Abstract
The contribution of HLA class II-restricted CD4+ T cell responses to HIV immune control is poorly defined. Here, we delineated novel peptide-DRB1 restrictions in functional assays and analyzed the host genetic effects of HLA-DRB1 alleles on HIV viremia in a large cohort of HIV controllers and progressors (n=1085). We found distinct stratifications in the effect of HLA-DRB1 alleles on HIV viremia, with DRB1*15:02 significantly associated with low viremia (P=0.003, q=0.04) and DRB1*03:01 significantly associated with high viremia (P=0.004, q=0.04). Interestingly, a sub-group of HLA-DRB1 alleles linked with low viremia showed the ability to promiscuously present a larger breadth of peptides with lower functional avidity when compared to HLA-DRB1 alleles linked with high viremia (p=0.018). Our data provide systematic evidence that HLA-DRB1 allele expression significantly impacts the durable control of HIV replication, an effect that appears to be mediated primarily by the protein-specificity of HIV-specific CD4+ T cell responses to Gag and Nef.
Control of viral infections is critically dependent on CD4+ T cell responses (1–3). CD4+ T cells provide help for the generation of high-affinity antibodies and induction of memory in CD8+ T cells and B cells (4), and may also directly exert anti-viral activity (2–3,5). Yet the role of virus-specific CD4+ T cell responses in HIV infection is less clear (6) since HIV preferentially infects HIV-specific CD4+ T cells (7). We have previously demonstrated that the breadth and specificity of HIV-specific CD4+ T cell responses is significantly associated with low viremia (8). However, little is known about the HLA class II restriction of these responses and how HLA class II alleles impact HIV control. The strongest genetic association with HIV control is undoubtedly mediated by HLA class I alleles such as B*57 (9–10), yet at least two studies (11–12) have suggested that HLA class II alleles, particularly at the DRB1 locus, may also exert an important influence on HIV control.
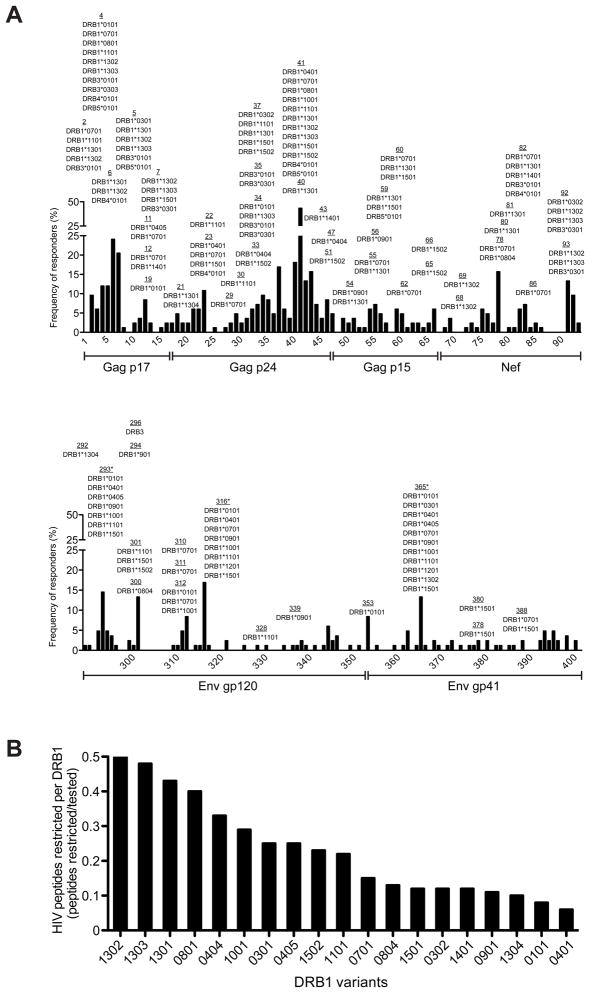
To define the role of DRB1-restricted HIV-specific CD4+ T cell responses on viral control, we tested 201 HIV-specific CD4+ T cell lines spanning 67 peptide specificities from 42 HIV-infected individuals in functional HLA-DRB1 restriction assays. We observed a high level of ‘promiscuity’, a characteristic feature of antigen-specific CD4+ T cell recognition in which a single peptide can be presented in the context of multiple HLA class II variants expressed by different individuals (13) (Fig 1a). Strikingly, of all peptides tested across the HIV proteome (Supplementary Table 1), 43% were restricted by two or more HLA-DR variants (Supplementary Table 2). OLP-41 in Gag p24, which is the most highly targeted CD4 peptide, showed the highest level of promiscuity with 12 distinct HLA-DR restrictions. We also evaluated the peptide-binding capacity (13) of five immunodominant CD4 peptides to 13 distinct HLA-DRB1 variants. These showed comparable or even higher peptide-binding promiscuity (Supplementary Table 3), thus confirming our in vitro restriction data. Furthermore, detailed analysis of OLP-41 revealed only limited variability in the minimal stimulatory epitopes presented by different HLA-DRB1 variants (Supplementary Figure S1), confirming that promiscuous HLA-DRB1 variants share largely overlapping epitope-binding registers (14–15). This high degree of peptide-binding promiscuity is likely mediated by the open conformation of HLA class II, which allows long CD4 peptides to extend beyond the HLA binding groove.
Figure 1. DRB alleles exhibit promiscuity and specificity in the number of HIV-specific CD4 T cell peptides presented.
(a) CD4 T cell responses to Gag, Nef and Env proteins are characterized by a high-degree of promiscuity in peptide-DRB restrictions. The HLA-DR restrictions are shown per peptide. Solid black bars indicate the observed percentage of HIV-infected individuals responding to each individual peptide within Gag (1–66), Nef (67–93) and Env sub-proteins gp120 (289–354) and gp41 (355–400). The asterisk (*) denotes three Env peptides for which we determined peptide promiscuity using a well-characterized peptide-HLA-DRB1 binding assay (13) as we were limited by insufficient sample availability of HIV chronic progressors who target these peptides.
(b) Marked differences exist in the number of HIV-specific CD4 peptides restricted by each DRB1 allele. The number of confirmed restrictions per DRB1 allele was normalized to the overall number of CD4 T cell lines tested to determine the overall fraction of HIV peptides restricted per DRB1 allele.
Importantly, despite the high degree of HLA-DRB1 binding promiscuity, we noted marked differences in the number of peptides restricted by each DRB1 variant (Fig. 1b). Variants such as DRB1*01:01 and *04:01 exhibited the lowest contribution, with only 3 confirmed peptides, while in contrast, DRB1*13:01, *13:02 and *13:03 restricted a total of 18, 11 and 8 HIV-specific CD4 peptides, respectively. This observation is intriguing since the expression of DRB1*13 variants, particularly DRB1*13:03, have been associated with lower HIV viral load (11–12).
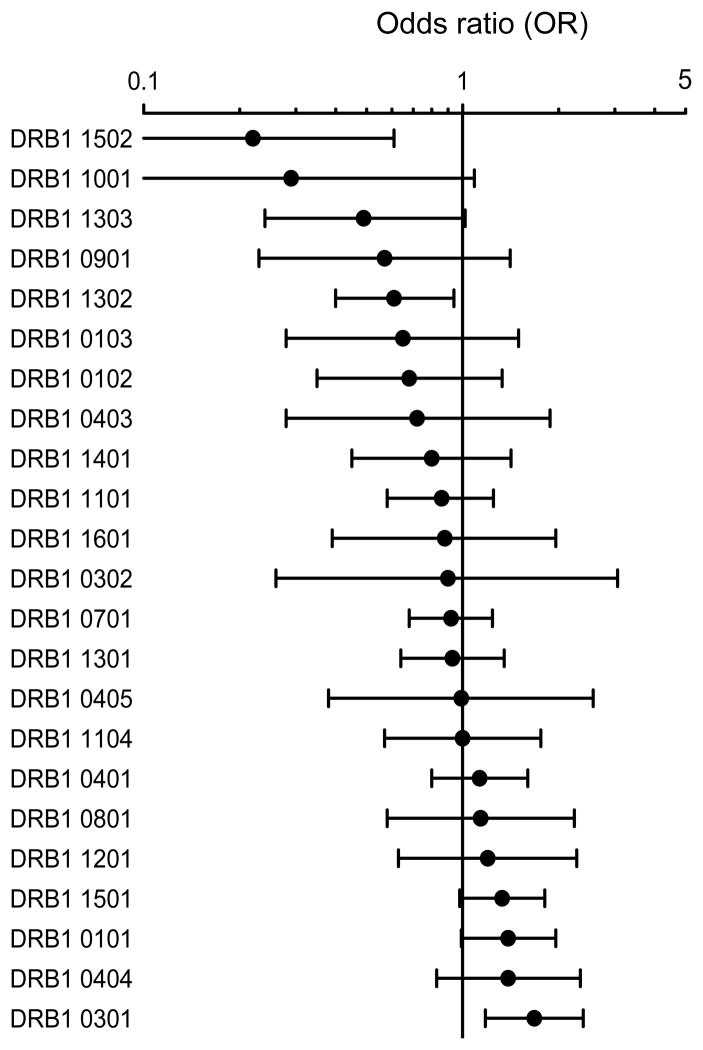
We next examined the relationship between HLA-DRB1 allele expression and viral load in a large cohort of 1085 treatment-naive HIV-infected individuals of European ancestry from the combined MGH and SCOPE cohorts. Subjects were categorized into two groups, HIV controllers (<2,000 HIV RNA copies/ml) and HIV progressors (>10,000 HIV RNA copies/ml), and we then evaluated the effect of individual HLA-DRB1 allele expression on mean viral load in a logistic regression model. An odds ratio (OR) was used to analyze the strength of the associations, with an OR of <1 indicative of HIV control, and an OR of >1 indicative of relatively poor HIV control. The well-defined HLA class I ‘protective’ alleles, HLA-B*57 and B*27, and the ‘risk’ allele B*35Px (9,16) were used as co-variables in this model to eliminate their confounding effects on HIV viremia. Interestingly, although we observed a wide range in the confidence intervals, a hierarchy of more protective to more hazardous DRB1 alleles was observed (Fig. 2). Specifically, subjects positive for HLA-DRB1*15:02 exhibited the lowest OR (0.22) and expression of this allele was significantly associated with control of HIV viremia (P=0.003, q=0.04). In contrast, HLA-DRB1*03:01 exhibited the highest OR (1.68) and was significantly associated with high viremia (P=0.004, q=0.04). We found distinct stratifications in the OR associated with other HLA-DRB1 alleles but we were underpowered to determine statistical differences in their association with HIV control after correction for multiple comparisons. While some of the HLA class II alleles associated with low viremia are relatively rare at the population level, we found no evidence of a ‘rare allele’ effect as suggested for HLA class I (17).
Figure 2. Association of HLA-DRB1 allele expression with HIV viral load at the population level.
Association of DRB1 alleles with differential odds ratios (OR) in a large cohort of antiretroviral therapy-naive chronically HIV-infected individuals of European ancestry (n=1085). The 1085 subjects were categorized into two groups; HIV controllers (mean viral load <2,000 HIV RNA copies/ml) and HIV progressors (mean viral load >10,000 HIV RNA copies/ml) to evaluate the effect of individual HLA-DRB1 allele expression on mean viral load in a logistic regression model. An odds ratio (OR) was used to analyze the strength of the associations. The OR per DRB1 allele (black circle) is shown with whiskers spanning 95% confidence intervals. The vertical black line indicates an OR of 1, with an OR of <1 indicative that DRB1 allele expression is associated with HIV control, and an OR of >1 indicative that DRB1 allele expression is associated with HIV progression. The analysis was adjusted for subjects expressing HLA class I B*57, B*27 and B*35px to eliminate their confounding effects on HIV viremia. Two HLA-DRB1 alleles remained statistically significant after multiple comparison, including HLA-DRB1*03:01 (p=0.004; q-value=0.04) associated with high OR and HLA-DRB1*15:02 (p=0.003; q-value=0.04) associated with low OR.
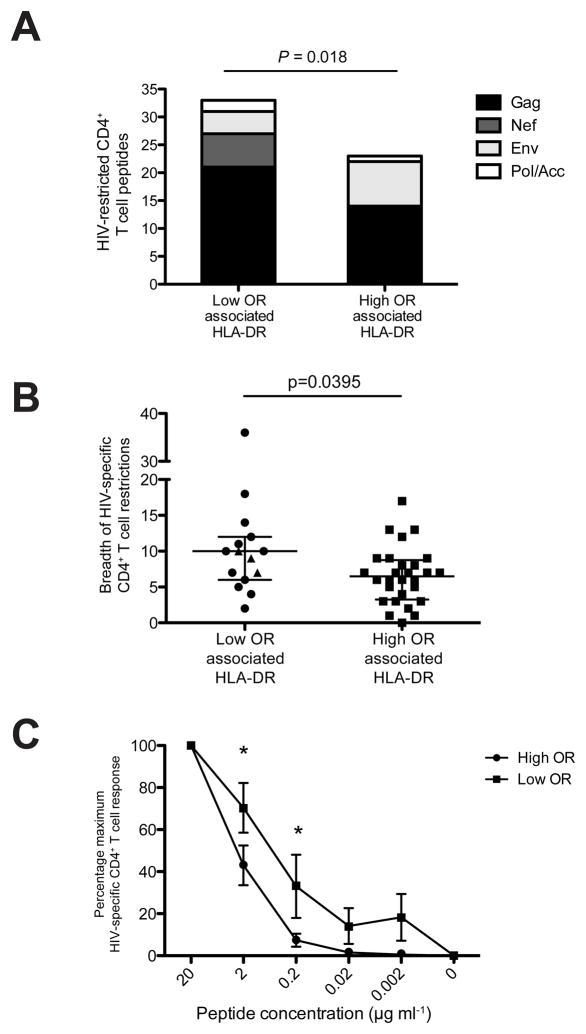
We previously described marked differences in the protein-specificity of HIV-specific CD4+ T cell responses between HIV controllers and high-viremia progressors (8). We therefore next investigated whether the DRB1 alleles linked with either low or high viral load in our genetic study (MGH and SCOPE cohort) were restricting for different HIV proteins in our initial functional screen. The cohorts used for the genetic and functional analyses were independent but did not differ in HLA-DRB1 frequencies (Supplementary Table 4, Supplementary Figure S2). We grouped the HIV-specific CD4+ T cell responses based on the upper quartile of DRB1 alleles linked with low viremia (DRB1*15:02, *10:01, *13:03, *09:01, *13:02) and lower quartile of DRB1 alleles linked with high viremia (DRB1*03:01, *04:04, *01:01, *15:01, *08:01). (DRB1*12:01 was not included due to a lack of patients expressing this allele in our functional cohort). A total of 26 HIV-infected subjects (17 HIV controllers and 9 progressors) were assessed for the sum of promiscuous peptide-DRB1 restrictions per protein. Strikingly, we observed that HLA-DRB1 alleles linked with low viremia showed a significantly greater number of peptide restrictions when compared to HLA-DRB1 alleles linked with high viremia (Fig. 3a, P=0.018 Fisher’s Exact Test). In particular, HLA-DRB1 alleles linked with low viremia were able to promiscuously present a total of 21 Gag peptides, yet only 14 Gag peptides were restricted by HLA-DRB1 alleles linked with high viremia. Moreover, Nef peptides were exclusively restricted by HLA-DRB1 alleles linked with low viremia. A caveat to this approach is that OR values can only be used as an approximate guide for stratifying alleles into the two groupings, given that many alleles exhibited wide confidence intervals in the precision of their OR estimates, and only two individual alleles were statistically significant after multiple comparisons. Nevertheless, the observed functional comparisons remained significant even after higher stringency selection of HLA-DRB1 alleles based on 10% significance prior to multiple comparisons (P=0.0148) (Supplementary Table 5).
Figure 3. HLA-DRB1 alleles linked with low viremia show a greater breadth of HIV-specific CD4 T cell responses and bind at lower functional avidity.
(a) DRB1 alleles associated with low OR showed a significantly higher number of confirmed peptide restrictions across the HIV proteome than DRB1 alleles associated with high OR (P=0.018 Fisher’s Exact Test, n=33/93 low OR vs. 22/108 high OR). The five DRB1 alleles associated with low viremia included DRB1*15:02, *10:01, *13:03, *09:01, *13:02 and the five DRB1 alleles associated with high viremia included DRB1*03:01, *04:04, *01:01, *15:01, *08:01. (DRB1*1201 was not included in the analysis due to lack of sample availability).
(b) HIV-infected subjects expressing DRB1 alleles associated with low OR demonstrated a significantly expanded breadth of total HIV-specific CD4 T cell responses (P=0.0395 Mann-Whitney test). The breadth of HIV-specific T cell responses was tested by fresh ex-vivo ELISPOT at the single-peptide level in 15 treatment-naive HIV-infected subjects expressing a DRB1 allele associated with low OR (black circles) and 28 subjects expressing a DRB1 allele associated with high OR (black squares). Three DRB1*1502 patients were only tested for Gag, Nef and gp120 peptides (black triangles) thus the breadth of responses in these individuals is likely to be underestimated. Individuals expressing >1 ‘protective’ or ‘non-protective’ allele were only counted once, and none of the subjects dually expressed both alleles. The median is shown with bars denoting interquartile range.
(c) HIV-specific CD4 T cell lines restricted by DRB1 alleles associated with low OR demonstrated a significantly lower functional avidity at 2 uM and 0.2uM denoted by an asterisk (*) (P<0.05 2-way ANOVA after Bonferroni correction). A total of 17 peptide-specific CD4 T cell lines were tested in duplicate against a serial dilution of their respective peptide, 9 from subjects expressing low OR associated HLA-DRB1 (black squares) and 8 from subjects expressing high OR associated HLA-DRB1 (black circles) with overlap in only 2/17 peptide specificities. Bars denote standard error. IFNy SFUs at 20 uM were normalized to 100%.
We next investigated whether subjects with HLA-DRB1 alleles linked to lower viremia have generally broader HIV-specific CD4+ T cell responses when compared to subjects that express DRB1 alleles linked with higher viremia. For this analysis, we evaluated the breadth of HIV-specific CD4+ T cell in a sub-cohort of 43 treatment-naive chronically HIV-infected subjects, with the exception of DRB1*15:02 subjects, for whom sample availability limited us to Gag, Nef and Env gp120 only (which is likely to under-estimate the total breadth). We detected no significant differences between the two groups in their mean HIV viral load or CD4 T cell count (Supplementary Figure S3). Strikingly, we observed that individuals with DRB1 alleles linked with low viremia exhibited significantly greater breadth of overall HIV-specific CD4 T cell responses directly ex vivo than individuals with DRB1 alleles linked with higher viremia (Fig. 3b, P=0.0395 Mann Whitney Test). The wide variability within each group is inherent to the fact that each individual expresses a diverse array of HLA class II alleles, thus some peptide-specific responses within the total breadth are likely restricted by an allele that is neither ‘protective’ nor ‘non-protective’. We also assessed the functional binding avidity of peptide-specific CD4+ T cell lines restricted in the context of HLA-DRB1 alleles linked with lower vs. higher viremia. Surprisingly, we observed that CD4+ T cells specific for HLA-DRB1 alleles linked with low viremia in general bind HIV epitopes with lower functional avidity compared to those alleles linked with high viremia (Fig. 3c, P<0.05 2-way ANOVA with Bonferroni correction). These data suggest an intriguing hypothesis in which HLA-DRB1 alleles linked with low viremia may bind at lower avidity and exhibit greater promiscuity in their peptide interactions. This difference in binding modality might give HIV-specific CD4+ T cell responses restricted by potentially ‘protective’ HLA-DRB1 alleles a survival advantage, where CD4+ T cells with lower binding avidity are less activated and thus less susceptible to infection or activation-induced cell death (18–19).
The contribution of HLA class II peptide presentation has been poorly defined in the control of human pathogens. Here, we provide the first systematic approach to the analysis of DRB1 restrictions, and show that DRB1 allele diversity is likely to impact immune containment of HIV infection on the population level. In particular, HLA-DRB1 alleles *15:02 and *03:01 were significantly associated with HIV control and progression, respectively, and remained significant even after correction for multiple comparisons. The strength of these DRB1-mediated effects was independent of HLA-B*57, B*27 and B*35Px, yet was markedly less than has been observed for such HLA class I alleles (9–10). Intriguingly, HLA class II DRB1*03:01 and *15:02 were also listed amongst the many HLA class I alleles implicated in influencing viral control in a GWAS analysis, yet only B*57 was independently associated with a protective affect (10). In sum, these data provide the first evidence linking HLA class II genetic associations with the functional responses of CD4+ T cells and point to an important role for HIV-specific CD4+ T cells in the control of HIV infection.
ONLINE METHODS
Study Subjects
A total of 1085 treatment-naive subjects with chronic HIV-1 infection were recruited at Massachusetts General Hospital (MGH) and the San Francisco General Hospital and the San Francisco Veterans Affairs Medical Center (two field sites comprising the UCSF SCOPE cohort). All MGH and SCOPE subjects utilized in this study were of Caucasian European ancestry and the analysis was statistically adjusted for those subjects expressing HLA class I -B*57, -B*27 and -B*35px. Overall, 594 subjects were classified as ‘HIV controllers’ (<2,000 HIV RNA copies/ml mean viral load) and 491 subjects were classified as ‘HIV progressors’ (>10,000 HIV RNA copies/ml mean viral load). Viral load measurements from three or more time points during chronic infection were used to calculate the mean viral load per subject. The overall mean viral load in this cohort was 40,472 HIV RNA copies/ml. In addition, an independent cohort of 42 treatment-naive HIV-infected subjects of Caucasian ancestry at MGH was analyzed for HLA-DRB1 restriction. These subjects were selected based upon prior delineation of their HIV-specific CD4 T cell responses at the peptide level (8), availability of frozen PBMC samples for further HLA class II restriction assays, and a similar HLA-DRB1 allele distribution in this independent cohort when compared to the MGH & SCOPE cohort. All subjects provided informed consent and each study was approved by the respective institutional review boards.
HLA typing
High-resolution 4-digit HLA class I and II genotyping was performed by sequence-specific PCR in accordance with standard procedures (12). Briefly, HLA class I genes were PCR amplified with primers spanning exons 2 and 3, while HLA class II DRB1 genes were identified by PCR amplification and sequencing of exon 2. ASSIGN 3.5 software developed by Conexio Genomics was used to interpret the sequencing results.
HIV-specific CD4 T cell lines
Frozen CD8-depleted peripheral blood mononuclear cell (PBMC) samples from a subset of 42 subjects with known CD4 T cell responses (8) were used to successfully generate a total of 201 peptide-specific CD4 T cell lines spanning 67 peptide specificities. The targeted peptides were evenly distributed among all expressed HIV proteins (Supplementary Table 1). In brief, CD8-depleted PBMC were thawed and simulated with 10 μg/mL of peptide at a concentration of 2 million cells on a 24-well plate in H10 medium (RPMI 1640 containing 10% heat inactivated fetal calf serum, 2 mM L-glutamine, 50 U of penicillin/ml, 50 ug of streptomycin/mL, and 10 mM HEPES) supplemented with 5 ng/mL recombinant interleukin-7 (IL-7) and 1 μg/mL nevirapine. The cells were incubated at 37 °C and 5% CO2. After 2 days, the cells were washed and fresh H10 medium supplemented with 100 U/mL recombinant interleukin-2 (IL-2) was added. The CD4 T cell lines were fed twice weekly with regular media exchanges.
HLA-DR Restriction Assay
After 14 days, the T cell lines were simultaneously assessed for their specificity and HLA-DR restriction using a large panel of L cell line (LCL) fibroblasts, each stably transfected with a single HLA-DR molecule, as previously described (13). Each peptide-specific CD4 T cell line (TCL) was systematically tested against two HLA-DRB1 expressing LCL that matched the heterozygous HLA-DRB1 typing of the subject from whom the TCL were generated. In addition, some peptide-specific CD4 T cell lines were also tested against HLA-DRB3/4/5 expressing LCL when in linkage disequilibrium). Each LCL was pulsed with 10 μg/mL peptide for 3 hours at 37 °C and 5% CO2 and washed four times to remove free peptide. We utilized clade B 2001 consensus sequence HIV overlapping peptides spanning the whole proteome in our assay. 10,000 peptide-pulsed LCL were then co-cultured in triplicate with each respective CD4 T cell line at a ratio of 1: 5 cells per well on a pre-coated IFNγ plate. As a negative control, each CD4 T cell line was co-cultured in triplicate with the appropriate LCL in the absence of peptide. As a positive control, phytohemagglutinin (Sigma) was added at 1.8 μg/mL. The plates were incubated overnight at 37°C and 5% CO2 and processed as previously described (8, 20). The AID ELISpot reader (Autoimmun Diagnostika GmbH, Strasbourg, Germany) was used to determine the number of spot-forming cells (SFC) per 50,000 of the CD4 T cell line. A HLA-DR restriction was only considered positive if it was at least ≥3 times the mean background and also ≥3 times the standard deviation of the negative control wells. If two overlapping peptides were experimentally restricted by the same HLA-DR allele they were counted as two independent responses. This is concordant with fine mapping data demonstrating that the minimal epitope recognized by a single HLA-DR may differ between overlapping peptides. However, a caveat to this approach is that overlapping peptides restricted by the same HLA-DR could also correspond to a single response.
OLP-41 Fine Mapping
For fine-mapping analysis, we evaluated the IFNγ responses of OLP-41-specific CD4 T cell lines against serial truncations of OLP-41 in the context of five HLA-DRB1 restrictions. Each OLP-41 specific CD4 T cell line was tested against 20 μM of the original 18-mer OLP-41 (YVDRFYKTLRAEQASQEV) and 14 serial truncations from the N- and C-terminus presented by the restricting HLA-DRB1 expressing LCL. Concordant with other CD4 T cell studies (14–15) the “minimal stimulatory epitope” was defined as the shortest peptide sequence triggering an IFNγ response ≥50% of the original 18-mer.
HLA-DRB1 Peptide Binding Assay
Immunodominant HIV-specific CD4 T cell peptides were tested for in vitro binding to a panel of 13 purified HLA-DRB1 molecules, as previously described (13). In brief, purified HLA-DRB1 molecules (5–500 nM) were incubated for 48 hours with different concentrations of unlabeled HIV peptide and 1–10 nM 125I-radiolabeled probe peptides. The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50 nM) was calculated, with a threshold of <1000 IC50 nM for binding.
Statistical Analysis
The 1085 subjects from the MGH and SCOPE cohorts were categorized into two groups; HIV controllers (mean viral load <2,000 HIV RNA copies/ml plasma) and HIV progressors (mean viral load >10,000 HIV RNA copies/ml plasma). The effect of individual HLA-DRB1 allele expression (with frequency ≥5) on mean viral load was evaluated with a logistic regression model. SAS 9.1 (SAS Institute) was used for statistical analyses. PROC FREQ was used to compute frequencies on each allele. PROC LOGISTIC was used to obtain odds ratios (OR), 95% confidence intervals, and two-sided p-values per DRB1 allele. PROC GLM was used for analysis of variance, and Bonferroni correction was performed for multiple comparisons. In order to eliminate the confounding effects of HLA-B*57, B*27 and B*35px, these factors were used as covariates in the logistical regression model. A Fisher’s exact test was used to assess significant differences in peptide restriction between DRB1 alleles associated with low and high viremia. A Mann-Whitney test was utilized to evaluate the breadth of ex vivo HIV-specific CD4 T cell responses in individuals expressing these two groups of DRB1 alleles. A two-way ANOVA with Bonferroni correction was used to analyze differences in functional avidity of peptide-specific CD4 T cell lines generated from subjects expressing HLA-DRB1 alleles associated with low vs. high viremia. Graphical presentation was performed using Graph Pad Prism 5.0.
Supplementary Material
Acknowledgments
We thank all individuals whose participation enabled this study. This study was funded by the National Institutes of Health (R01 AI091450-01, R01 AI094602-01) and the Harvard University Centre for AIDS Research (P-30-AI060354). H. Streeck is funded by a cooperative agreement (R01 AI091450-01, R01 AI094602-01). S. Ranasinghe is supported by a Harvard CFAR Scholar Award (NIH/NIAID 5P30AI060354-09) and the MGH ECOR Fund for Medical Discovery. This project has also been funded in part with federal funds from the Intramural Research Program of the National Institute of Health, National Cancer Institute, Center for Cancer Research, under contract HHSN261200800001E. The UCSF SCOPE cohort was supported by the Centers for AIDS Research at UCSF (PO AI27763), CFAR Network of Integrated Systems (R24 AI067039), the UCSF CTSI (UL1 RR024131), and NIAID (RO1 AI087145, K24 AI069994). The MGH cohort was supported by the Mark and Lisa Schwartz Foundation and the Collaboration of AIDS Vaccine Discovery of the Bill and Melinda Gates Foundation. This research has been supported in part by The International HIV Controllers Study (IHCS), funded by the Bill and Melinda Gates Foundation, the AIDS Healthcare Foundation and the Harvard University Center for AIDS Research (CFAR), an NIH funded program (P30 AI060354), which is supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, FIC, and OAR. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
AUTHOR CONTRIBUTIONS
S.R designed the study, performed experiments, analyzed data, and wrote the manuscript; S.C., I.D., R.L., D.S., G.K., M.L., B.K., G.A., contributed to in vitro studies and experimental design; J.S and A.S generated the HLA-DR transfected L-cells and conducted peptide-DRB1 binding assays; Y.Q., X.G., and M.C., performed high resolution HLA-typing and analyzed HLA-DRB1 associations with viral control; S.D. and B.W. provided clinical samples from HIV-infected subjects enrolled in the SCOPE and MGH cohorts; M.C. and B.W. gave intellectual input; H.S conceived and designed the study, analyzed data, wrote the manuscript and was responsible for the overall study.
References
- 1.Aubert RD, et al. Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proc Natl Acad Sci U S A. 2011;108:21182–21187. doi: 10.1073/pnas.1118450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;29;18(2):274–80. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 3.Soghoian DZ, et al. HIV-Specific Cytolytic CD4 T Cell Responses During Acute HIV-1 Infection Predict Disease Outcome. Sci Transl Med. 2012;4:123ra25. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitmore JK. Induction and function of virus-specific CD4+ T cell responses. Virology. 2011;411(2):216–228. doi: 10.1016/j.virol.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soghoian DZ, Streeck H. Cytolytic CD4+ T cells in viral immunity. Expert Rev Vaccines. 2010;9(12):1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streeck H, D’Souza MP, Littman DR, Crotty S. Harnessing CD4(+) T cell responses in HIV vaccine development. Nat Med. 2013;19:143–149. doi: 10.1038/nm.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douek DC, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417(6884):95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 8.Ranasinghe S, et al. HIV-specific CD4 T cell responses to different viral proteins have discordant associations with viral load and clinical outcome. J Virol. 2012;86:277–283. doi: 10.1128/JVI.05577-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrington M, Walker BD. Immunogenetics of spontaneous control of HIV. Annu Rev Med. 2012;63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International HIV Controller Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra U, et al. Role for HLA class II molecules in HIV-1 suppression and cellular immunity following antiretroviral treatment. J Clin Invest. 2001;107(4):505–517. doi: 10.1172/JCI11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julg B, et al. Possession of HLA Class II DRB1*1303 Associates with Reduced Viral Loads in Chronic HIV-1 Clade C and B Infection. J Infect Dis. 2011;203:803–809. doi: 10.1093/infdis/jiq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southwood S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160(7):3363–3373. [PubMed] [Google Scholar]
- 14.Gerlach JY, et al. Minimal T-cell-stimulatory sequences and spectrum of HLA restriction of immunodominant CD4+ T-cell epitopes within hepatitis C virus NS3 and NS4 proteins. J Virol. 2005;79(19):12425–33. doi: 10.1128/JVI.79.19.12425-12433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulze zur Wiesch J, et al. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol. 175(6):3603–13. doi: 10.4049/jimmunol.175.6.3603. [DOI] [PubMed] [Google Scholar]
- 16.Gao X, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344(22):1668–75. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 17.Trachtenberg E, et al. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9(7):928–25. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 18.Mallone R, et al. Functional avidity directs T-cell fate in autoreactive CD4+ T cells. Blood. 2005;106(8):2798–805. doi: 10.1182/blood-2004-12-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caserta S, et al. Reduced Functional Avidity Promotes Central and Effector Memory CD4 T Cell Responses to Tumor-Associated Antigens. J Immunol. 2010;185(11):6545–6554. doi: 10.4049/jimmunol.1001867. [DOI] [PubMed] [Google Scholar]
- 20.Streeck H, Frahm N, Walker BD. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat Protoc. 2009;4(4):461–469. doi: 10.1038/nprot.2009.7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.