycin C (MMC) for 24 h (b). Whole-cell lysates were prepared at the indicated time points, resolved, and immunoblotted with anti-FANCD2, anti-p53, anti-p21, and anti-α-tubulin antibodies. *, non-specific band.
The p21Cip/Waf1 protein is required for efficient DNA damage-inducible FANCD2 and FANCI monoubiquitination
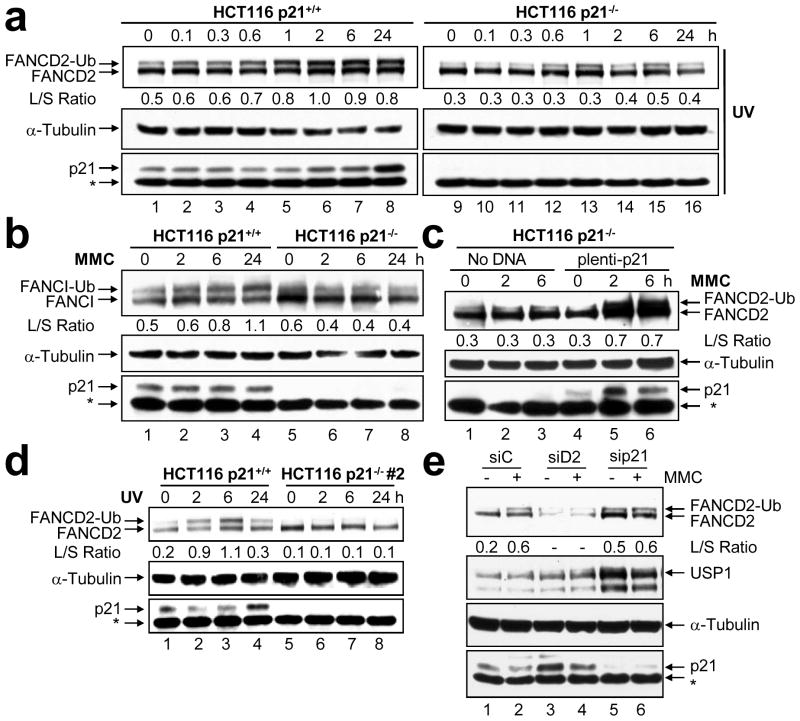
To examine the role of p21 in the activation of the FA-BRCA pathway, HCT116 p21+/+ and p21−/− cells (Waldman et al., 1995) were exposed to UV-C irradiation, MMC, and VP-16, and FANCD2 monoubiquitination was again assessed by immunoblotting. As expected, treatment of p21+/+ cells with all three DNA-damaging agents resulted in robust accumulation of monoubiquitinated FANCD2. However, DNA damage-inducible FANCD2 monoubiquitination was markedly attenuated in the absence of p21 (Figure 2a, Supplementary Figure 2a–c). For example, an approximate 2-fold increase in the FANCD2-Ub:FANCD2 ratio was observed in the p21+/+ cells 2 hours following exposure to UV-C irradiation (Figure 2a, lane 6, Supplementary Figure 2a). Under the same conditions, no appreciable change in the FANCD2-Ub:FANCD2 ratio was observed for the p21−/− cells (Figure 2a, lane 14, Supplementary Figure 2a). Similar results were observed following exposure to MMC and VP-16 (Supplementary Figures 2b and c). These results strongly suggest that the p21 protein is required for the activation of the FA-BRCA pathway following cellular exposure to a wide range of DNA damaging agents. Several studies have demonstrated that the monoubiquitination of the FANCD2 and FANCI proteins is coupled (Sims et al., 2007; Smogorzewska et al., 2007). For example, the monoubiquitination of the FANCI protein is dependent on the presence and monoubiquitination of FANCD2 (Sims et al., 2007; Smogorzewska et al., 2007). Therefore, we next examined FANCI monoubiquitination in the p21+/+ and p21−/− cells. As expected, in the p21+/+ cells, we observed a time-dependent increase in monoubiquitinated FANCI following exposure to MMC (Figure 2b, lanes 2–4). In contrast, levels of monoubiquitinated FANCI failed to increase in the p21−/− cells following exposure to MMC (Figure 2b, lanes 6–8).
Figure 2.
p21 is required for DNA damage-inducible FANCD2 and FANCI monoubiquitination. (a) HCT116 p21+/+ and p21−/− cells were treated with 20 J/m2 UV-C irradiation (UV) and 100 nM mitomycin C (MMC) for 24 h (b). Whole cell lysates were prepared at the indicated time points, resolved, and immunoblotted with anti-FANCD2, anti-FANCI, anti-α-tubulin, and anti-p21 antibodies. (c) Transfection of HCT116 p21−/− cells with pLenti6.2-p21 restores DNA damage-induced FANCD2 monoubiquitination. Cells were treated with 100 nM MMC. (d) A second independently isolated HCT116 p21−/− clone demonstrates markedly attenuated DNA damage-inducible FANCD2 monoubiquitination following treatment with 20 J/m2 UV-C irradiation. *, non-specific band. (e) U2OS cells were transfected with control non-targeting (siC), FANCD2 (siD2), or p21 (sip21) siRNAs. Seventy-two hours post-transfection, cells were untreated (−) or treated (+) with 200 nM mitomycin C (MMC). Whole-cell lysates were prepared 6 h later, resolved, and immunoblotted with anti-FANCD2, anti-USP1, anti-p21, and anti-α-tubulin antibodies. The L/S ratio is the ratio of FANCD2-Ub (long isoform) to FANCD2 (short isoform).
Two approaches were taken to confirm that the observed effects were a specific consequence of the absence of p21. First, we transiently transfected the p21−/− cells with pLenti6.2-p21, treated the cells with MMC and analyzed FANCD2 monoubiquitination by immunoblotting. Importantly, transient expression of p21 was sufficient to restore DNA damage-inducible FANCD2 monoubiquitination in the p21−/− cells (Figure 2c, Supplementary Figure 2d). Next, we analyzed FANCD2 monoubiquitination in a second independently isolated p21−/− clone. Once again, DNA damage-inducible FANCD2 monoubiquitination was markedly attenuated in the absence of p21 (Figure 2d, lanes 6–8). These results strongly support a specific role for p21 in promoting this posttranslational modification.
To verify our findings using an alternative approach, we transiently depleted p21 protein from U2OS cells using siRNA, and assayed both spontaneous and DNA damage-inducible FANCD2 monoubiquitination. Transient p21 depletion led to an increase in basal levels of both nonubiquitinated (>2-fold) and monoubiquitinated FANCD2 (Figure 2e). Nevertheless, similar to that observed for the HCT116 p21−/− cells, DNA damage-inducible FANCD2 monoubiquitination was markedly attenuated upon transient depletion of p21. For example, a 4-fold increase in the FANCD2-Ub:FANCD2 ratio was observed for a control non-targeting siRNA, while no appreciable increase in the FANCD2-Ub:FANCD2 ratio was observed upon p21 depletion, with very similar results being obtained for three independent p21 siRNA oligonucleotides (Figure 2e, lanes 5 and 6, and results not shown).
The p21Cip/Waf1 protein is required for efficient DNA damage-inducible FANCD2 nuclear foci formation
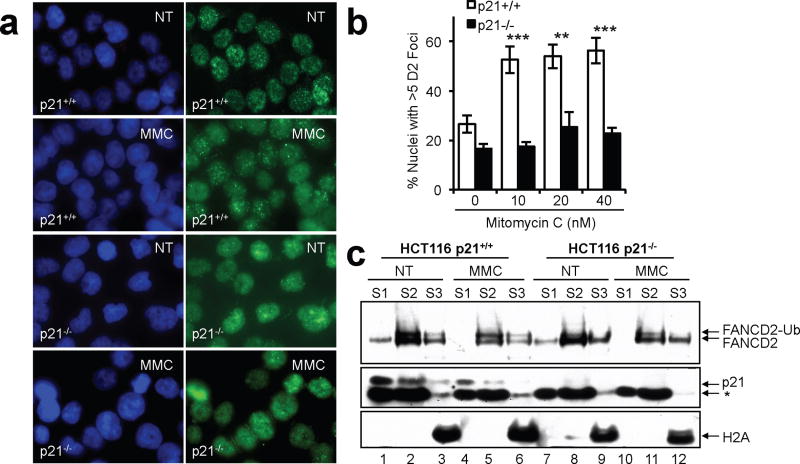
Monoubiquitinated FANCD2 accumulates in discrete nuclear foci following exposure to DNA damaging agents (Garcia-Higuera et al., 2001; Howlett et al., 2005; Taniguchi et al., 2002). Therefore, we next examined the influence of p21 on FANCD2 nuclear foci formation. p21+/+ and p21−/− cells were exposed to MMC for one cell cycle and FANCD2 nuclear foci formation was analyzed by immunocytochemistry. No differences in the number of nuclei displaying >5 discrete FANCD2 nuclear foci were observed between p21+/+ and p21−/− cells in the absence of DNA damage (Figures 3a and b). However, the ability of FANCD2 to assemble into discrete nuclear foci following exposure to MMC was markedly attenuated in p21−/− cells, compared with p21+/+ cells. For example, 16 h following exposure to 10 nM MMC an approximate 2-fold increase in nuclei displaying >5 discrete FANCD2 nuclear foci was observed in p21+/+ cells, while no appreciable induction was observed for p21−/− cells (p < 0.0001) (Figures 3a and b). Similar results were observed following UV-C irradiation (results not shown). We also examined the subcellular localization of FANCD2 in the p21+/+ and p21−/− cells. Monoubiquitinated FANCD2 was enriched in the soluble nuclear (S2) and chromatin (S3) fractions of p21+/+ cells, but not p21−/− cells (Figure 3c). Nevertheless, nonubiquitinated FANCD2 remained competent for chromatin localization in the absence of p21 (Figure 3c, lanes 9 and 12). Chromatin localization of nonubiquitinated FANCD2 has previously been described (Alpi et al., 2007; Howlett et al., 2009).
Figure 3.
In the absence of p21 FANCD2 localizes to chromatin yet fails to assemble into DNA damage-inducible nuclear foci. (a) HCT116 p21+/+ and p21−/− cells were untreated (NT) or exposed to mitomycin C (MMC) for 16 h. Cells were fixed and immunostained with a polyclonal antibody against FANCD2 (green) and counterstained with DAPI (blue). (b) The percentage of nuclei with > 5 FANCD2 foci were counted and plotted. At least 300 nuclei were scored per cell line and treatment. Error bars represent the standard errors of the means. **, p < 0.01; ***, p < 0.001. (c) Cells were incubated in the absence and presence of 60 nM MMC for 18 h, fractionated into cytoplasmic (S1), soluble nuclear (S2), and chromatin-associated fractions (S3), and immunoblotted with antibodies to FANCD2, p21, and H2A. *, non-specific band.
The role of p21 in the regulation of FANCD2 monoubiquitination is cell cycle-independent
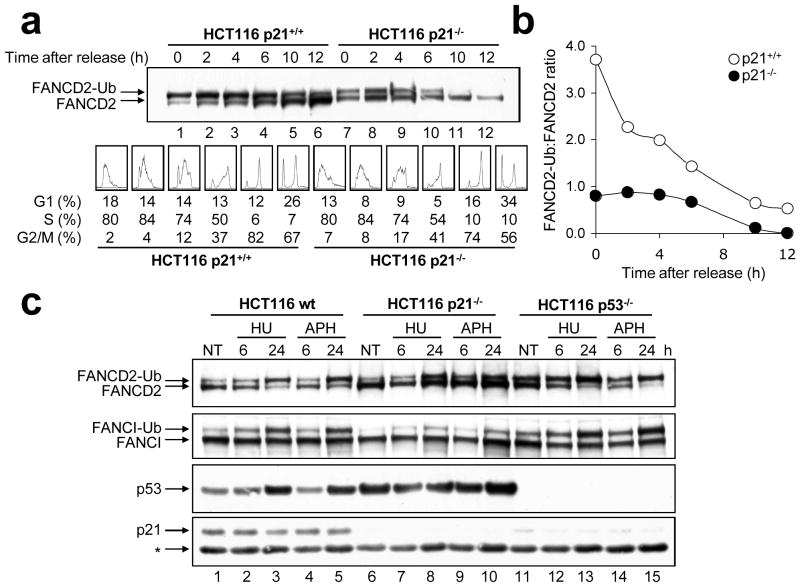
Together with p53, the p21 protein is well known to play a major role in cell cycle checkpoint regulation (Bunz et al., 1998; Waldman et al., 1995). To determine if the effect of p21 on DNA damage-inducible FANCD2/I monoubiquitination was a consequence of aberrant cell cycle progression, p21+/+ and p21−/− cells were synchronized in early S-phase by double-thymidine block, released, and FANCD2 monoubiquitination was examined as cells progressed through the cell cycle. Previous studies have demonstrated that the FANCD2 protein is subject to monoubiquitination during S-phase of the cell cycle (Taniguchi et al., 2002). Consistent with this study, in the p21+/+ cells, monoubiquitinated FANCD2 was the major isoform present upon release from double-thymidine arrest (Figure 4a, lane 1). In contrast, double-thymidine arrest-induced FANCD2 monoubiquitination was severely impaired in the absence of p21. For example, upon release from double-thymidine arrest, a ~5-fold greater FANCD2-Ub:FANCD2 ratio was observed for the p21+/+ cells, compared with the p21−/− cells (Figure 4a, lanes 1 and 7). Pronounced differences in the FANCD2-Ub:FANCD2 ratio between the p21+/+ and p21−/− cells persisted during S-phase and throughout the cell cycle. Furthermore, for the p21−/− cells, at no point during the cell cycle did levels of monoubiquitinated FANCD2 exceed those of nonubiquitinated FANCD2 (Figures 4a and b). These results indicate that abrogated FANCD2/I monoubiquitination is not a general consequence of irregular cell cycle progression in the absence of p21.
Figure 4.
p21-dependent FANCD2/I monoubiquitination is cell cycle independent. (a) HCT116 p21+/+ and p21−/− cells were synchronized via double thymidine block, released into thymidine-free media and pellets collected for immunoblotting with anti-FANCD2 (top panel) and FACS analysis (bottom panel) at the indicated time points. (b) Band intensities from (a) were quantified using ImageJ software and plotted. (c) HCT116 wild type, p21−/− and p53−/− cells were untreated (NT) or treated with hydroxyurea (HU) and aphidicolin (APH), whole cell lysates were prepared, and resolved proteins immunoblotted with anti-FANCD2, anti-FANCI, anti-p53, and anti-p21 antibodies. *, non-specific band. For (b), while the band intensities for a single experiment are shown, this experiment was repeated multiple times with very similar findings.
Next, we examined the effects of the DNA replication inhibitors hydroxyurea (HU) and aphidicolin (APH) on FANCD2/I monoubiquitination in wild type, p21−/− and p53−/− cells. HU inhibits the deoxyribonucleotide reductase enzyme leading to depletion of cellular dNTP pools, while APH is a specific inhibitor of DNA polymerase α: both agents are potent inducers of FANCD2 monoubiquitination (Howlett et al., 2005). Similar to that observed above, robust HU- and APH-induced FANCD2/I monoubiquitination was observed for both the wild type and p53−/− cells. In contrast, HU- and APH-induced FANCD2/I monoubiquitination was again severely attenuated in the absence of p21 (Figure 4c, Supplementary Figures 3a–c).
p21 regulates the transcriptional repression of the USP1 deubiquitinating enzyme following exposure to DNA damaging agents
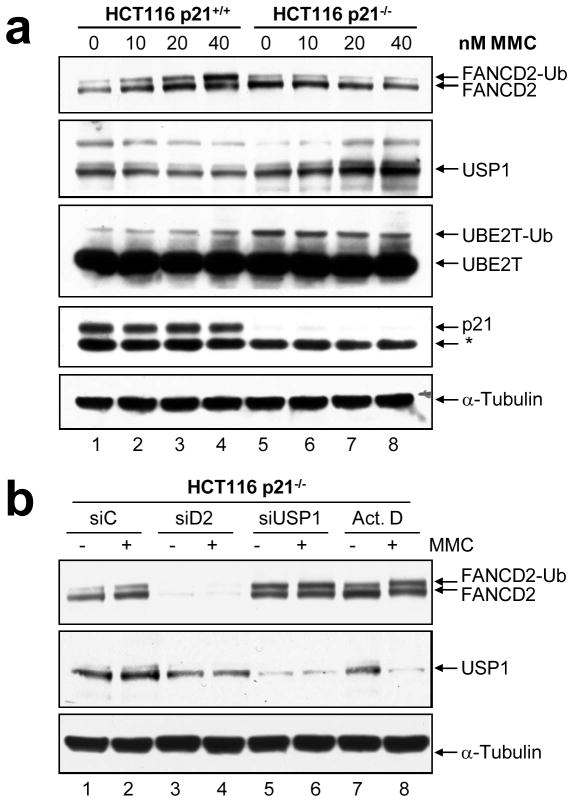
We next sought to determine if the failure to activate the monoubiquitination of FANCD2 and FANCI in the absence of p21 was a direct consequence of altered regulation of the USP1 deubiquitinating enzyme and/or the UBE2T ubiquitin-conjugating enzyme. The regulation of the FANCL E3 ubiquitin ligase in vitro could not be assessed because of the lack of a suitable commercially available antibody. p21+/+ and p21−/− cells were exposed to a range of MMC concentrations for one cell cycle and USP1 and UBE2T protein expression was examined by immunoblotting. Consistent with previous observations following UV-C irradiation (Cohn et al., 2007), we observed a dose-dependent decrease in USP1 protein levels following exposure to MMC in the p21+/+ cells, which correlated with the accumulation of monoubiquitinated FANCD2 (Figure 5a, lanes 1–4, Supplementary Figure 4a). However, in contrast, in the p21−/− cells, USP1 protein levels exhibited a dose-dependent increase following MMC exposure, coincident with a failure to activate FANCD2 monoubiquitination (Figure 5a, lanes 5–8 and Supplementary Figure 4a). Increased USP1 levels were also observed upon siRNA-mediated p21 depletion (see Figure 2e, lanes 5 and 6). Furthermore, while basal levels of the UBE2T enzyme did not differ significantly between p21+/+ and p21−/− cells, we observed a persistent and marked increase in the levels of monoubiquitinated UBE2T in the p21−/− cells (Figure 5a, lanes 5–8).
Figure 5.
Altered regulation of the USP1 and UBE2T enzymes in p21-null cells. (a) HCT116 p21+/+ and p21−/− cells were incubated in the absence or presence of the indicated concentrations of mitomycin C (MMC) for 24 h, whole-cell lysates prepared, and FANCD2, USP1, UBE2T, p21, and α-tubulin protein expression was analyzed by immunoblotting. (b) HCT116 p21−/− cells were transfected with control (non-targeting) (siC), FANCD2 (siD2), and USP1 (siUSP1) siRNAs. Seventy-two hours later, cells were untreated (−) or treated (+) with 40 nM mitomycin C (MMC) for 24 h, in the presence or absence of 4 μM actinomycin D (Act. D). Whole-cell lysates were prepared, and FANCD2, USP1, and α-tubulin protein expression was analyzed by immunoblotting.
To confirm that attenuated DNA damage-inducible FANCD2 monoubiquitination was a consequence of a failure to downregulate USP1, we depleted USP1 from the HCT116 p21−/− cells using siRNA and examined FANCD2 monoubiquitination. Importantly, USP1 depletion led to a complete restoration of MMC-inducible FANCD2 monoubiquitination in the p21−/− cells (Figure 5b, lanes 5 and 6, Supplementary Figure 4b). For example, a 3-fold increase in the FANCD2-Ub:FANCD2 ratio was observed for MMC-treated HCT116 p21−/− transfected with USP1 siRNA, compared with a control non-targeting siRNA (Figure 5b, lanes 2 and 6). Furthermore, treatment of the p21−/− cells with the transcription inhibitor actinomycin D led to a striking reduction in USP1 protein levels following exposure to MMC, and a dramatic restoration of DNA damage-inducible FANCD2 monoubiquitination in the p21−/− cells (Figure 5b, lane 8, Supplementary Figure 4b). Taken together, these results demonstrate that p21-dependent DNA damage-inducible FANCD2 monoubiquitination is mediated primarily via the regulation of the transcriptional repression of the USP1 gene.
p21−/− cells hypersensitive to the clastogenic effects of mitomycin C
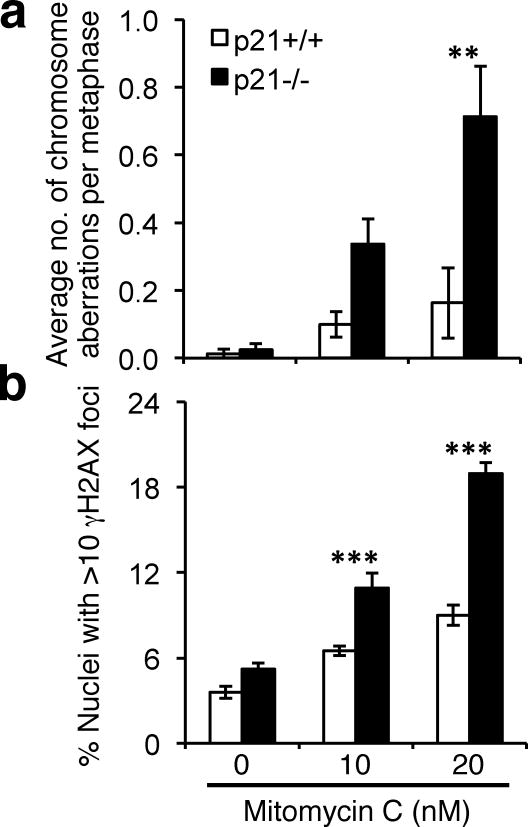
Hypersensitivity to the clastogenic effects of DNA crosslinking agents, such as MMC, is a hallmark of FA patient cells (Auerbach, 1993). The failure to activate both S-phase and DNA damage-inducible FANCD2/I monoubiquitination prompted us to next examine the functional role of p21 in the cellular response to DNA crosslinking agents. p21+/+ and p21−/− cells were incubated in the absence and presence of 10 and 20 nM MMC for 16 h, metaphase chromosomes were prepared, and chromosome aberrations were scored. Pronounced differences in the average number of metaphase chromatid gaps and breaks and radial chromosome formations were observed between MMC-treated p21+/+ and p21−/− cells. For example, a >4-fold increased frequency of chromosome aberrations was observed for p21−/− cells compared with p21+/+ cells, following exposure to 20 nM MMC (p = 0.003) (Figure 6a and Supplementary Figure 5). Furthermore, these findings were corroborated using immunocytochemistry for the phosphorylated H2AX variant (γH2AX) on interphase cells. Here, we observed a greater than 2-fold increase in nuclei harboring >10 discrete γH2AX foci for p21−/− cells, compared with p21+/+ cells, 24 h following exposure to 20 nM MMC (p < 0.0001) (Figure 6b). Taken together these findings strongly support an important functional role for the p21 protein in the cellular response to DNA crosslinking agents.
Figure 6.
p21−/− cells display elevated mitomycin C-inducible chromosome aberrations and γH2AX nuclear foci formation. (a) HCT116 p21+/+ and p21−/− cells were incubated in the absence and presence of 10 and 20 nM MMC for one cell cycle, metaphase spreads were prepared and chromosome aberrations, including chromosome and chromatid gaps and breaks and radial formations, were scored. At least 100 metaphases were scored per treatment. (b) HCT116 p21+/+ and p21−/− cells were treated as for (a) and the number of nuclei displaying >10 discrete γH2AX foci recorded. Error bars represent standard errors of the means and all experiments were performed in triplicate with similar findings. **, p < 0.01; ***, p < 0.001
Discussion
Recent reports have described an important role for monoubiquitinated FANCD2 in the recruitment of the FAN1 (for FANCD2-associated nuclease) protein to damaged chromatin, where it is thought to function in a nucleolytic step of homologous recombination DNA repair (HR) (Kratz et al., 2010; Liu et al., 2010; MacKay et al., 2010; Smogorzewska et al., 2010). Our understanding of the cellular factors regulating FANCD2/I monoubiquitination upstream of FAN1 nucleolytic activity, however, remains largely incomplete. Inactivating mutations of any of the eight members of the core FA complex, which account for >90% of FA patients, lead to complete abrogation of FANCD2/I monoubiquitination. Additional proteins with important roles in DNA replication, including ATR, CHK1, HCLK2, PCNA and RPA, have also been demonstrated to affect DNA damage-inducible FANCD2 monoubiquitination (Andreassen et al., 2004; Collis et al., 2007; Howlett et al., 2009; Wang et al., 2007). This is consistent with studies demonstrating an important function for the FA-BRCA pathway in the DNA replication stress response (Chan et al., 2009; Howlett et al., 2005; Naim and Rosselli, 2009). In this study, we describe a novel role for the p21 protein in the regulation of FANCD2/I monoubiquitination that is independent of G1- to S-phase regulation. Two independently isolated HCT116 cell lines harboring homozygous deletions at the p21 locus displayed markedly attenuated S-phase and DNA damage-inducible FANCD2/I monoubiquitination and nuclear foci formation, while similarly derived HCT116 p53−/− cells remained competent for FANCD2/I monoubiquitination. Furthermore, defective DNA damage-inducible FANCD2 monoubiquitination was rescued by the transient introduction of a p21 transgene. These results argue strongly against the possibility that a coincident acquired somatic mutation is responsible for the observed phenotype. In addition, using an RNA interference approach, p21 knockdown recapitulated the effects observed for the p21-nullizygous cells, and resulted in abrogation of DNA damage-inducible FANCD2 monoubiquitination. Interestingly, increased basal levels of nonubiquitinated and monoubiquitinated FANCD2 were observed upon transient p21 depletion. The inhibition of cyclin-CDK activity by p21 leads to pRb hypophosphorylation and repression of E2F-mediated transcription (Abukhdeir and Park, 2008). Previous studies in primary human keratinocytes have demonstrated that the FANCD2 gene harbors E2F transcription factor consensus sequences, and that FANCD2 protein expression can be modulated by ectopic expression of both E2F and active pRb (Hoskins et al., 2008). Thus, the increased FANCD2 protein levels observed upon transient p21 depletion may reflect increased E2F-mediated FANCD2 expression. Similarly, over the course of these studies we have examined Fancd2 protein expression in primary p21+/+, p21+/−, and p21−/− murine embryonic fibroblasts (MEFs), and could not detect appreciable levels of Fancd2 protein in primary p21+/+ and p21+/− MEFs. In contrast, robust levels of Fancd2 protein were detected in the p21−/− MEFs, precluding a comparison of DNA damage-inducible Fancd2 monoubiquitination between these cells (results not shown). Collectively, these results suggest that p21 may play both a transcriptional and posttranslational regulatory role in the FA-BRCA pathway. The increased basal levels of monoubiquitinated FANCD2 observed upon transient p21 depletion may simply be a consequence of increased total FANCD2 protein levels, or may reflect the presence of increased spontaneous DNA damage upon acute and incomplete p21 depletion. A similar observation of increased basal levels of monoubiquitinated FANCD2 yet persistent hypersensitivity to MMC has been demonstrated upon transient depletion of the CHK1 kinase (Wang et al., 2007). Nevertheless, in both cases the absence of p21 precluded the accumulation of monoubiquitinated FANCD2/I following cellular exposure to DNA damaging agents.
Several lines of evidence firmly demonstrate that this newly discovered function for p21 is independent of G1- to S-phase transition, nor is it an indirect consequence of altered cell cycle progression: First, the G1-S checkpoint is abrogated in both the p53−/− and p21−/− cells used in this study (Waldman et al., 1995). However, we clearly demonstrate that DNA damage-inducible FANCD2/I monoubiquitination is intact in the p53−/− cells, yet markedly attenuated in the p21−/− cells. p53−/− cells express low but readily detectable levels of p21, as the p21/CDKN1A gene is also subject to p53-independent transcriptional regulation through the action of several transcription factors, including E2F1, SP1 and 3, and AP2 (reviewed in (Abbas and Dutta, 2009)). Consistent with these findings, we have observed robust DNA damage-inducible FANCD2 monoubiquitination in numerous p53-defective (and G1-S checkpoint-defective) transformed cell lines including HeLa, MDA-MB-231, NCI-H1703, SW900, and T47D. Second, as a consequence of an abrogated G1-S checkpoint, asynchronous populations of p21−/− cells, as well as p53−/− cells, have an approximately 2-fold increased number of S-phase cells, compared with their wild-type counterparts (see Supplementary Figure 3a). Previous studies have demonstrated that, even in the absence of exogenous DNA damage, FANCD2 undergoes monoubiquitination as cells traverse unperturbed S-phase (Taniguchi et al., 2002). Therefore, one would expect to observe much greater levels of monoubiquitinated FANCD2 in the p21−/− cells, simply as a function of increased S-phase progression. Again, this is clearly not what we have observed. Third, double-thymidine block-mediated cell cycle synchronization, followed by release, revealed marked differences in the levels of monoubiquitinated FANCD2 between the p21+/+ and p21−/− cells as these cells progressed through the cell cycle. Indeed, for the p21−/− cells, at no stage throughout the cell cycle did levels of monoubiquitinated FANCD2 exceed those of nonubiquitinated FANCD2. In addition, direct chemical inhibition of DNA synthesis with HU and APH failed to result in the accumulation of appreciable levels of monoubiquitinated FANCD2 in the absence of p21, while robust FANCD2/I monoubiquitination was observed in wild type and p53−/− cells. These agents are among the most potent inducers of FANCD2/I monoubiqutination (Howlett et al., 2005; Taniguchi et al., 2002). Taken together, these findings demonstrate that the role of p21 in the regulation of FANCD2/I monoubiquitination is not merely a consequence of an impaired G1- to S-phase checkpoint, or altered cell cycle progression.
Instead, we demonstrate that p21 is required for the transcriptional repression of the USP1 gene following exposure to DNA damaging agents. The USP1 deubiquitinating enzyme undergoes autocleavage and proteasomal degradation, as well as transcriptional repression, following exposure to UV-C irradiation, thereby facilitating the accumulation of monoubiquitinated FANCD2/I (Cohn et al., 2007; Huang et al., 2006). Consistent with these findings, we observed a dose-dependent decrease in USP1 protein levels in the p21+/+ cells following exposure to MMC. In contrast, in the p21−/− cells, USP1 protein levels increased under the same conditions. Similarly, the transient depletion of p21 using siRNA led to an increase in basal levels of USP1 protein. Importantly, we demonstrate that DNA damage-inducible FANCD2 monoubiquitination in the p21−/− cells can be rescued by depleting USP1 using siRNA and by inhibiting transcription. Our results suggest that, in the absence of p21, constitutive cyclin-CDK activity leads to pRb hyperphosphorylation and elevated E2F-mediated transcriptional activation of the USP1 gene, precluding the accumulation of monoubiquitinated FANCD2/I. In addition, we also observed elevated constitutive UBE2T monoubiquitination in the absence of p21. Previous studies have established that UBE2T undergoes monoubiquitination in vivo, and that monoubiquitination inactivates UBE2T ubiquitin-conjugating activity (Machida et al., 2006). Collectively, our results indicate that, in the absence of p21, persistent USP1 expression, and possibly constitutive inhibitory UBE2T monoubiquitination, precludes the DNA damage-inducible accumulation of monoubiquitinated FANCD2/I.
Upregulation of FANCD2/I monoubiquitination occurs in response to a wide spectrum of DNA damaging agents, as well as during S-phase of the cell cycle (Garcia-Higuera et al., 2001; Howlett et al., 2005; Taniguchi et al., 2002). Similarly, p53-mediated transcriptional upregulation of p21 occurs following cellular exposure to DNA damaging agents, e.g. ionizing radiation (Di Leonardo et al., 1994; el-Deiry et al., 1994; el-Deiry et al., 1993). However, recent studies have demonstrated that the p21 protein is targeted for proteolytic degradation via CRLCdt2-mediated ubiquitination, following exposure to low fluences of UV irradiation (Abbas et al., 2008; Bendjennat et al., 2003; Kim et al., 2008). Consistent with these findings, in our studies, while we generally observed robust p21 accumulation following exposure to VP-16, we did not observe significant accumulation of p21 following exposure to low doses of UV-C irradiation or MMC. Instead, we typically observed an initial decrease in p21 protein levels followed by p21 accumulation at later time points, suggesting that p21 may also be subject to ubiquitin-mediated proteolysis following exposure to DNA crosslinking agents.
In this study we also demonstrate that, like FA patient cells, p21−/− cells demonstrate elevated levels of chromosome aberrations, including radial formations, and γH2AX nuclear foci formation following exposure to MMC, indicative of persistent unrepaired DNA strand breaks (Rogakou et al., 1998). We propose that these observations reflect a bias toward nonconservative, error-prone DNA repair and damage avoidance pathways in the absence of p21, for example nonhomologous DNA end joining (NHEJ) and translesion DNA synthesis (TLS). Indeed, an important role for p21 in the negative regulation of TLS has recently been established (Avkin et al., 2006). Recent studies have also firmly established that the FA proteins play a central role in preventing the promiscuous activity of NHEJ proteins during ICL repair (Adamo et al., 2010; Pace et al., 2010), consistent with the long-held view that the FA pathway promotes error-free conservative HR DNA repair (Garcia-Higuera et al., 2001; Nakanishi et al., 2005; Wang et al., 2004). The role of p21 in the regulation of NHEJ and HR has yet to be described.
In summary, we have uncovered a novel role for the p21 cyclin-dependent kinase inhibitor in the regulation of the monoubiquitination of FANCD2 and FANCI and in the activation of a major tumor suppressor pathway that is independent of G1- to S-phase progression. Further, we propose that p21 may play a broader role in DNA repair pathway determination following exposure to DNA crosslinking agents, a hypothesis that is currently under investigation in our laboratory.
Materials and methods
Cell culture, immunoblotting, immunocytochemistry, and antibodies
HCT116 and U2OS cells were grown in McCoy’s 5A and DMEM media, respectively, supplemented with 12% FBS, L-glutamine and penicillin/streptomycin. Cellular subfractionation, immunoblotting, and immunocytochemistry were performed as previously described (Howlett et al., 2009; Howlett et al., 2005). The following antibodies were used: rabbit polyclonal antisera against FANCD2 (NB100-182, Novus Biologicals), FANCI (Dr. Patrick Sung, Yale University), H2A (07-146; Millipore), p21 (N-20; Santa-Cruz), UBE2T (A301-874A; Bethyl Laboratories) and USP1 (Dr. Tony Huang, New York University), and mouse monoclonal sera against α-tubulin (MS-581-PO, Neomarkers), γH2AX (JBW301; Millipore), and p53 (DO-1; Santa Cruz).
Cell-cycle synchronization and analysis
Cells were synchronized by double thymidine block and cell pellets harvested for immunoblotting and FACS analysis as described previously (Howlett et al., 2009; Taniguchi et al., 2002). Flow cytometric analysis was performed on a Becton Dickson FACS Caliber instrument and cell cycle stage distribution was determined using ModFit LT software.
Chromosome breakage assay
Cells were treated with MMC for 16 hours, and harvested for chromosome preparations as described previously (Arlt et al., 2004). Metaphase chromosomes were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software.
siRNA Experiments
Cells were plated in 6-well dishes at a density of 250,000 cells per well in growth medium lacking penicillin/streptomycin. The following day, cells were transfected with 20 nM (Ambion silencer select) or 200 nM (Qiagen) siRNAs, using Lipofectamine 2000 (Invitrogen). Seventy-two hours post-transfection, cells were incubated in the absence or presence of MMC for a fixed period, and harvested for immunoblotting. siRNA sequences are provided in supplementary methods.
Supplementary Material
Acknowledgments
We thank members of the Howlett laboratory, Paul R. Andreassen, Matthew Stoner, and Patrick Sung for helpful discussions. We thank Tony T. Huang and Patrick Sung for the anti-USP1 and anti-FANCI antibodies, respectively. We thank Bert Vogelstein for cells. This work was supported by a Leukemia Research Foundation New Investigator grant (NGH), RI-INBRE grant P20RR016457-09 from the National Center for Research Resources (NGH), and National Institutes of Health/National Heart, Lung and Blood Institute grant R21HL095991-01 (NGH).
Footnotes
Conflict of interest
There are no competing financial interests in relation to the work described.
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–14. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–30. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–63. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 Is Required for Common-Fragile-Site Stability via Its G2/M Checkpoint Function. Mol Cell Biol. 2004;24:6701–9. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21:731–3. [PubMed] [Google Scholar]
- Avkin S, Sevilya Z, Toube L, Geacintov N, Chaney SG, Oren M, et al. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol Cell. 2006;22:407–13. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, et al. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–60. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chen U, Chen S, Saha P, Dutta A. p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc Natl Acad Sci U S A. 1996;93:11597–602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–97. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–51. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Harper JW, O’Connor PM, Velculescu VE, Canman CE, Jackman J, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–74. [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–62. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Harney JA, Shimamura A, Howlett NG. Fanconi anemia: a multi-age cancer susceptibility syndrome. Pediatric Health. 2008;2:175–187. [Google Scholar]
- Hoskins EE, Gunawardena RW, Habash KB, Wise-Draper TM, Jansen M, Knudsen ES, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008;27:4798–808. doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Harney JA, Rego MA, Kolling FW, Glover TW. Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J Biol Chem. 2009;284:28935–42. doi: 10.1074/jbc.M109.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–94. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–6. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–19. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–6. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25:5875–84. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–70. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–8. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D’Andrea AD, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–5. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–23. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Podust VN, Podust LM, Goubin F, Ducommun B, Hubscher U. Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry. 1995;34:8869–75. doi: 10.1021/bi00027a039. [DOI] [PubMed] [Google Scholar]
- Prives C, Gottifredi V. The p21 and PCNA partnership: a new twist for an old plot. Cell Cycle. 2008;7:3840–6. doi: 10.4161/cc.7.24.7243. [DOI] [PubMed] [Google Scholar]
- Rego MA, Kolling FW, Howlett NG. The Fanconi anemia protein interaction network: casting a wide net. Mutat Res. 2009;668:27–41. doi: 10.1016/j.mrfmmm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–7. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–41. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–20. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–9. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–90. [PubMed] [Google Scholar]
- Wang X, Andreassen PR, D’Andrea AD. Functional Interaction of Monoubiquitinated FANCD2 and BRCA2/FANCD1 in Chromatin. Mol Cell Biol. 2004;24:5850–62. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27:3098–108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe M, Sonoda E, Hochegger H, Takeda S. Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair (Amst) 2004;3:1175–85. doi: 10.1016/j.dnarep.2004.03.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.