Abstract
Viral infection of mammalian host results in the activation of innate immune responses. Toll-like receptors (TLRs) have been shown to mediate the recognition of many types of pathogens, including viruses. The genomes of viruses possess unique characteristics that are not found in mammalian genomes, such as high CpG content and double-stranded RNA. These genomic nucleic acids serve as molecular signatures associated with viral infections. Here we show that TLR7 recognizes the single-stranded RNA viruses, vesicular stomatitis virus and influenza virus. The recognition of these viruses by plasmacytoid dendritic cells and B cells through TLR7 results in their activation of costimulatory molecules and production of cytokines. Moreover, this recognition required intact endocytic pathways. Mice deficient in either the TLR7 or the TLR adaptor protein MyD88 demonstrated reduced responses to in vivo infection with vesicular stomatitis virus. These results demonstrate microbial ligand recognition by TLR7 and provide insights into the pathways used by the innate immune cells in the recognition of viral pathogens.
The mammalian innate immune system recognizes the presence of invading pathogens by a family of receptors belonging to the Toll-like receptors (TLRs) (1, 2). Viral infection triggers an array of immune responses that effectively limit the replication and spread of viral infection. A predominant antiviral effector is the type I IFN system consisting of IFNα, IFNβ, IFNω, and IFNτ (3). Because replication of the viral genomes and virion synthesis occur intracellularly, a TLR responsible for viral recognition must be expressed in an appropriate intracellular compartment of the infected cells. Several characteristics of the viral genome, such as double-stranded RNA (dsRNA) and high CpG content, can serve as molecular signatures that can be distinguished by the host as nonself.
The host–virus interactions that lead to the secretion of type I IFNs by the infected cells are likely to involve pattern recognition through TLRs. Although most types of cells can produce IFNα and IFNβ on viral infection, plasmacytoid dendritic cells (pDCs) are particularly adept at secreting very high levels of type I IFNs in response to certain viruses (4). Recently, pDCs have been shown to recognize the dsDNA genome of the herpes simplex virus (HSV) type 1 and type 2 containing high CpG content through the TLR9/MyD88 pathway (5, 6). On the other hand, the recognition of RNA viruses is thought to involve TLR3, which recognizes dsRNA (7) that is present either as a part of their genomic structures or is generated during viral RNA replication. The dsRNA also triggers other antiviral pathways involving dsRNA-dependent PKR, oligoadenylate synthase, and RNase L (3). Not surprisingly, most RNA viruses have evolved strategies to sequester dsRNA by a variety of mechanisms to avoid activation of these antiviral pathways.
Despite the evasion mechanisms used by many viruses, pDCs remain capable of recognizing viruses and secreting high levels of type I IFNs. Influenza virus encodes NS1 protein, which binds to dsRNA and inhibits various antiviral pathways in infected cells (8). However, pDCs can still secrete high levels of IFNα on infection with influenza virus (9–11), whereas classical DCs can only do so when the NS1 protein is deleted (12). The molecular pathways involved in the pDC activation of antiviral immunity against single-stranded RNA (ssRNA) viruses are largely unknown.
In this study, we demonstrate the requirement for TLR7/MyD88 in the recognition of the ssRNA viruses, vesicular stomatitis virus (VSV) and influenza. Through the generation of TLR7-deficient mice, we determined that TLR7 is required for responsiveness to both VSV and influenza and, further, that recognition of these ssRNA viruses requires endosomal acidification. Finally, the in vivo ability of VSV to stimulate IFNα secretion depended on the functional expression of MyD88 and TLR7. These results present evidence for the requirement of TLR7 in the generation of immunity against ssRNA viruses and suggest an important role for this receptor in the recognition of a wide range of human pathogenic viruses.
Methods
Viruses. VSV-GFP (13) was propagated and assayed on BHK cells. In brief, confluent BHK cells were infected with VSV and incubated with virus until complete infection had occurred (≈1 day). Cells were centrifuged at 1,500 rpm for 5 min and supernatant was collected and saved. Virus was purified by layering on a 60%/10% sucrose gradient and centrifuging for 1.5 h at 27,000 rpm at 4°C. Purified virus was isolated from the interface and concentrated by centrifuging at 35,000 × g for 1 h at 4°C. The pellet was resuspended in sterile PBS and stored at –80°C. A standard plaque assay on BHK cells was performed on all purified virus stocks before use. Influenza virus (strain A/WSN/33) was a kind gift from P. Cresswell (Yale University). VSV-respiratory syncytial virus (RSV)-F (14) and VSV-GFP (13) were kindly provided by J. Kahn and J. Rose (Yale University), respectively. Sendai virus (Cantell) was purchased from American Type Culture Collection.
Animals. TLR7–/– mice were generated by the VelociGene approach as described (15). A pZEN6 cassette was constructed, in which a reporter LacZ gene was placed in tandem repeat with a neomycin resistance gene flanked by loxP sites that allowed for positive selection in both bacterial and mammalian cells. The pZEN6 cassette was ligated to double-stranded oligonucleotides and used for the generation of a bacterial artificial chromosomebased targeting vector. The upstream nucleotide (291 bp) corresponded to the promoter of the TLR7 gene, whereas the downstream oligonucleotide started at the amino acid 803 of the TLR7 gene. The 170-kb bacterial artificial chromosome-targeting vector was electroporated into the CJ7 embryonic stem cell line. β-Galactosidase activity was detected histochemically by 5-bromo-4-chloro-3-indolyl β-d-galactoside staining, as described (16). MyD88–/– (17), TLR3–/– (7), and TLR9–/– (18) mice (all on the B6x129 F2 background) had been described. Male and female B6x129 F2 mice were obtained from The Jackson Laboratory. Mice received i.v. tail vein injections of purified VSV-GFP (5 × 107 plaque-forming units (pfu) per mouse) in 150-μl volumes while restrained, and blood was collected at the indicated time points. All procedures used in this study complied with federal guidelines and institutional policies by the Yale animal care and use committee.
Antibodies. The following antibodies were used for the identification of pDCs and other cell types: anti-B220 (RA3–6B2), anti-CD69 (H1.2F3), and anti-CD86 (GL1), all purchased from BD Pharmingen. In addition, 120G8, a rat monoclonal antibody against pDCs (19) a kind gift from G. Trinchieri (Schering-Plough, Dardilly, France) was biotinylated and used for fluorescence-activated cell sorter staining.
Cell Preparation and Purification. Bone marrow cells were prepared from the femurs and tibiae of mice. A single cell suspension was prepared, and red blood cells were lysed. Total bone marrow populations were stimulated in the presence of the indicated reagents in 96-well, round-bottomed tissue culture plates at 1 × 106 cells in 200 μl of medium per well for 18 h. Similarly, total splenocytes were prepared by harvesting spleens, preparing a single-cell suspension, and lysing red blood cells. Splenocytes were stimulated in 96-well, round-bottomed tissue culture plates at 1 × 106 cells in 200 μl of medium per well for 18 h. For fluorescence-activated cell sorting of pDCs and non-pDCs, bone marrow cells were stained with biotin-conjugated 120G8 followed by phycoerythrin-conjugated streptavidin and CyChrome-conjugated anti-B220. Cells were washed extensively and sorted by using a FACSVantage (BD Biosciences). Sorted cells were incubated with various stimuli at 2 × 105 cells per well in 200 μl of medium. Eighteen hours later, cytokine levels were measured by using ELISA.
Bone Marrow Dendritic Cell Preparation and Stimulation. Bone marrow cells were isolated as described above and cultured in RPMI medium 1640 supplemented with 10% FCS/10 mM Hepes/50 μM β-mercaptoethanol/100 units/ml penicillin/100 μg/ml streptomycin and granulocyte-macrophage-colony-stimulating factor. Media were replaced every 2 days. On day 5, bone marrow DCs were stimulated with 1 μg/ml R848. On day 7, culture supernatants were harvested, and IL-6, IL-12 p40, and tumor necrosis factor α (TNFα) levels were measured by ELISA.
Peritoneal Macrophage Cell Stimulation. Macrophages were isolated from the peritoneal cavity of WT, TLR7–/–, TLR9–/–, and MyD88–/– mice 4 days after peritoneal injection with 3 ml of 4% thioglycollate medium. The cells were inoculated at a density of 1 × 106 cells per ml and plated into 96-well plates at 200 μl per well. After 2–3 h incubation, the cells were washed and stimulated with 1 μg/ml R848. After 48 h, supernatants were harvested, and IL-6, IL-12 p40, and TNF-α production was measured by ELISA.
In Vitro pDC Stimulation. The following reagents were used to stimulate cells in vitro: poly(I:C) (Amersham Pharmacia Biotech) at 25 μg/ml, R-848 (InvivoGen, San Diego) at 1 μg/ml, and stimulatory CpG 2084 (TCCTGACGTTGAAGT with a phosphodiester backbone; synthesized by Invitrogen) at 5 μg/ml. Purified VSV, influenza virus, or VSV-RSV-F was used at a final concentration of 5 × 106 pfu/ml. Sendai virus was used at 1.4 × 106 chicken embryo ID50/ml. To prevent endosome acidification, chloroquine or bafilomycin A1 (Sigma) was used before in vitro stimulation. In brief, bone marrow cells were preincubated with the indicated concentrations of chloroquine for 2 h or 10–500 nM bafilomycin A1 for 30 min at 37°C before the addition of various stimulants. Cells were then incubated for an additional 18 h at 37°C, and cell viability was confirmed by trypan blue exclusion assays. Supernatants were analyzed for cytokine production by ELISA.
ELISA Measurement of Cytokines. The levels of mouse IFNα were determined by ELISA as described (6). In brief, 96-well plates were coated with rat anti-mouse IFNα (mAb F-18, HyCult Biotechnology, Uden, The Netherlands) in carbonate buffer overnight. Nonspecific binding was blocked and plates were incubated with different concentrations of samples. Some wells were incubated with known concentrations of recombinant mouse IFNα standard in duplicate (HyCult Biotechnology). After an overnight incubation, plates were sequentially incubated with polyclonal rabbit anti-mouse IFNα (PBL Biomedical Laboratories, New Brunswick, NJ) for 1 h and with horseradish peroxidase-conjugated donkey anti-rabbit F(ab′)2 (Jackson ImmunoResearch) for 1 h. Plates were developed by using substrate solution TMB (eBioscience, San Diego), and the reaction was stopped with 2 N H2SO4. Optical density at 450 nm was measured and the concentration of IFNα was calculated from the standard curve. Concentrations of IL-12p40 were measured by using the OptEIA kit (BD PharMingen) according to the manufacturer's instructions, and TNFα and IL-6 were measured by using the eBioscience kits according to manufacturer's instructions.
Results
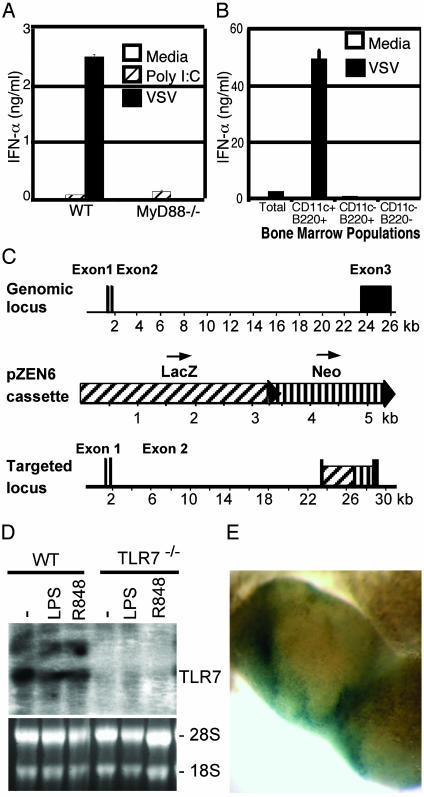
MyD88 Is Required for VSV Recognition. To first determine whether ssRNA virus recognition occurs through a TLR, bone marrow cells of mice deficient for the TLR adaptor protein, MyD88, were stimulated with VSV. MyD88 is an obligate adaptor protein downstream of many TLRs (20). To track the infection of cells by VSV, a recombinant VSV was used in which the virus contains an extra G gene fused to GFP at its cytoplasmic domain (13). The VSV-G/GFP virus has replication capacity equivalent to that of WT virus (13). The MyD88–/– bone marrow cells infected with VSV secreted significantly reduced IFNα levels compared with those of WT mice (Fig. 1A). Similar observations were made with nonrecombinant WT VSV (data not shown). On further analysis, pDCs were found to be the only bone marrow cell type capable of secreting IFNα in response to VSV (Fig. 1B). In contrast, a synthetic dsRNA, poly(I:C), elicited comparable responses from both WT and MyD88–/– cells (Fig. 1 A). These results suggested that a receptor that requires MyD88 activation is involved in VSV recognition.
Fig. 1.
Generation of TLR7–/– mice. (A) Total bone marrow cell suspensions were prepared from WT or MyD88–/– mice and cultured for 18 h with media (open), 25 μg/ml poly(I:C) (cross-hatched), or 5 × 106 pfu/ml VSV (filled). IFNα levels were measured from culture supernatants by ELISA. (B) pDCs were purified from the bone marrow of WT mice by flow cytometric cell sorting and cultured with media (open) or 5 × 106 pfu/ml VSV (filled) for 18 h. IFNα levels were measured from culture supernatants by ELISA. (C) Structure of the TLR7 gene, the pZEN6 reporter cassette, and the targeted mutated locus. Black arrow heads denote loxP sites. (D) Northern blot analysis of RNA from WT and TLR7–/– bone marrow-derived macrophages. RNA was prepared from cells that were cultured for 4 h with media (–), 0.1 mg/ml lipopolysaccharide, or 100 nM R848. Ethidium bromide staining of the RNA gel is included as control (lower gel). (E) Whole-mount view of reporter gene expression (LacZ reporter, in blue) in mesenteric lymph nodes from TLR7–/– mice.
Generation and Characterization of TLR7–/– Mice. The TLRs expressed by mouse pDCs include TLR1, TLR2, TLR5, TLR6, TLR7, TLR8, and TLR9 (21, 22). Because the entry and infection by VSV occur through endocytosis (23), we focused on the TLRs that are localized within the lysosomes. Recently, mouse TLR7 and TLR9 have been shown to localize within the endosomes and require endosomal maturation for signal transduction through MyD88 (24, 25). To examine whether TLR7 participates in the recognition of ssRNA viruses, we generated TLR7-deficient mice by a high-throughput and automated approach, termed VelociGene, that uses targeting vectors based on bacterial artificial chromosomes (Fig. 1C) (15). Gene-targeted TLR7-deficient (TLR7–/–) mice were born at the expected Mendelian ratio and grew to be healthy in specific-pathogen-free conditions. Northern blot analysis showed that TLR7 transcripts were absent in TLR7–/– mice (Fig. 1D). To determine the natural expression of the TLR7 gene, the expression of the lacZ reporter gene that was introduced under the TLR7 promoter in the TLR7–/– mice was determined by β-galactosidase assay on whole organs. Within the mesenteric lymph nodes, the site of TLR7 expression was confined to the perifollicular regions (Fig. 1E), consistent with the location of pDCs.
To demonstrate the functional deficiency and the specificity of the TLR7–/– mice, pDCs isolated from the bone marrow of these mice were stimulated with a known TLR7 ligand, R848 (26). IL-12 and IL-6 secretion from pDC isolated from the TLR7–/– mice was abolished, whereas responses to TLR9 ligands, synthetic CpG oligonucleotide or HSV-2, were intact (Fig. 2A). Further, responses to R848 by macrophages and bone marrow-derived DCs were completely eliminated in TLR7–/– mice (Fig. 2B).
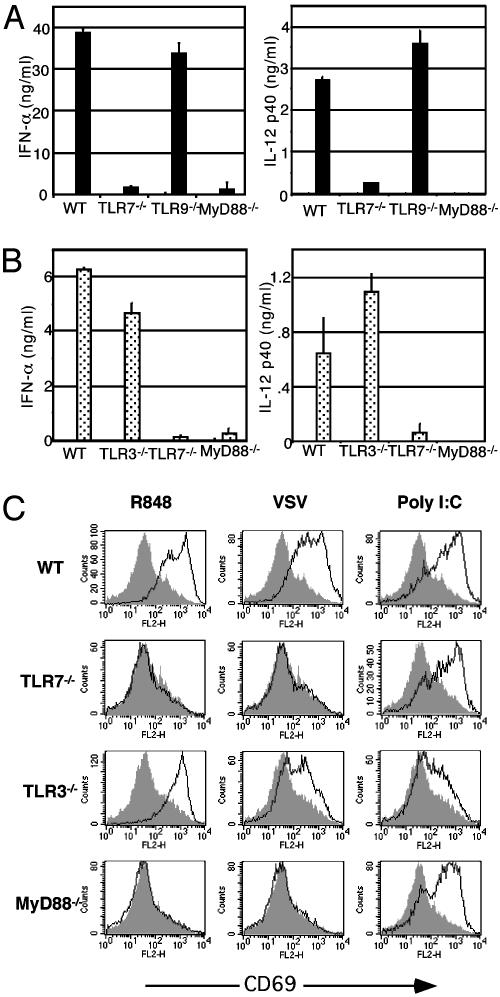
Fig. 2.
TLR7–/– mice display a functional deficiency in cytokine responses to R848. (A) Total bone marrow cell suspensions were prepared from WT or TLR7–/– mice and cultured for 18 h with media, 1 μg/ml R848, 5 μg/ml CpG, or 5 × 106 pfu/ml HSV-2. (B) Bone marrow-derived DCs and peritoneal macrophages were prepared from WT, TLR7–/–, TLR9–/–, and MyD88–/– mice and stimulated with 1 μg/ml R848 for 48 h. Cytokines were measured by ELISA.
TLR7 Is Required for Responsiveness to VSV and Influenza. We next examined the requirement of TLR7 in IFNα responses by pDC to VSV infection. The pDCs isolated from the bone marrow of TLR7–/– mice, like those from the MyD88–/– mice, secreted significantly reduced IFNα levels compared with WT mice in response to VSV infection in vitro. On the other hand, TLR9–/– pDCs elicited normal levels of IFNα and IL-12 when infected with VSV (Fig. 3A). These differences could not be accounted for by a difference in viral replication in these pDCs (Table 1). To extend our observation to other ssRNA viruses, we also tested the ability of bone marrow pDCs to respond to influenza virus. Only the WT and TLR3–/– pDCs secreted IFNα in response to inf luenza infection, whereas TLR7–/– and MyD88–/– cells failed to secrete IL-12 or IFNα (Fig. 3B). These results indicate that two distinct ssRNA viruses, VSV, a rhabdovirus, and influenza, an orthomyxovirus, are both recognized through the TLR7/MyD88 pathway.
Fig. 3.
TLR7 is required for responsiveness to VSV and influenza. (A) pDCs were purified from the bone marrow of WT, TLR7–/–, TLR9–/–, or MyD88–/– mice by flow cytometric cell sorting and infected with 5 × 106 pfu/ml VSV for 18 h. IFNα and IL-12 p40 levels were measured from culture supernatants by ELISA. (B) Total bone marrow cells were prepared from WT, TLR3–/–, TLR7–/–, and MyD88–/– mice and cultured with media or 5 × 106 pfu/ml influenza for 18 h. IFNα and IL-12 p40 levels were measured from culture supernatants by ELISA. (C) Splenocytes from WT, TLR3–/–, TLR7–/–, and MyD88–/– mice were cultured for 18 h with 1 μg/ml R848, 25 μg/ml poly(I:C), or 5 × 106 pfu/ml VSV (black lines) or medium alone (shaded area). Cells were stained and analyzed by flow cytometry. Histograms show CD69 expression levels in gated, B220+ cell populations.
Table 1. pDCs display low levels of VSV infection after 18 h of culture.
| GFP+ cells % | |
|---|---|
| WT | 0.61 |
| TLR7-/- | 0.64 |
| TLR9-/- | 0.40 |
| MyD88-/- | 0.44 |
FACS-sorted pDCs from WT, TLR7-/-, TLR9-/-, and MyD88-/- bone marrow were cultured for 18 h with 5 × 106 pfu/ml VSV-GFP. Cells were collected and analyzed by flow cytometry to quantify the infected GFP+ cells.
TLR3 Is Not Required for Responsiveness to VSV. Both human and mouse pDCs lack the expression of TLR3 (21, 27). Accordingly, TLR3–/– pDCs secreted WT levels of IFNα on infection with VSV (data not shown) and influenza (Fig. 3B). To test whether TLR3+ cells use this receptor in VSV recognition, we stimulated splenic B cells with VSV. The analysis of the expression of an activation marker CD69 on splenic B cells revealed that TLR3–/– cells were activated by VSV infection comparable with that of WT B cells (Fig. 3C). In contrast, in the absence of TLR7, B cells failed to up-regulate CD69 after infection with VSV (Fig. 3C). However, B cells responded to poly(I:C) activation in a TLR3-dependent manner (Fig. 3C), indicating functional TLR3 expression on these cells. Similar results were obtained when other activation markers were examined (data not shown). These results showed that TLR7, and not TLR3, plays a critical role in ssRNA virus recognition.
pDC Recognition of Viruses Requires Endosomal Acidification. TLR7 has been shown to bind to guanine nucleoside analogues (28), suggesting that the viral ligand might be the genomic RNA. To examine whether the viral genomic RNA can trigger TLR7, VSV virions were purified through sucrose density gradient, and RNA was isolated. High concentrations of naked VSV RNA did not elicit activation of pDCs (data not shown), likely because of rapid degradation by ribonucleases. Because the innate immune system cannot distinguish between the purified single-stranded viral RNA genome and the endogenous mammalian self-RNA, we reasoned that the intracellular location of the viral RNA must play an important role in viral recognition. A VSV virion binds to a cellular receptor and enters the cell by receptor-mediated endocytosis. On fusion with lysosomes, the acidic environment allows fusion of the viral membrane and the membrane of the lysosome, releasing the viral nucleocapsid into the cytosol (29). However, this fusion process is not entirely efficient, leading to the degradation of VSV virions within the lysosome, as evidenced by the fact that VSV-infected cells release degraded viral material after 15 min of infection (23). Because TLR7 recognition of guanosine analogue loxoribine and resiquimod R848 both occur within the lysosomal compartment (25), we hypothesized that the viral RNA recognition takes place within the lysosome and that endocytically introduced RNA could trigger TLR7 activation in pDCs.
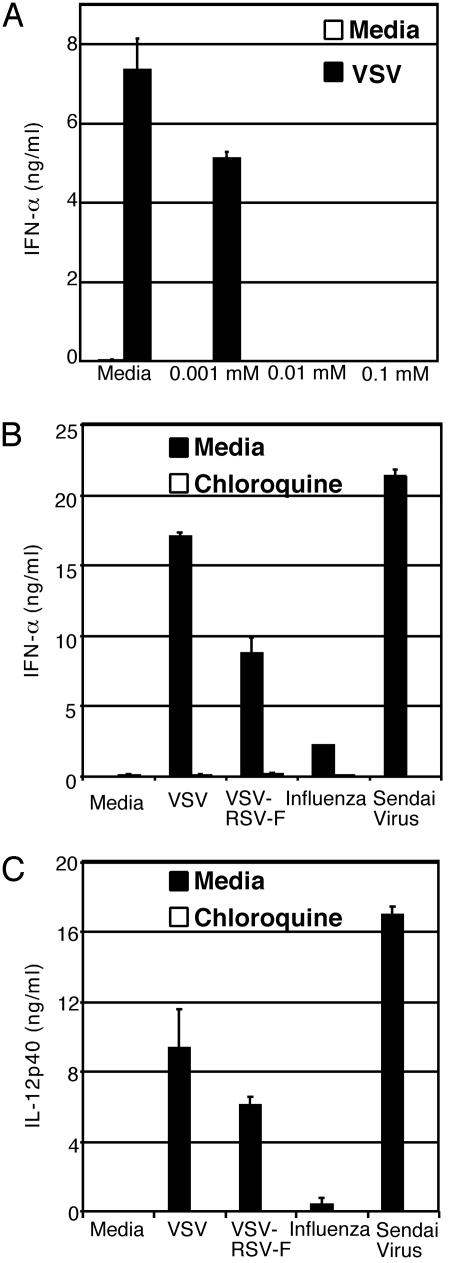
To test this hypothesis, pDCs from WT mice were stimulated with VSV in the presence of an inhibitor of lysosomal acidification, chloroquine (Fig. 4A). Both IL-12 and IFNα secretion from pDCs was inhibited in a dose-dependent manner by chloroquine, indicating that the acidification of the lysosomes is important for cellular responses to VSV. A similar inhibition was observed with another inhibitor of lysosomal acidification, Bafilomycin A (data not shown). A caveat of this experiment is that VSV infection also requires acidification of the lysosomes (23). To circumvent this complication and to test the hypothesis that endocytic uptake of RNA is necessary to trigger TLR7 activation, we used a recombinant VSV that expresses the fusion (F) protein of the RSV, VSV-RSV-F. This virus expresses both RSV-F and VSV-G proteins and mediates fusion at both the plasma membrane in a pH-independent manner (by RSV-F protein; ref. 30) and through the pH-dependent endocytic pathway (by VSV-G protein; ref. 14). Infection of bone marrow cells with VSV-RSV-F virus resulted in reduced but significant secretion of IL-12 and IFNα (Fig. 4 B and C). Moreover, treatment of cells with chloroquine resulted in complete reduction of IL-12 and IFNα secretion from cells infected with either WT VSV or VSV-RSV-F (Fig. 4 B and C). These observations also applied to influenza infection (Fig. 4 B and C). Because the plasma membrane fusion by the VSV-RSV-F virus may not be as efficient as that used by viruses that exclusively fuse with the plasma membrane, we further examined the requirement for acidification of endosomes in pDC activation by using Sendai virus. The members of the Paramyxoviridae, such as the Sendai virus, fuse exclusively with the plasma membrane and release the viral genome into the cytoplasm. Our results demonstrated that pDC recognition of Sendai virus also occurs in acidified endosomes (Fig. 4 B and C). These results indicated that endocytic acidification plays a critical role in ssRNA viral recognition by pDCs regardless of the mechanisms used by the viruses to enter a target cell.
Fig. 4.
Recognition of RNA viruses requires endosomal acidification. WT bone marrow cells were pretreated for 2 h with media alone or chloroquine at the indicated concentrations. (A) After the addition of 5 × 106 pfu/ml VSV, cells were cultured for an additional 18 h, and supernatants were collected and IFNα levels were measured by ELISA. (B and C) Chloroquine (0.1 mM) or media-treated cells were stimulated with 5 × 106 pfu/ml VSV, VSV-RSV-F, or influenza, or 1.4 × 106 chicken embryo ID50/ml Sendai virus for an additional 18 h and IFNα (B) and IL-12 p40 (C) were measured by ELISA.
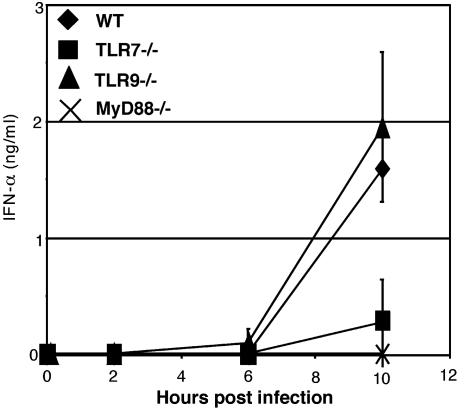
In Vivo IFNα Production in Response to VSV Requires MyD88 and TLR7. Finally, to demonstrate in vivo relevance of TLR7-mediated VSV recognition, serum IFNα levels were measured in mice systemically infected with VSV as described (31). Although WT and TLR9–/– mice secreted similar levels of IFNα, TLR7–/– mice had severely diminished levels of IFNα in their sera (Fig. 5). Mice deficient in MyD88 completely failed to secrete IFNα. These results demonstrated that TLR7 is important in the innate antiviral response against a ssRNA virus such as VSV in vivo.
Fig. 5.
In vivo IFNα production in response to VSV requires MyD88 and TLR7. WT, TLR7–/–, TLR9–/–, and MyD88–/– mice infected i.v. with 5 × 107 pfu of VSV. Sera were collected at the indicated time points and assayed for IFNα by ELISA.
Discussion
It is well established that TLRs play a crucial role in the recognition of microbial pathogens, thereby inducing innate immune responses in mammalian hosts. Recent studies have demonstrated the TLR9/MyD88 dependence of pDC recognition of the DNA viruses HSV-1 and HSV-2 (5, 6), and this study highlights a similar MyD88 dependence of pDC recognition of the ssRNA viruses. However, in the ssRNA viruses, TLR7, rather than TLR3 or TLR9, is required for viral recognition and the subsequent cytokine production by pDCs and activation of B cells.
Whereas IFNβ production induced by the dsRNA analogue poly(I:C) depends on the adaptor molecule TRIF in a MyD88-independent fashion (32, 33), our data here demonstrated complete dependence on MyD88 in type I IFN secretion from pDCs. These results indicate that ssRNA viruses are recognized through a pathway distinct from that used for dsRNA viruses. Further, our results indicate that even if dsRNA was to be generated during VSV or influenza infection, it does not engage the TLR3-TRIF pathway for activation. The exact viral ligand that triggers TLR7 is unknown. The known abilities of TLR7 to recognize ribonucleoside analogues suggests that viral genomic RNA or RNA species generated during viral replication can bind to TLR7 and transduce signals by MyD88. Although we cannot formally rule out viral proteins in TLR7 activation, such ligands must be present in both influenza and VSV, which are two viruses belonging to distinct families that do not share obvious homologies in any of the encoded proteins except the M protein, which is weakly homologous (34).
Many viruses use endocytosis as a means to gain entry into mammalian host cells. The pDCs have been known for many years to be the type I IFN-producing cells (35), owing to their unparalleled ability to secrete high levels of type I IFNs in response to viruses. These cells are able to survey the environment for the presence of viruses by endocytic uptake of the virions, regardless of whether their natural mode of entry is fusion at the plasma membrane (e.g., HSV, VSV-RSV-F, and Sendai virus) or at the endosomal membrane (e.g., VSV and influenza). Our results suggest that the single-stranded oligoribonucleotides introduced into the endosomal but not cytoplasmic compartment triggers TLR7 activation in mammalian cells, irrespective of the entry mechanisms used by the viruses. Further, the introduction of viral RNA genomes by VSV-RSV-F and Sendai virus into the cytoplasm is not sufficient to induce cytokine secretion from pDCs. Thus, the endocytic location of the viral RNA serves as a molecular recognition signature for RNA viruses, and the strategic localization of TLR7 within the lysosome is important in this pathway of viral detection. By rapidly producing high levels of type I IFNs, pDCs rarely become infected with viruses taken up through the endocytic pathways. Unlike myeloid cells, pDCs do not possess efficient phagocytic abilities to take up large particles or apoptotic cells (36). Thus, the details of viral uptake and intracellular processing by the pDCs resulting in their potent antiviral functions will require further investigation.
Our demonstration of TLR7 mediating viral recognition provides a molecular target for both acute and chronic viral infections in humans. It will be important to determine whether certain viruses, in particular, those that establish chronic infections, have developed evasion mechanisms against this recognition pathway. The use of TLR7 agonists for antiviral vaccines and immunological interventions to clear existing viral infections provide promising approaches in the search for new antiviral treatments.
Acknowledgments
We thank J. S. Kahn and P. Cresswell for providing viruses, G. Trinchieri for providing an antibody, and J. K. Rose for providing both technical and intellectual expertise on VSV and a critical review of the manuscript. This work was supported by grants from the National Institutes of Health (to A.I. and R.A.F.) and Wyeth-Lederle Vaccine Young Investigator Award (to A.I.). A.S. is supported by the James Hudson Brown–Alexander Brown Cox postdoctoral fellowship. L.A. was supported by National Institutes of Health Grants AI053279 and AI055740 (to R.A.F.). R.A.F. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: TLR, Toll-like receptor; VSV, vesicular stomatitis virus; DC, dendritic cell; pDC, plasmacytoid DC; ssRNA, single-stranded RNA; pfu, plaque-forming unit; HSV, herpes simplex virus; dsRNA, double-stranded RNA; TNFα, tumor necrosis factor α; RSV, respiratory syncytial virus.
References
- 1.Akira, S. (2001) Adv. Immunol. 78, 1–56. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov, R. & Janeway, C., Jr. (2000) Immunol. Rev. 173, 89–97. [DOI] [PubMed] [Google Scholar]
- 3.Katze, M. G., He, Y. & Gale, M., Jr. (2002) Nat. Rev. Immunol. 2, 675–687. [DOI] [PubMed] [Google Scholar]
- 4.Colonna, M., Krug, A. & Cella, M. (2002) Curr. Opin. Immunol. 14, 373–379. [DOI] [PubMed] [Google Scholar]
- 5.Krug, A., Luker, G. D., Barchet, W., Leib, D. A., Akira, S. & Colonna, M. (2004) Blood 103, 1433–1437. [DOI] [PubMed] [Google Scholar]
- 6.Lund, J., Sato, A., Akira, S., Medzhitov, R. & Iwasaki, A. (2003) J. Exp. Med. 198, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732–738. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Sastre, A. (2002) Microb. Infect. 4, 647–655. [DOI] [PubMed] [Google Scholar]
- 9.Cella, M., Facchetti, F., Lanzavecchia, A. & Colonna, M. (2000) Nat. Immunol. 4, 305–310. [DOI] [PubMed] [Google Scholar]
- 10.Nakano, H., Yanagita, M. & Gunn, M. D. (2001) J. Exp. Med. 194, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asselin-Paturel, C., Boonstra, A., Dalod, M., Durand, I., Yessaad, N., Dezutter-Dambuyant, C., Vicari, A., O'Garra, A., Biron, C., Briere, F. & Trinchieri, G. (2001) Nat. Immunol. 2, 1144–1150. [DOI] [PubMed] [Google Scholar]
- 12.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P. & Reis e Sousa, C. (2003) Nature 424, 324–328. [DOI] [PubMed] [Google Scholar]
- 13.Dalton, K. P. & Rose, J. K. (2001) Virology 279, 414–421. [DOI] [PubMed] [Google Scholar]
- 14.Kahn, J. S., Schnell, M. J., Buonocore, L. & Rose, J. K. (1999) Virology 254, 81–91. [DOI] [PubMed] [Google Scholar]
- 15.Valenzuela, D. M., Murphy, A. J., Frendewey, D., Gale, N. W., Economides, A. N., Auerbach, W., Poueymirou, W. T., Adams, N. C., Rojas, J., Yasenchak, J., et al. (2003) Nat. Biotechnol. 21, 652–659. [DOI] [PubMed] [Google Scholar]
- 16.Schlaeger, T. M., Qin, Y., Fujiwara, Y., Magram, J. & Sato, T. N. (1995) Development (Cambridge, U.K.) 121, 1089–1098. [DOI] [PubMed] [Google Scholar]
- 17.Adachi, O., Kawai, T., Takeda, K., Matsumoto, M., Tsutsui, H., Sakagami, M., Nakanishi, K. & Akira, S. (1998) Immunity 9, 143–150. [DOI] [PubMed] [Google Scholar]
- 18.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K. & Akira, S. (2000) Nature 408, 740–745. [DOI] [PubMed] [Google Scholar]
- 19.Asselin-Paturel, C., Brizard, G., Pin, J. J., Briere, F. & Trinchieri, G. (2003) J. Immunol. 171, 6466–6477. [DOI] [PubMed] [Google Scholar]
- 20.Muzio, M., Ni, J., Feng, P. & Dixit, V. M. (1997) Science 278, 1612–1615. [DOI] [PubMed] [Google Scholar]
- 21.Edwards, A. D., Diebold, S. S., Slack, E. M., Tomizawa, H., Hemmi, H., Kaisho, T., Akira, S. & Reis e Sousa, C. (2003) Eur. J. Immunol. 33, 827–833. [DOI] [PubMed] [Google Scholar]
- 22.Boonstra, A., Asselin-Paturel, C., Gilliet, M., Crain, C., Trinchieri, G., Liu, Y. J. & O'Garra, A. (2003) J. Exp. Med. 197, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matlin, K. S., Reggio, H., Helenius, A. & Simons, K. (1982) J. Mol. Biol. 156, 609–631. [DOI] [PubMed] [Google Scholar]
- 24.Ahmad-Nejad, P., Hacker, H., Rutz, M., Bauer, S., Vabulas, R. M. & Wagner, H. (2002) Eur. J. Immunol. 32, 1958–1968. [DOI] [PubMed] [Google Scholar]
- 25.Heil, F., Ahmad-Nejad, P., Hemmi, H., Hochrein, H., Ampenberger, F., Gellert, T., Dietrich, H., Lipford, G., Takeda, K., Akira, S., et al. (2003) Eur. J. Immunol. 33, 2987–2997. [DOI] [PubMed] [Google Scholar]
- 26.Hemmi, H., Kaisho, T., Takeuchi, O., Sato, S., Sanjo, H., Hoshino, K., Horiuchi, T., Tomizawa, H., Takeda, K. & Akira, S. (2002) Nat. Immunol. 3, 196–200. [DOI] [PubMed] [Google Scholar]
- 27.Jarrossay, D., Napolitani, G., Colonna, M., Sallusto, F. & Lanzavecchia, A. (2001) Eur. J. Immunol. 31, 3388–3393. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J., Chuang, T. H., Redecke, V., She, L., Pitha, P. M., Carson, D. A., Raz, E. & Cottam, H. B. (2003) Proc. Natl. Acad. Sci. USA 100, 6646–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White, J., Matlin, K. & Helenius, A. (1981) J. Cell Biol. 89, 674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh, E. E. & Hruska, J. (1983) J. Virol. 47, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barchet, W., Cella, M., Odermatt, B., Asselin-Paturel, C., Colonna, M. & Kalinke, U. (2002) J. Exp. Med. 195, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., et al. (2003) Nature 424, 743–748. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K. & Akira, S. (2003) Science 301, 640–643. [DOI] [PubMed] [Google Scholar]
- 34.Rose, J. K., Doolittle, R. F., Anilionis, A., Curtis, P. J. & Wunner, W. H. (1982) J. Virol. 43, 361–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perussia, B., Fanning, V. & Trinchieri, G. (1985) Nat. Immun. Cell Growth Regul. 4, 120–137. [PubMed] [Google Scholar]
- 36.Stent, G., Reece, J. C., Baylis, D. C., Ivinson, K., Paukovics, G., Thomson, M. & Cameron, P. U. (2002) Cytometry 48, 167–176. [DOI] [PubMed] [Google Scholar]