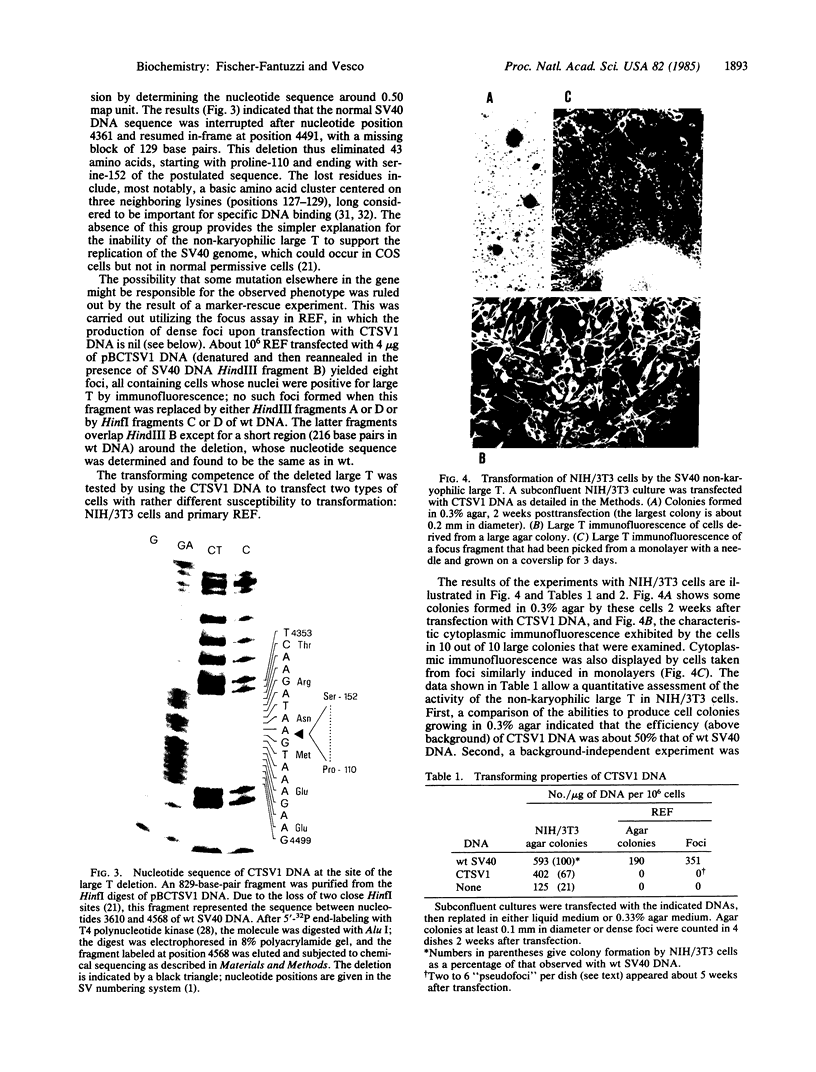
Abstract
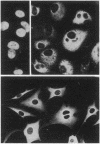
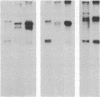
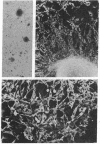
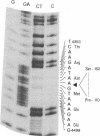
We have characterized a simian virus 40 (SV40) mutant, derived from the viral DNA insertion present in simian cell transformants, which carries a deletion affecting the NH2-terminal region of the SV40 large tumor antigen. This mutant protein is 6% smaller than normal, has lost the typical nuclear localization of the SV40 large tumor antigen, and accumulates in the cytoplasm. The deletion begins at nucleotide position 4490 of the SV40 DNA and ends in-frame at nucleotide position 4362. The missing 43 amino acids begin with proline-110 and end with serine-152 of the predicted sequence; they include a cluster of basic residues, presumably important for the viral origin-DNA binding, and most of the phosphorylation sites present in the NH2-terminal half of the molecule. The protein can still be phosphorylated considerably in vivo. This mutant viral genome is replication-defective but has conserved the competence to transform established cells, such as NIH/3T3 cells. Transfection of cloned mutant DNA into such cells resulted in the production of full transformants. Full transformants were not produced in similar transfections carried out in primary rat embryo fibroblasts, although some primary transfectants expressing the non-karyophilic large tumor antigen might be considered minimally transformed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carmichael G., Schaffhausen B. S., Mandel G., Liang T. J., Benjamin T. L. Transformation by polyoma virus is drastically reduced by substitution of phenylalanine for tyrosine at residue 315 of middle-sized tumor antigen. Proc Natl Acad Sci U S A. 1984 Feb;81(3):679–683. doi: 10.1073/pnas.81.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. S., Pater M. M., Hutchinson N. I., di Mayorca G. Transformation by purified early genes of simian virus 40. Virology. 1984 Mar;133(2):341–353. doi: 10.1016/0042-6822(84)90400-8. [DOI] [PubMed] [Google Scholar]
- Clark R., Peden K., Pipas J. M., Nathans D., Tjian R. Biochemical activities of T-antigen proteins encoded by simian virus 40 A gene deletion mutants. Mol Cell Biol. 1983 Feb;3(2):220–228. doi: 10.1128/mcb.3.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C. E., Murphy D., Lovett M., Rigby P. W. A fragment of the SV40 large T-antigen gene transforms. Nature. 1982 Sep 2;299(5878):59–61. doi: 10.1038/299059a0. [DOI] [PubMed] [Google Scholar]
- Colby W. W., Shenk T. Fragments of the simian virus 40 transforming gene facilitate transformation of rat embryo cells. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5189–5193. doi: 10.1073/pnas.79.17.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Crawford L. V., Berg P. Simian virus 40 mutants with deletions at the 3' end of the early region are defective in adenovirus helper function. J Virol. 1979 Jun;30(3):683–691. doi: 10.1128/jvi.30.3.683-691.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosman D. J., Tevethia M. J. Characterization of a temperature-sensitive, DNA-positive, nontransforming mutant of simian virus 40. Virology. 1981 Jul 30;112(2):605–624. doi: 10.1016/0042-6822(81)90306-8. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. Nucleocytoplasmic segregation of proteins and RNAs. Cell. 1983 Apr;32(4):1021–1025. doi: 10.1016/0092-8674(83)90285-4. [DOI] [PubMed] [Google Scholar]
- Deppert W., Walter G. Domains of simian virus 40 large T-antigen exposed on the cell surface. Virology. 1982 Oct 15;122(1):56–70. doi: 10.1016/0042-6822(82)90377-4. [DOI] [PubMed] [Google Scholar]
- Fantuzzi L. F., Vesco C. Molecular cloning and restriction mapping of a simian virus 40 deletion mutant derived from simian transformants expressing a non-karyophilic T antigen. FEBS Lett. 1984 May 7;170(1):125–130. doi: 10.1016/0014-5793(84)81383-6. [DOI] [PubMed] [Google Scholar]
- Fey G., Lewis J. B., Grodzicker T., Bothwell A. Characterization of a fused protein specified by the adenovirus type 2-simian virus 40 hybrid Ad2+ND1 dp2. J Virol. 1979 Apr;30(1):201–217. doi: 10.1128/jvi.30.1.201-217.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Fantuzzi L., Vesco C. Cold-sensitive growth of simian virus 40 in semipermissive variants of CV1 cells. J Virol. 1982 Sep;43(3):791–799. doi: 10.1128/jvi.43.3.791-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Fantuzzi L., Vesco C. Murine cell complementation of a cold-sensitive defect for simian virus 40 replication in simian cells. Mol Cell Biol. 1982 Sep;2(9):1155–1157. doi: 10.1128/mcb.2.9.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanti N., Jonak G. J., Soprano K. J., Floros J., Kaczmarek L., Weissman S., Reddy V. B., Tilghman S. M., Baserga R. Characterization and biological activity of cloned simian virus 40 DNA fragments. J Biol Chem. 1981 Jun 25;256(12):6469–6474. [PubMed] [Google Scholar]
- Gluzman Y., Ahrens B. SV40 early mutants that are defective for viral DNA synthesis but competent for transformation of cultured rat and simian cells. Virology. 1982 Nov;123(1):78–92. doi: 10.1016/0042-6822(82)90296-3. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R. R., Mercer W. E., Liu H. T., Baserga R. Transforming potential of deletion mutants of the SV40 T antigen coding gene in Syrian hamster cells. Virology. 1984 Apr 15;134(1):220–229. doi: 10.1016/0042-6822(84)90287-3. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lanford R. E., Butel J. S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984 Jul;37(3):801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- Lanford R. E., Butel J. S. Inhibition of nuclear migration of wild-type SV40 tumor antigen by a transport-defective mutant of SV40-adenovirus 7 hybrid virus. Virology. 1980 Sep;105(2):303–313. doi: 10.1016/0042-6822(80)90032-x. [DOI] [PubMed] [Google Scholar]
- Lanford R. E., Butel J. S. Intracellular transport of SV40 large tumor antigen: a mutation which abolishes migration to the nucleus does not prevent association with the cell surface. Virology. 1982 May;119(1):169–184. doi: 10.1016/0042-6822(82)90074-5. [DOI] [PubMed] [Google Scholar]
- Linzer D. I., Levine A. J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979 May;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- Melero J. A., Stitt D. T., Mangel W. F., Carroll R. B. Identification of new polypeptide species (48-55K) immunoprecipitable by antiserum to purified large T antigen and present in SV40-infected and -transformed cells. Virology. 1979 Mar;93(2):466–480. doi: 10.1016/0042-6822(79)90250-2. [DOI] [PubMed] [Google Scholar]
- Morrison B., Kress M., Khoury G., Jay G. Simian virus 40 tumor antigen: isolation of the origin-specific DNA-binding domain. J Virol. 1983 Jul;47(1):106–114. doi: 10.1128/jvi.47.1.106-114.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C., Graessmann A., Graessmann M. Mapping of early SV40-specific functions by microinjection of different early viral DNA fragments. Cell. 1978 Oct;15(2):579–585. doi: 10.1016/0092-8674(78)90026-0. [DOI] [PubMed] [Google Scholar]
- Pintel D., Bouck N., di Mayorca G. Separation of lytic and transforming functions of the simian virus 40 A region: two mutants which are temperature sensitive for lytic functions have opposite effects on transformation. J Virol. 1981 May;38(2):518–528. doi: 10.1128/jvi.38.2.518-528.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C., Barnet B., Scheller A., Khoury G., Jay G. Discrete regions of simian virus 40 large T antigen are required for nonspecific and viral origin-specific DNA binding. J Virol. 1982 Jul;43(1):73–82. doi: 10.1128/jvi.43.1.73-82.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M., Cowie A., Carr A., Glaichenhaus N., Kamen R., Cuzin F. The roles of individual polyoma virus early proteins in oncogenic transformation. Nature. 1982 Dec 23;300(5894):713–718. doi: 10.1038/300713a0. [DOI] [PubMed] [Google Scholar]
- Risser R., Pollack R. Factors affecting the frequency of transformation of rat embryo cells by simian virus 40. Virology. 1979 Jan 15;92(1):82–90. doi: 10.1016/0042-6822(79)90216-2. [DOI] [PubMed] [Google Scholar]
- Ruley H. E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983 Aug 18;304(5927):602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Scheidtmann K. H., Echle B., Walter G. Simian virus 40 large T antigen is phosphorylated at multiple sites clustered in two separate regions. J Virol. 1982 Oct;44(1):116–133. doi: 10.1128/jvi.44.1.116-133.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L., Danna K. J. Simian virus 40 deletion mutants that transform with reduced efficiency. Mol Cell Biol. 1983 Mar;3(3):484–489. doi: 10.1128/mcb.3.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staufenbiel M., Deppert W. Different structural systems of the nucleus are targets for SV40 large T antigen. Cell. 1983 May;33(1):173–181. doi: 10.1016/0092-8674(83)90346-x. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Mutant of simian virus 40 large T-antigen that is defective for viral DNA synthesis, but competent for transformation of cultured rat cells. J Virol. 1982 Jun;42(3):854–864. doi: 10.1128/jvi.42.3.854-864.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P., Robb J. A., Widmer C., Ozer H. L. Altered protein metabolism in infection by the late tsB11 mutant of simian virus 40. J Virol. 1974 Oct;14(4):997–1007. doi: 10.1128/jvi.14.4.997-1007.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornow J., Cole C. N. Nonviable mutants of simian virus 40 with deletions near the 3' end of gene A define a function for large T antigen required after onset of viral DNA replication. J Virol. 1983 Sep;47(3):487–494. doi: 10.1128/jvi.47.3.487-494.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderame M. F., Kohtz D. S., Pollack R. E. 94,000- and 100,000-molecular-weight simian virus 40 T-antigens are associated with the nuclear matrix in transformed and revertant mouse cells. J Virol. 1983 May;46(2):575–583. doi: 10.1128/jvi.46.2.575-583.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roy F., Fransen L., Fiers W. Improved localization of phosphorylation sites in simian virus 40 large T antigen. J Virol. 1983 Jan;45(1):315–331. doi: 10.1128/jvi.45.1.315-331.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Eb A. J., Graham F. L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65(1):826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]