Abstract
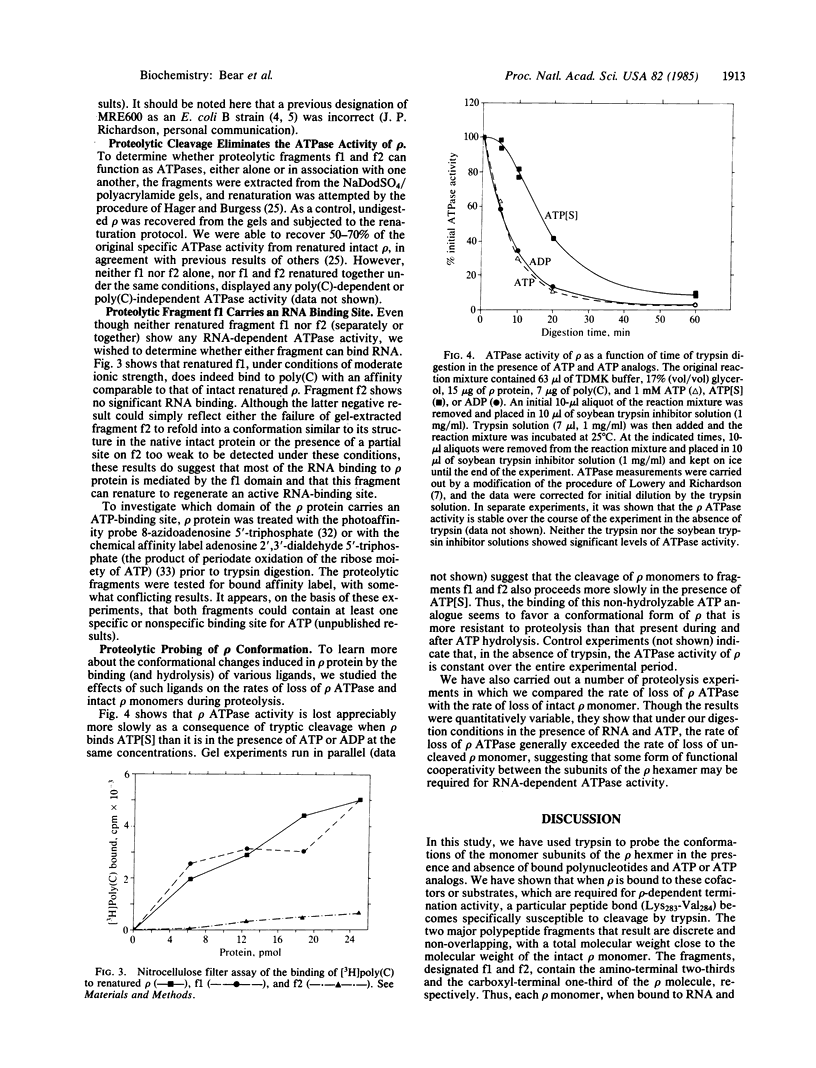
Limited tryptic digestion of Escherichia coli transcription termination factor rho [an RNA-dependent nucleoside triphosphatase (NTPase)] yields predominantly two fragments (f1 and f2) when the protein is bound to both poly(C) and ATP. The apparent molecular masses of the two fragments are 31 kDa for f1 and 15 kDa for f2, adding up to the molecular mass of the intact rho polypeptide chain (46 kDa). Sequence analysis of the amino termini demonstrates that f1 is derived from the amino-terminal portion of rho and that the trypsin cleavage that defines f2 occurs at lysine-283. These results suggest that, in the liganded (activated) form, the native rho protein monomer is organized into two distinct structural domains that are separable by a single proteolytic cleavage. The f1 fragment, purified from NaDodSO4/polyacrylamide gels and renatured, binds poly(C) but the f2 fragment does not; neither regains any ATPase activity. ATP- and polynucleotide-dependent changes in the rate of proteolysis and in the character of the fragments produced suggest that rho undergoes a series of conformational transitions as a consequence of RNA binding, NTP binding and NTP hydrolysis. The rate of loss of rho ATPase activity and of intact rho monomers is slower in the presence of adenosine 5'-[gamma-thio]triphosphate than in the presence of either ATP or ADP, indicating that the hydrolysis of ATP may result in different conformational effects than does the binding of this ligand. These findings are discussed within the context of recent models of rho-dependent transcription termination.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K., Yasuda S., Kornberg A. Mechanism of dnaB protein action. I. Crystallization and properties of dnaB protein, an essential replication protein in Escherichia coli. J Biol Chem. 1981 May 25;256(10):5247–5252. [PubMed] [Google Scholar]
- Brown S., Albrechtsen B., Pedersen S., Klemm P. Localization and regulation of the structural gene for transcription-termination factor rho of Escherichia coli. J Mol Biol. 1982 Dec 5;162(2):283–298. doi: 10.1016/0022-2836(82)90527-7. [DOI] [PubMed] [Google Scholar]
- Clertant P., Cuzin F. Covalent affinity labeling by periodate-oxidized [alpha-32P]ATP of the large-T proteins of polyoma and SV40 viruses. J Biol Chem. 1982 Jun 10;257(11):6300–6305. [PubMed] [Google Scholar]
- Das A., Merril C., Adhya S. Interaction of RNA polymerase and rho in transcription termination: coupled ATPase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4828–4832. doi: 10.1073/pnas.75.10.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel D., Richardson J. P. Conformational alterations of transcription termination protein rho induced by ATP and by RNA. Nucleic Acids Res. 1984 Oct 11;12(19):7389–7400. doi: 10.1093/nar/12.19.7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files J. G., Weber K. Limited proteolytic digestion of lac repressor by trypsin. Chemical nature of the resulting trypsin-resistant core. J Biol Chem. 1976 Jun 10;251(11):3386–3391. [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Procedure for purification of Escherichia coli ribonucleic acid synthesis termination protein rho. Biochemistry. 1981 Mar 17;20(6):1640–1645. doi: 10.1021/bi00509a036. [DOI] [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Stabilization of the hexameric form of Escherichia coli protein rho under ATP hydrolysis conditions. J Mol Biol. 1982 Mar 25;156(1):203–219. doi: 10.1016/0022-2836(82)90467-3. [DOI] [PubMed] [Google Scholar]
- Galluppi G. R., Richardson J. P. ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J Mol Biol. 1980 Apr 15;138(3):513–539. doi: 10.1016/s0022-2836(80)80016-7. [DOI] [PubMed] [Google Scholar]
- Guarente L. P., Beckwith J. Mutant RNA polymerase of Escherichia coli terminates transcription in strains making defective rho factor. Proc Natl Acad Sci U S A. 1978 Jan;75(1):294–297. doi: 10.1073/pnas.75.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Restoration of termination by RNA polymerase mutations is rho allele-specific. J Mol Biol. 1979 Apr 5;129(2):295–304. doi: 10.1016/0022-2836(79)90283-3. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hong S. L., Barker H. A. Aerobic metabolism of 3,5-diaminohexanoate in a Brevibacterium. Purification of 3,5-diaminohexanoate dehydrogenase and degradation of 3-keto-5-aminohexanoate. J Biol Chem. 1973 Jan 10;248(1):41–49. [PubMed] [Google Scholar]
- Howard B. H., de Crombrugghe B. ATPase activity required for termination of transcription by the Escherichia coli protein factor rho. J Biol Chem. 1976 Apr 25;251(8):2520–2524. [PubMed] [Google Scholar]
- Janin J., Wodak S. J. Structural domains in proteins and their role in the dynamics of protein function. Prog Biophys Mol Biol. 1983;42(1):21–78. doi: 10.1016/0079-6107(83)90003-2. [DOI] [PubMed] [Google Scholar]
- Krakow J. S., Pastan I. Cyclic adenosine monophosphate receptor: loss of cAMP-dependent DNA binding activity after proteolysis in the presence of cyclic adenosine monophosphate. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2529–2533. doi: 10.1073/pnas.70.9.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowery-Goldhammer C., Richardson J. P. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc Natl Acad Sci U S A. 1974 May;71(5):2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesi termination factor p. I. Enzymatic properties and effects of inhibitors. J Biol Chem. 1977 Feb 25;252(4):1375–1380. [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor p. II. Influence of synthetic RNA homopolymers and random copolymers on the reaction. J Biol Chem. 1977 Feb 25;252(4):1381–1385. [PubMed] [Google Scholar]
- Lowey S., Slayter H. S., Weeds A. G., Baker H. Substructure of the myosin molecule. I. Subfragments of myosin by enzymic degradation. J Mol Biol. 1969 May 28;42(1):1–29. doi: 10.1016/0022-2836(69)90483-5. [DOI] [PubMed] [Google Scholar]
- Moise H., Hosoda J. T4 gene 32 protein model for control of activity at replication fork. Nature. 1976 Feb 12;259(5543):455–458. doi: 10.1038/259455a0. [DOI] [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983 Aug 10;258(15):9565–9574. [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Specificity of release by Escherichia coli transcription termination factor rho of nascent mRNA transcripts initiated at the lambda PR. J Biol Chem. 1984 Jul 10;259(13):8664–8671. [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Nakayama N., Arai N., Kaziro Y., Arai K. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem. 1984 Jan 10;259(1):88–96. [PubMed] [Google Scholar]
- Pinkham J. L., Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983 Jun 11;11(11):3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter R. L., Haley B. E. Photoaffinity labeling of nucleotide binding sites with 8-azidopurine analogs: techniques and applications. Methods Enzymol. 1983;91:613–633. doi: 10.1016/s0076-6879(83)91054-6. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Hurwitz J. The dnaB gene product of Escherichia coli. I. Purification, homogeneity, and physical properties. J Biol Chem. 1978 Jun 10;253(11):4043–4050. [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. Binding of rho factor to Escherichia coli RNA polymerase mediated by nusA protein. J Biol Chem. 1984 Dec 25;259(24):15000–15002. [PubMed] [Google Scholar]
- Sharp J. A., Galloway J. L., Platt T. A kinetic mechanism for the poly(C)-dependent ATPase of the Escherichia coli transcription termination protein, rho. J Biol Chem. 1983 Mar 25;258(6):3482–3486. [PubMed] [Google Scholar]
- Sharp J. A., Platt T. Rho-dependent termination and concomitant NTPase activity requires a specific, intact RNA region. J Biol Chem. 1984 Feb 25;259(4):2268–2273. [PubMed] [Google Scholar]
- Suryanarayana T., Subramanian A. R. Functional domains of Escherichia coli Ribosomal Protein S1. Formation and characterization of a fragment with ribosome-binding properties. J Mol Biol. 1979 Jan 5;127(1):41–54. doi: 10.1016/0022-2836(79)90458-3. [DOI] [PubMed] [Google Scholar]
- Wade H. E., Robinson H. K. Magnesium ion-independent ribonucleic acid depolymerases in bacteria. Biochem J. 1966 Nov;101(2):467–479. doi: 10.1042/bj1010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Platt T., Weber K. Amino-terminal sequence analysis of proteins purified on a nanomole scale by gel electrophoresis. J Biol Chem. 1972 May 25;247(10):3242–3251. [PubMed] [Google Scholar]
- Williams K. R., Spicer E. K., LoPresti M. B., Guggenheimer R. A., Chase J. W. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983 Mar 10;258(5):3346–3355. [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]