Abstract
Lipopolysaccharide is a highly acylated saccharolipid located on the outer leaflet of the outer membrane of Gram-negative bacteria. Lipopolysaccharide is critical to maintaining the barrier function preventing the passive diffusion of hydrophobic solutes such as antibiotics and detergents into the cell. Lipopolysaccharide has been considered an essential component for outer membrane biogenesis and cell viability based on pioneering studies in the model Gram-negative organisms Escherichia coli and Salmonella. With the isolation of lipopolysaccharide-null mutants in Neisseria meningitides, Moraxella catarrhalis, and most recently in Acinetobacter baumannii, it has become increasingly apparent that lipopolysaccharide is not an essential outer membrane building block in all organisms. We suggest the accumulation of toxic intermediates, misassembly of essential outer membrane porins, and outer membrane stress response pathways that are activated by mislocalized lipopolysaccharide may collectively contribute to the observed strain-dependent essentiality of lipopolysaccharide.
Introduction
Gram-positive bacteria contain a cytoplasmic membrane surrounded by a layer of peptidoglycan; in contrast, Gram-negative bacteria contain a cytoplasm surrounded by what appears to be three layers: an inner membrane, a layer of peptidoglycan and an outer membrane [1,2]. The outer membrane (OM) of Gram-negative bacteria is an asymmetric bilayer with an inner leaflet consisting of phospholipids and an outer leaflet consisting of lipopolysaccharide (LPS). Much of what we know about LPS derives from early work beginning in the 1960s on Escherichia coli and Salmonella typhimurium. Using a newly developed analytical technique to allow separation of the inner membrane (IM) from the OM [3], Osborn and coworkers established that LPS fractionates to the OM [4]. Remarkably, LPS was subsequently shown to be localized exclusively on the outer leaflet of the OM [5–7]. At the same time, the site of (bio)synthesis of LPS was determined to take place at the inner membrane [8]. Work done by Osborn, Raetz, and others first established steps in the biosynthesis of LPS [9–14] and, more recently, the details of LPS transport from its site of synthesis to the cell surface have begun to be uncovered [15]. LPS was shown to be essential and LPS-defective mutants were hypersusceptible to antibiotics [16–18]. The picture that has emerged from the sum of these studies is that individual LPS molecules interact with one another on the cell surface mediated through divalent cations to form a permeability barrier, which prevents entry of small hydrophobic compounds, such as antibiotics, bile salts and detergents, and thus allows Gram-negative bacteria to survive in harsh environments [19–22]. Because the proper assembly of LPS on the cell surface is required to create an effective permeability barrier, genes involved in biosynthesis and assembly (biogenesis) of LPS have become targets for the design of novel classes of antibiotics [23,24].
Historically, interest in developing inhibitors of LPS biosynthesis was predominantly based on the view that LPS was an essential structural component necessary to create an OM. LPS is a large detergent-like molecule comprising three regions; a highly acylated di-GlcNAc backbone (lipid A) connected to a polysaccharide containing repeating sugars (O-antigen) linked through a highly conserved oligosaccharide Kdo/heptose core (Figure 1A). The minimal LPS structure supporting a functional OM and cell viability in enteric bacteria was shown to be Kdo2-lipid A [14,25]. However, our understanding of the importance of LPS in the physiology of Gram-negative bacteria became clouded by the discovery that certain genera do not require LPS to assemble an OM and survive. Remarkably, certain strains of Neisseria can live when their lpxA gene encoding the first enzyme in LPS biosynthesis is inactivated, thus depleting these organisms of LPS [26]. One early hypothesis to explain how these strains of Neisseria could survive was that capsular polysaccharide was a structural substitute for LPS and became essential in these LPS-deficient strains [27]. However, the ability to construct double mutants lacking both lpxA and capsule expression in N. meningitidis disproved this theory [28]. Subsequently viable strains of Moraxella and Acinetobacter completely lacking LPS were isolated and characterized [29,30]. Taken together, these studies called into question the generality of the conclusion drawn from the classic experiments in E. coli and Salmonella. Clearly the assumption that LPS is simply required as a structural component of the outer leaflet of the OM in all Gram-negative bacteria cannot be correct. The essentiality of LPS varies considerably, depending not only on the genera of Gram-negative bacteria but also on the species, and in some cases, even on the particular strain background. This review will consider alternate explanations to account for strain-dependent LPS essentiality in Gram-negative bacteria and discuss the underlying implications for developing antibiotics targeting LPS.
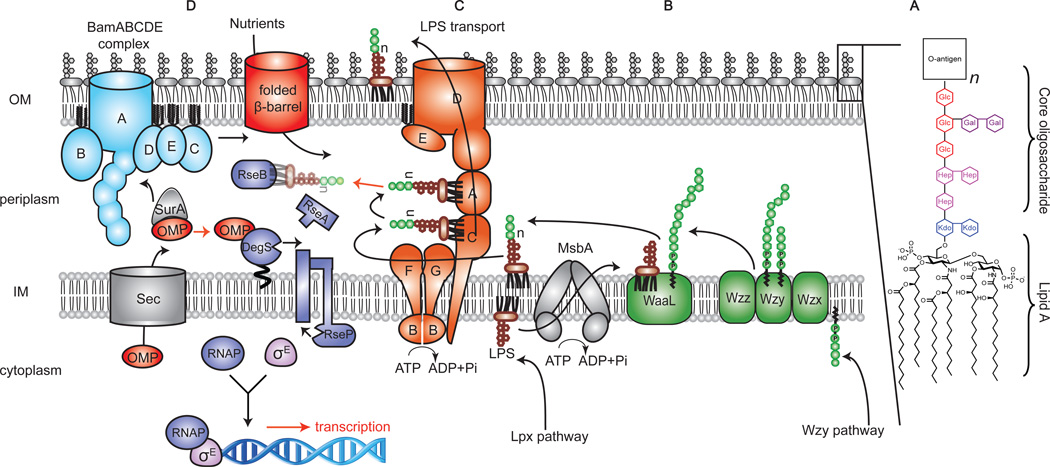
Figure 1.
Pathways that mediate outer membrane (OM) biogenesis and maintain OM integrity. (A) LPS is a complex glycolipid that consists of three regions: lipid A, a core oligosaccharide and an O-antigen polysaccharide. (B) O-antigen is assembled through the Wzy-dependent pathway. The O-antigen repeat unit is assembled in the cytoplasm and flipped to the periplasmic face of the IM by Wzx; O-antigen is polymerized by Wzy and the chain length is modulated by Wzz. (C) LPS is biosynthesized in the cytoplasm, flipped to the periplasmic face of the inner membrane (IM) and transported through the periplasm to the outer leaflet of the OM via the Lpt pathway. Ligation to O-antigen depends on the particular strain background. (D) Mislocalized or misfolded OMPs and LPS loss or defects initiate the σE envelope stress response pathway. LPS loss or defects affect porin assembly; misassembled porin binds to DegS, degrading RseA and initiating the σE stress response. LPS signal could directly bind to RseB, subjecting RseA to proteolysis by DegS to activate the σE stress response.
LPS synthesis and assembly pathways in E. coli
The LPS biosynthesis/transport pathway spans three compartments of Gram-negative bacteria [14]. In E. coli, the Kdo2-lipid A domain is synthesized inside the cytoplasm [11–13]. After sequential addition of sugars to produce the lipid A-oligosaccharide core at the cytoplasmic membrane, this molecule is flipped onto the periplasmic face of the inner membrane by an ABC transporter (MsbA) before the O-antigen is added [31–36]. In the canonical (Wzy-dependent) O-antigen pathway in E. coli, O-antigen biosynthesis begins in the cytoplasm with the sequential addition of three to five monosaccharides onto the undecaprenyl monophosphate carrier lipid (Und-P) to make the O-antigen repeat subunit. This oligosaccharide subunit is transported to the periplasmic face of the IM by Wzx and polymerized en bloc by Wzy to form a mature O-antigen polysaccharide chain containing as many as 40 to 200 repeat units. This polysaccharide must then be transferred to the lipid A-core acceptor by the O-antigen ligase WaaL [14] prior to transit through the periplasm to the cell surface via the Lpt pathway [36] (Figure 1B). The Lpt pathway consists of seven proteins that form a trans-envelope structure containing an IM complex (LptB/F/G/C) required to extract LPS from the IM, a bridge (LptA) between the IM and the OM to permit transit across the aqueous periplasmic compartment, and an OM translocon (LptD/E) to allow the large detergent-like LPS molecule to pass through the OM to its final destination on the cell surface [37–54] (Figure 1C).
The loss of LPS biosynthesis from a given organism has deep-seeded consequences for the assembly of other components of the cell envelope. Syntheses of O-antigen, peptidoglycan, secondary cell wall polymers and outer-membrane proteins (OMPs) are impacted by the absence of LPS [14,15]. While LPS itself maybe not be required for viability, the extent to which the essential functions of the cell envelope are compromised by the loss of LPS could ultimately determine whether LPS is essential in any given strain.
Inhibition of LPS biosynthesis could cause accumulation of cell envelope components in inappropriate compartments
Inhibition of LPS biosynthesis (e.g. LpxC deletion, the first committed step of LPS biosynthesis) depletes levels of the oligosaccharide lipid A core within the IM. The lack of oligosaccharide lipid A core acceptor available for O-antigen transfer can potentially cause unligatable Und-PP O-antigen precursors to accumulate. Accumulation of such precursors has been shown to be toxic in Salmonella, leading to the suggestion that undecaprenyl sequestration influences essential Und-P dependent pathways [55]. Both O-antigen and peptidoglycan biogenesis utilize this membrane-bound carrier for addition of nucleotide sugars. Because undecaprenyl levels are limited in bacterial membranes, blocking transfer of Und-PP O-antigen to the lipid A core leads to sequestration of Und-P, thereby depleting the Und-P pool available to other essential pathways such as peptidoglycan biosynthesis. This toxicity would be predicted to be highly dependent on the strain background.
There are many different factors which may contribute to whether a bacterial strain is susceptible to accumulation of the Und-PP O-antigen. For instance, some O-antigen serotypes utilize a second distinct biosynthetic pathway, the ABC transporter-dependent pathway [14]. The O-antigen homopolymer is assembled on a single Und-P carrier in the cytoplasm prior to transport and ligation to the oligosaccharide-lipid A acceptor on the periplasmic face of the IM [14,56]. Those strains that utilize this ABC pathway would clearly be less susceptible to the accumulation of O-antigen because the demand for the carrier lipid is far less. Secondly, different strains of bacteria modify their lipid A core with O-antigen to varying extents [57]. Some bacteria tend to cap a larger portion of lipid A-oligosaccharides with O-antigen, producing smooth LPS, whereas other organisms tend to have predominantly underivatized lipid A-oligosaccharide core present on the surface (rough LPS) [14]. The extent to which lipid A-oligosaccharides are end-capped with O-antigen (smooth to rough ratio) reflects the flux through the pathway, which in turn is related to the usage of Und-P carrier. A final factor contributing to the sensitivity of a given strain to O-antigen accumulation and the resulting Und-P sequestration is whether cellular mechanisms exist to process the stalled intermediates. Ordinarily, after the O-antigen polysaccharide is transferred to the lipid A-core via WaaL, the newly released Und-PP is recycled back to its active monophosphoryl form Und-P via pyrophosphatases, which liberates carriers enabling the next round of O-antigen and peptidoglycan biosynthesis [58]. It is possible that certain pyrophosphatases can also cleave the O-antigen precursors when its biosynthesis or ligation stalls and allow the Und-P lipid carrier to be recycled. Indeed, in certain E. coli strains with group 4 capsules, a fraction of O-antigen is normally released by hydrolysis to form an extracellular capsule polysaccharide layer [59,60]. This discussion is simply meant to illustrate that there might be many strain-specific mechanisms to relieve the buildup of O-antigen intermediates that would otherwise result in toxicity due to sequestration of the lipid carrier.
Inhibition of LPS biosynthesis could affect the assembly and function of membrane proteins
In addition to LPS, the outer membrane of Gram-negative bacteria contains two major classes of proteins: lipoproteins and integral membrane proteins of β-barrel structure. The exact function of most membrane β-barrel proteins is not known, but many are believed to form pores (porins) in the membrane to provide nonspecific channels across the OM to allow entry of nutrients, which are generally small and hydrophilic [22,61]. It is believed that LPS facilitates porin assembly and function by acting as a molecular chaperone [35]. For example, the porins OmpC and OmpF depend on LPS for trimerization [62–64] and for maintaining proper channel gating function [65], while the protease OmpT requires LPS for its proteolytic activity [66]. While the complement of essential OMPs has only been defined in a limited number of species, there are two outer-membrane β-barrel proteins known to be essential in E. coli. One, LptD is a component of the heterodimeric OM translocon responsible for LPS transport and assembly on the cell surface [38,39,41,49–51]. In those strains of Neisseria where LPS is not essential, LptD becomes non-essential as well [39]. The other BamA is an essential component of the five-protein complex responsible for assembling all OMPs [67–70]. In fact, there are some endosymbionts that have evolved minimal genomes and do not contain genes involved in LPS biogenesis pathway (either Lpx or Lpt) [71,72]. However BamA is generally found to be essential even in minimal genomes, suggesting some β-barrel proteins must be present to permit passage of metabolites across the outer membrane. Clearly, different strains of bacteria have unique nutrient requirements and hence may depend on a specific repertoire of porins for essential nutrient uptake. In the case where these porins depend on LPS for folding/function, LPS would become indispensable.
The loss of LPS could also disrupt the structure and function of the inner membrane. It is possible that inhibition of LPS biosynthesis leads to accumulation of glycolipid (e.g., Und-PP O-antigen, Und-PP enterobacterial common antigen [73,74], or other secondary cell envelope polymers [75]) intermediates. One could imagine many scenarios through which these accumulated glycolipid intermediates could compromise IM functions. The accumulation of glycolipid intermediates could influence the functions of IM proteins. For example, various essential IM proteins are involved in peptidoglycan biosynthesis [76]. Since both the O-antigen and Lipid II contain an Und-PP activating group, accumulation of Und-PP O-antigen could compete with Lipid II inhibiting cell wall synthesis. Additional glycolipids could also influence the proper assembly of inner-membrane proteins by affecting the lipid environment in the IM. Simply by affecting IM bilayer packing, these accumulated glycolipids could also create physical defects in the IM and cause problems by dissipation of membrane potential. Here again, the presence of strain-specific mechanisms to relieve the buildup of O-antigen intermediates (or other secondary cell envelope polymers) would determine whether a given Gram-negative organism would be susceptible to LPS deletion.
Inhibition of LPS biosynthesis could trigger stress response pathways causing inhibition of growth
Inhibition of growth could result directly from the loss of LPS or from the cellular response to the loss – activation of an alternative genetic program in response to the stress of LPS deletion. It has been shown that the accumulation of mistargeted and/or misfolded outer membrane proteins in the periplasm is detected by a sensor protein, DegS, initiating a proteolytic cascade that results in activation of the σE -dependent envelope stress response system [77,78] (Figure 1D). The σE -transcription factor up-regulates both the expression of genes involved in the targeting and assembly of OMPs, as well as for genes encoding proteases in order to clear misfolded substrates [79]. Literally hundreds of genes are turned on in order to restore the intracellular trafficking of OMPs. Because various OMPs require LPS to fold, inhibition of LPS biosynthesis could also cause misfolding of porins initiating the envelope stress response resulting in growth stasis until the intracellular levels of LPS can be restored [80–82]. A recent report has suggested that there are surveillance systems that directly detect mistargeted LPS in the periplasm of E. coli, and triggers the envelope stress response by activating σE (Figure 1D) [83]. The fact that bacteria respond to LPS and OMP defects or loss by activating a quality control mechanism further emphasizes the essentiality of LPS in maintaining OM integrity. In E. coli and Salmonella, a particularly stringent OM stress response has presumably evolved in order to allow these organisms to colonize the gut, where a high concentration of detergent-like molecules (e.g. bile salts) must be tolerated [84]. Hence, LPS may, in part, be required to prevent growth stasis triggered by stress response systems. Although relatively less is known about stress response systems in other Gram-negative organisms, it is conceivable that such OM quality control surveillance systems may not be as prominent as their counterparts in E. coli and Salmonella [35]. These bacteria may continue to grow and divide in an OM compromised state, whereas other strains would cease growing due to stress response signaling.
Conclusion
For several decades LPS was thought to be an essential structural component of the OM of Gram-negative bacteria just as amino acids are essential to the structures of proteins. However, with the discovery of LPS-deficient organisms it is now clear that the essentiality of LPS to Gram-negative bacteria is more complex. It seems reasonable that LPS was selected in Nature because when combined in an asymmetric bilayer with phospholipids it produces an unusual membrane that prevents the passage of toxic hydrophobic molecules into the cell. However, at this point, maintaining a proper LPS permeability barrier is tangential to its essentiality. Having become so heavily integrated into the cell envelope physiology, its removal may affect other metabolic processes through indirect means. Whether it is possible to delete LPS from a given organism will depend on the cellular context and perhaps even how it is removed. While LPS may be non-essential in some organisms, its loss is not inconsequential. Strains lacking LPS are less virulent [85] and much more susceptible to antibiotics that normally do not penetrate the OM [29,30,86]. It is still believed that compounds which interfere with the functions of LPS synthesis, transport, or assembly, will have the potential to function on their own as antibiotics as well as to potentiate the entry of other existing antibiotics normally excluded by the OM. In fact, a better understanding of the importance of LPS biogenesis on bacterial physiology could provide clues as to the specific vulnerabilities of a given Gram-negative pathogen to inhibition of LPS at different steps in synthesis and assembly.
Highlights.
We discuss the cellular role of lipopolysaccharide within Gram-negative bacteria.
We propose explanations for why lipopolysaccharide is essential in certain organisms.
We consider implications for developing lipopolysaccharide-targeting antibiotics.
Acknowledgments
This work was supported by the Blavatnik Biomedical Accelerator Fund and the National Institutes of Health (NIH) grant AI081059 to D.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kellenberger E, Ryter A. Cell wall and cytoplasmic membrane of Escherichia coli. J Biophys Biochem Cytol. 1958;4:323–326. doi: 10.1083/jcb.4.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bladen HA, Mergenhagen SE. Ultrastructure of Veillonella and Morphological Correlation of an Outer Membrane with Particles Associated with Endotoxic Activity. J Bacteriol. 1964;88:1482–1492. doi: 10.1128/jb.88.5.1482-1492.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miura T, Mizushima S. Separation by density gradient centrifugation of two types of membranes from spheroplast membrane of Escherichia coli K12. Biochim Biophys Acta. 1968;150:159–161. doi: 10.1016/0005-2736(68)90020-5. Developed sucrose density/gradient centrifugation technique to separate the IM from the OM.
- 4. Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. Analyzed the composition of the OM and established LPS localization to this membrane.
- 5.Muhlradt PF, Golecki JR. Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur J Biochem. 1975;51:343–352. doi: 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 6. Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. Classic paper, which established that the outer leaflet of the OM is devoid of phospholipids, leading to the hypothesis that the OM is an asymmetric bilayer.
- 7.Funahara Y, Nikaido H. Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J Bacteriol. 1980;141:1463–1465. doi: 10.1128/jb.141.3.1463-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Osborn MJ, Gander JE, Parisi E. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. J Biol Chem. 1972;247:3973–3986. Established that the site of (bio)synthesis of LPS is in the IM. Pointed out that this observation raises the question of how to transport LPS across three cellular compartments -- still the most important unanswered question in the field.
- 9.Rick PD, Fung LW, Ho C, Osborn MJ. Lipid A mutants of Salmonella typhimurium. Purification and characterization of a lipid A precursor produced by a mutant in 3-deoxy-D-mannooctulosonate-8-phosphate synthetase. J Biol Chem. 1977;252:4904–4912. [PubMed] [Google Scholar]
- 10.Rick PD, Osborn MJ. Lipid A mutants of Salmonella typhimurium. Characterization of a conditional lethal mutant in 3-deoxy-D-mannooctulosonate-8-phosphate synthetase. J Biol Chem. 1977;252:4895–4903. [PubMed] [Google Scholar]
- 11.Crowell DN, Anderson MS, Raetz CR. Molecular cloning of the genes for lipid A disaccharide synthase and UDP-N-acetylglucosamine acyltransferase in Escherichia coli. J Bacteriol. 1986;168:152–159. doi: 10.1128/jb.168.1.152-159.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly TM, Stachula SA, Raetz CR, Anderson MS. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J Biol Chem. 1993;268:19866–19874. [PubMed] [Google Scholar]
- 13.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CR, Hyland SA, Anderson MS. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem. 1995;270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 14.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamaki S, Sato T, Matsuhashi M. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J Bacteriol. 1971;105:968–975. doi: 10.1128/jb.105.3.968-975.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanderson KE, MacAlister T, Costerton JW, Cheng KJ. Permeability of lipopolysaccharide-deficient (rough) mutants of Salmonella typhimurium to antibiotics, lysozyme, and other agents. Can J Microbiol. 1974;20:1135–1145. doi: 10.1139/m74-176. [DOI] [PubMed] [Google Scholar]
- 18.Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1993;37:2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macgregor DR, Elliker PR. A comparison of some properties of strains of Pseudomonas aeruginosa sensitive and resistant to quaternary ammonium compounds. Can J Microbiol. 1958;4:499–503. doi: 10.1139/m58-054. [DOI] [PubMed] [Google Scholar]
- 20.Leive L. A Nonspecific Increase in Permeability in Escherichia Coli Produced by Edta. Proc Natl Acad Sci U S A. 1965;53:745–750. doi: 10.1073/pnas.53.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leive L. The barrier function of the gram-negative envelope. Ann N Y Acad Sci. 1974;235:109–129. doi: 10.1111/j.1749-6632.1974.tb43261.x. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, et al. Antibacterial agents that inhibit lipid A biosynthesis. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. Most studied antibiotic target in LPS biosynthesis.
- 24. Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. First report of a small molecule that targets a component (LptD) of OM biogenesis (LPS assembly) machinery.
- 25.Gronow S, Brade H. Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J Endotoxin Res. 2001;7:3–23. [PubMed] [Google Scholar]
- 26. Steeghs L, den Hartog R, den Boer A, Zomer B, Roholl P, van der Ley P. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–450. doi: 10.1038/33046. Defined and characterized a strain of Gram-negative bacteria completely lacking LPS. This paper first called into question the generality of LPS essentiality.
- 27.Steeghs L, de Cock H, Evers E, Zomer B, Tommassen J, van der Ley P. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 2001;20:6937–6945. doi: 10.1093/emboj/20.24.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bos MP, Tommassen J. Viability of a capsule- and lipopolysaccharide-deficient mutant of Neisseria meningitidis. Infect Immun. 2005;73:6194–6197. doi: 10.1128/IAI.73.9.6194-6197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng D, Hong W, Choudhury BP, Carlson RW, Gu XX. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun. 2005;73:7569–7577. doi: 10.1128/IAI.73.11.7569-7577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. This paper determined that LPS is not essential in Acinetobacter baumannii, which along with Peng et al.’s study in Moraxella raised the question of what it is that makes LPS essential for some organisms, but not others.
- 31.Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol Microbiol. 1993;7:69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 32.Clementz T, Bednarski JJ, Raetz CR. Function of the htrB high temperature requirement gene of Escherchia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J Biol Chem. 1996;271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an Essential ABC Family Transporter, in Lipid A and Phospholipid Biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 34.Doerrler WT, Gibbons HS, Raetz CR. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 35.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 36.Liechti G, Goldberg JB. Outer membrane biogenesis in Escherichia coli, Neisseria meningitidis, and Helicobacter pylori: paradigm deviations in H. pylori. Front Cell Infect Microbiol. 2012;2:29. doi: 10.3389/fcimb.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. Clever selection. Discovery of the first gene (originally known as Imp -- Increased membrane permeability) in the LPS transport/assembly pathway.
- 38. Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. Established the function of LptD (previously called Imp) in OM biogenesis.
- 39. Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci U S A. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. Identified the functional role of Imp (LptD) as the outer-membrane transporter of LPS to the cell surface by knocking out LptD in Neisseria. This could be done because in the particular strain LPS was not essential.
- 40.Serina S, Nozza F, Nicastro G, Faggioni F, Mottl H, Deho G, Polissi A. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res Microbiol. 2004;155:692–701. doi: 10.1016/j.resmic.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran AX, Trent MS, Whitfield C. The LptA protein of Escherichia coli is a periplasmic lipid A-binding protein involved in the lipopolysaccharide export pathway. J Biol Chem. 2008;283:20342–20349. doi: 10.1074/jbc.M802503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol. 2008;190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sperandeo P, Deho G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Narita S, Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chimalakonda G, Ruiz N, Chng SS, Garner RA, Kahne D, Silhavy TJ. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freinkman E, Chng SS, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci U S A. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freinkman E, Okuda S, Ruiz N, Kahne D. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry. 2012;51:4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012;338:1214–1217. doi: 10.1126/science.1228984. This paper established that Lpt proteins directly transport LPS. Observed intermediates in LPS tansport opening the door for mechanistic studies.
- 54.Chng SS, Xue M, Garner RA, Kadokura H, Boyd D, Beckwith J, Kahne D. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337:1665–1668. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuasa R, Levinthal M, Nikaido H. Biosynthesis of cell wall lipopolysaccharide in mutants of Salmonella. V. A mutant of Salmonella typhimurium defective in the synthesis of cytidine diphosphoabequose. J Bacteriol. 1969;100:433–444. doi: 10.1128/jb.100.1.433-444.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenfield LK, Whitfield C. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res. 2012;356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 57.Burrows LL, Lam JS. Effect of wzx (rfbX) mutations on A-band and B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa O5. J Bacteriol. 1999;181:973–980. doi: 10.1128/jb.181.3.973-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatar LD, Marolda CL, Polischuk AN, van Leeuwen D, Valvano MA. An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl phosphate recycling. Microbiology. 2007;153:2518–2529. doi: 10.1099/mic.0.2007/006312-0. [DOI] [PubMed] [Google Scholar]
- 59.Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 60.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 61.Danese PN, Silhavy TJ. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 62.Ried G, Hindennach I, Henning U. Role of lipopolysaccharide in assembly of Escherichia coli outer membrane proteins OmpA, OmpC, and OmpF. J Bacteriol. 1990;172:6048–6053. doi: 10.1128/jb.172.10.6048-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen K, Nikaido H. In vitro trimerization of OmpF porin secreted by spheroplasts of Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:743–747. doi: 10.1073/pnas.87.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sen K, Nikaido H. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J Bacteriol. 1991;173:926–928. doi: 10.1128/jb.173.2.926-928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindler H, Rosenbusch JP. Matrix protein in planar membranes: clusters of channels in a native environment and their functional reassembly. Proc Natl Acad Sci U S A. 1981;78:2302–2306. doi: 10.1073/pnas.78.4.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kramer RA, Brandenburg K, Vandeputte-Rutten L, Werkhoven M, Gros P, Dekker N, Egmond MR. Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT. Eur J Biochem. 2002;269:1746–1752. doi: 10.1046/j.1432-1327.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 67.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 68.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 70.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rick PD, Wolski S, Barr K, Ward S, Ramsay-Sharer L. Accumulation of a lipid-linked intermediate involved in enterobacterial common antigen synthesis in Salmonella typhimurium mutants lacking dTDP-glucose pyrophosphorylase. J Bacteriol. 1988;170:4008–4014. doi: 10.1128/jb.170.9.4008-4014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rick PD, Hubbard GL, Kitaoka M, Nagaki H, Kinoshita T, Dowd S, Simplaceanu V, Ho C. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology. 1998;8:557–567. doi: 10.1093/glycob/8.6.557. [DOI] [PubMed] [Google Scholar]
- 75.Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, Woodard RW. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J Biol Chem. 2007;282:7790–7798. doi: 10.1074/jbc.M611034200. [DOI] [PubMed] [Google Scholar]
- 76.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 78.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 79.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tam C, Missiakas D. Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol Microbiol. 2005;55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- 81.Klein G, Lindner B, Brabetz W, Brade H, Raina S. Escherichia coli K-12 Suppressor-free Mutants Lacking Early Glycosyltransferases and Late Acyltransferases: minimal lipopolysaccharide structure and induction of envelope stress response. J Biol Chem. 2009;284:15369–15389. doi: 10.1074/jbc.M900490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, Gross CA. Signal integration by DegS and RseB governs the σE- mediated envelope stress response in Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science. 2013;340:837–841. doi: 10.1126/science.1235358. Established that off-pathway intermediates of LPS acts as a separate activating signal to initiate the σE envelope stress response.
- 84.Prouty AM, Brodsky IE, Falkow S, Gunn JS. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology. 2004;150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 85.Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, Montgomery JI, Reilly U, Barbacci EG, Hujer K, et al. Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. MBio. 2012;3 doi: 10.1128/mBio.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, Nation RL, Owen RJ, Wong S, Spelman D, Franklin C. Antibiograms of multidrug-resistant clinical Acinetobacter baumannii: promising therapeutic options for treatment of infection with colistin-resistant strains. Clin Infect Dis. 2007;45:594–598. doi: 10.1086/520658. [DOI] [PubMed] [Google Scholar]