Abstract
IL-15 and the IL-15 receptor (IL-15R)α chain are essential for normal development of naive CD8 T cells, intestinal intraepithelial lymphocytes (IEL), and natural killer (NK)/NK/T cells. However, whether IL-15Rα expression by these subsets is necessary for their production and which cell type needs to produce IL-15 to drive development are unknown. We analyzed the requirements for IL-15 and IL-15Rα expression by bone marrow-derived or parenchymal cells for mediating lymphocyte subset development. Naive CD8 T cell development required IL-15Rα expression by both bone marrow-derived and parenchymal cells, whereas memory-phenotype CD8 T cells required IL-15Rα expression only by hematopoietic cells. In contrast and surprisingly, the development of IEL subsets, particularly CD8ααThy1–Vγ5+ T cell antigen receptor γδ and the CD8αα Thy1– T cell antigen receptor αβ IEL populations, depended completely on parenchymal cell expression of IL-15Rα and IL-15 but not IL-15Rβ. In the case of NK and NK/T cell generation and maturation, expression of IL-15 and IL-15Rα by both parenchymal and hematopoietic cells was important, although the latter played the greatest role. These results demonstrated dichotomous mechanisms by which IL-15 regulated lymphoid development, interacting with distinct cell types depending on the developmental pathway.
Generation of a normal lymphoid compartment is mediated at multiple levels including the production and maturation of cellular precursors in the primary lymphoid organs, the bone marrow (BM) and the thymus; the release of mature or semi-mature cells from these sites; and the maintenance of mature cell types in the periphery by means of homeostatic mechanisms, which control the size of peripheral pools of the various lymphoid subsets (1, 2). Cytokines, particularly those of the common γ chain (γC) family, play critical roles in these processes.
IL-15 is a γC cytokine and a member of the four α-helix bundle cytokine family. IL-15 is requisite for the generation or maintenance of specific hematopoietic lineages. In the absence of IL-15 or IL-15 receptor (IL-15R)α, defects are observed in naive and memory CD8 T cells, intestinal intraepithelial lymphocytes (IEL), and natural killer (NK) and NK/T lineages (3, 4). Depending on the lymphoid lineage or stage of differentiation, IL-15 can have various roles in development and homeostasis. IL-15 can act to increase survival, induce proliferation, and/or drive differentiation. Although it is clear that IL-15 is important for the development of these hematopoietic lineages, the mechanisms of IL-15-mediated actions are not well defined.
The receptor for IL-15 is composed of an IL-15Rα chain, capable of binding IL-15 with high affinity in the absence of other receptor subunits, the IL-15/IL-2 receptor β chain, and the γC chain (5, 6). Although soluble IL-15 can bind the IL-15R complex and induce signals in a manner similar to other cytokines and cytokine receptors (7, 8), recent reports have identified a mechanism by which the IL-15/IL-15Rα system may operate to generate signals. Dubois et al. (9) initially demonstrated that, in vitro, IL-15Rα+ cells could bind IL-15 and “transpresent” IL-15 to cells lacking IL-15Rα. Plate-bound IL-15Rα can also serve as a platform to present IL-15 to CD8 memory T cells in vitro (10). In vivo, IL-15Rα-deficient CD8 memory T cells and NK cells are able to respond to IL-15 (10, 11). Indeed, recent reports suggest that transpresentation of IL-15 specifically by BM-derived cells is a major mechanism that mediates memory CD8 T cell proliferation in vivo (10, 12). For survival of NK cells in vivo, IL-15Rα expression by BM-derived cells is important (11), although whether parenchymal cell IL-15Rα expression is also involved is not clear. These findings suggest that IL-15Rα expression on opposing cell types is important for lymphoid homeostasis and that transpresentation of IL-15 may be the underlying mechanism for delivery of IL-15-mediated signals. However, whether this scenario applies to development of other IL-15-dependent lineages is not known. Thus, we undertook an analysis of the role of IL-15Rα as well as IL-15 expression in the development of peripheral CD8 T cells, intestinal IEL subsets, NK cells, and NK/T cells. Surprisingly, the results demonstrated that, depending on the lymphoid cell type affected and the tissue, expression of IL-15Rα by either BM-derived cells, parenchymal cells, or both was required to drive IL-15-mediated development.
Materials and Methods
Mice. C57BL/6J (Ly5.1) and C57BL/6-IL-2/15Rβ+/– mice (13) were purchased from The Jackson Laboratory, and C57BL/6J (Ly5.2) mice were purchased from Charles River Breeding Laboratories through the National Cancer Institute program. C57BL/6-IL-15–/– mice (4) and C57BL/6-IL-15Rα–/– mice (3) were generously provided by Jacques Peschon (Immunex, Seattle) and by Averil Ma (University of Chicago, Chicago), respectively. All mice were maintained under specific pathogen-free conditions at the University of Connecticut Health Center.
Generation of BM Chimeras. BM cells were obtained from femurs and tibias of IL-15–/–, IL-15Rα–/–, and control mice. To remove T cells present in the BM suspension, BM cells were treated with anti-Thy1 mAb (T24) (14) followed by incubation with low-ToxM rabbit complement (Cedarlane Laboratories) for 1 h at 37°C. Recipients were irradiated with 1,000 rad and injected i.v. with 5 × 106 BM cells. In some experiments, BM from mice congenic at the Ly5 locus were used to distinguish donor-derived cells from residual hosts cells. Chimeras were analyzed 8–12 weeks later.
Isolation of Lymphocyte Populations. Spleens were homogenized in HBSS-HGPG [Hanks' balanced salt solution (HBSS) with Hepes, l-glutamine, penicillin/streptomycin, and gentamycin sulfate], filtered through Nitex nylon mesh, and treated with Tris-ammonium chloride to lyse red blood cells. To obtain lymphocytes from liver, anesthetized mice were perfused with PBS containing 75 units/ml heparin until the tissues were cleared of blood. Lymphocytes were isolated from liver as described in ref. 15. IEL were isolated as described in ref. 16.
Immunofluorescence Staining. Antibodies specific for the following molecules and coupled to the indicated fluorochromes were used: CD8α-peridinin chlorophyll protein, CD8β-FITC, T cell antigen receptor (TCR)γδ-phycoerythrin (GL-3), TCRαβ (H57)-allophycocyanin (APC), CD4(GK1.5)-FITC, Vγ2-FITC, Thy1–APC, NK1.1 (PK136)-phycoerythrin, CD3 (145–2C11)-FITC, and CD11b (M1.70)-APC were purchased from Becton Dickinson, and anti-Vγ5 mAb was described in ref. 17 and was used as a FITC conjugate here. Anti-Vγ1 mAb was received from P. Pereira (Instit Pasteur, Paris) (18) and was biotinylated. Streptavidin-APC was purchased from Jackson ImmunoResearch. Ly49 expression was measured by using a mixture of two rat anti-mouse NK mAb (LAK6/99 and LAK6/292, conjugated to APC) that, combined, recognize the LY49A, -C, -D, -E, -G, and -I molecules. The specificity of these mAb was determined by crosscompetition and transfection studies (H.L.A., unpublished observations). Relative fluorescence intensities were measured with a FACSCalibur (Becton Dickinson). Data were analyzed by using winmdi software (Joseph Trotter, The Scripps Clinic, La Jolla, CA) or FlowJo (Tree Star, San Carlos, CA). Statistics were performed by using the Tukey–Kramer multiple comparisons test or the Student t test.
Results
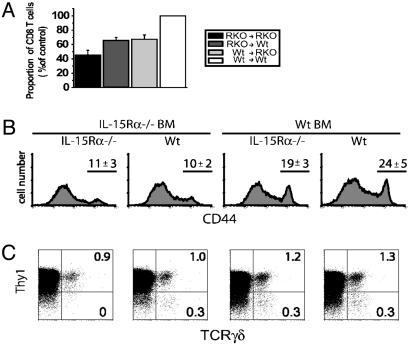
Expression of IL-15Rα by BM-Derived or Parenchymal Cells Mediates Splenic CD8 TCRαβ and TCRγδ T Cell Development. To determine whether expression of IL-15Rα by BM-derived (radiation-sensitive) or non-BM-derived (radiation-resistant) cells contributes to the development of specific T cell subsets, various combinations of IL-15Rα-deficient BM chimeras were generated and analyzed for development of specific T cell populations. CD8 TCRαβ T cells require IL-15 and IL-15Rα expression for normal development (3, 4). In IL-15Rα–/– and IL-15–/– mice, the percentage and number of CD8 T cells are decreased by ≈50% (3, 4), and this defect was recapitulated in the IL-15Rα–/– →IL-15Rα–/– chimeras (Fig. 1A). The percentage of naive CD8 T cells (CD44low) in the wild-type (Wt)→IL-15Rα–/– chimeras and IL-15Rα–/–→Wt chimeras was greater than in the IL-15Rα–/–→IL-15Rα–/– chimeras, but neither of these mixed BM chimeras achieved normal levels of CD8 T cells that were present in control chimeras (P < 0.01) (Fig. 1 A). In a comparison of the two mixed BM chimeras, there was no significant difference in the percentage of naive CD8 T cells generated in chimeras with IL-15Rα expression limited to BM-derived cells and chimeras with IL-15Rα expression limited to the parenchyma. Therefore, the presence of IL-15Rα in either the BM-derived or non-BM-derived compartment partially rescues CD8 T cell development, suggesting that both cell types participated in generating normal naive CD8 T cell numbers. In contrast, naive CD8 T cell development was defective only when BM-derived cells, but not parenchymal cells, lacked IL-15Rβ (data not shown).
Fig. 1.
Development of naive and memory-phenotype TCRαβ CD8 T cells. TCRγδ T cells in the spleen have differential requirements for IL-15Rα expression. (A) The proportion of CD44lowCD8 TCRαβ T cells present in the spleen of each IL-15Rα–/– chimera as compared to the control chimera (Wt→ Wt). The proportions of CD8 T cells in the mixed BM chimeras (IL-15Rα–/–→ Wt and Wt→ IL-15Rα–/–) were significantly different from the IL-15Rα–→ IL-15Rα–/– and control chimeras (Wt→ Wt) (P < 0.05, n = 10–11) but were not significantly different from each other. (B) Representative histograms of CD44 expression on CD8 T cells. Numbers indicate the average percentage (±SD) of CD44high (memory-phenotype) CD8 T cells present in the indicated chimeras. (C) TCRγδ and Thy1 expression on CD3+CD11b– cells from the spleen of each set of chimeras. Numbers in the dot plots represent the percentage of cells in quadrant. For all fluorescence-activated cell sorter plots, the log scale is four decades.
Even in the absence of overt antigen exposure, the CD8 T cell population contains cells with naive and memory phenotypes (CD44low and CD44high, respectively). The memory-phenotype CD8 T cells are exquisitely sensitive to IL-15 and are deficient in IL-15Rα–/– and IL-15–/– mice (3, 4, 19). Recently it was shown that antigen-specific memory CD8 T cells require IL-15Rα expression by BM-derived cells to undergo basal proliferation in vivo (10, 12). Therefore, the percentage of CD44high cells was determined in the various chimeras. The percentage of memory-phenotype CD8 T cells was decreased to a similar level in the complete absence of IL-15Rα (IL-15Rα–/–→IL-15Rα–/– chimeras) as when IL-15Rα was absent from only the BM-derived cells (Fig. 1B). In contrast, normal percentages of memory-phenotype CD8 T cells were present in chimeras with IL-15Rα expression limited to the BM-derived cells and in the control chimeras. Thus, the development (or maintenance) of memory-phenotype CD8 T cells had a similar requirement for IL-15Rα expression by BM-derived cells as did antigen-specific memory CD8 T cells for IL-15-mediated basal proliferation.
Development of TCRγδ T cells in the intestine also requires IL-15Rα and IL-15 (3, 4), but whether this extends to splenic γδ T cells has not been tested. The small population of splenic Thy1+ γδ T cells was largely unaffected by the absence of IL-15Rα. However, the Thy1– TCRγδ cells in spleen were absent in the IL-15Rα–/–→IL-15Rα–/– chimeras (Fig. 1C). Interestingly, similar to CD8 TCRαβ T cells, expression of IL-15Rα by either hematopoietic or parenchymal cells reversed this defect.
IL-15Rα is acting either as part of the IL-15R complex to facilitate a signal or to present IL-15 to the developing cells. In either case, IL-15 is needed for both functions. Thus, we asked whether the developing CD8 T cells had a preferential requirement for IL-15 expression by BM or parenchymal cells. To this end, mixed BM chimeras were generated by using IL-15–/– mice as a BM source or as BM recipients in different combinations. The percentage of conventional CD8 TCRαβ T cells in the spleen was partially restored when IL-15 was expressed by hematopoietic or parenchymal cells (data not shown). In addition, the percentage of memory-phenotype CD8 T cells was increased in chimeras that expressed IL-15 in BM-derived cells (data not shown). There was little effect on the percentages of splenic Thy1+TCRγδ T cells in the various IL-15–/– chimeras; however, the Thy1– TCRγδ cells required expression of IL-15 in at least the BM-derived or non-BM-derived compartments (data not shown).
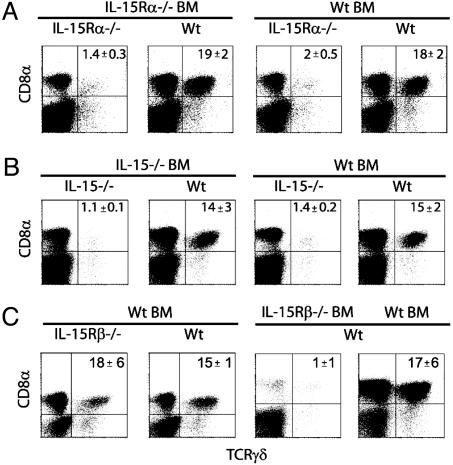
Parenchymal Cell Expression of IL-15Rα and IL-15, but Not IL-15Rβ, Is Required for the Generation of TCRγδ IEL. Specific subsets of IEL, such as CD8ααTCRγδ and CD8ααTCRαβ IEL subsets, depend heavily on IL-15 and IL-15Rα for development (3, 4). To determine whether IEL subsets required specific cell types to express IL-15Rα, IL-15Rα–/– chimeric mice were examined for the presence of IEL subsets. Total numbers of IEL recovered from IL-15Rα–/–→IL-15Rα–/– chimeras and Wt→IL-15Rα–/– chimeras were decreased by 30% (data not shown). Among the IL-15Rα–/– chimeras, TCRγδ IEL were severely deficient in IL-15Rα–/–→IL-15Rα–/– chimeras and Wt→IL-15Rα–/– chimeras (Fig. 2A), demonstrating an absolute requirement for IL-15Rα expression by parenchymal cells. In contrast, TCRγδ IEL were present at similar levels in the IL-15Rα–/–→Wt chimeras and in Wt→Wt chimeras. This indicates that expression of IL-15Rα limited to the parenchyma was sufficient for normal γδ T cell development. To determine requirements for IL-15 expression, IEL from IL-15–/– chimeras were also examined for the ability to allow TCRγδ T cell development. Following the same trend as the IL-15Rα–/– chimera, γδ T cells were virtually absent in chimeras that lacked IL-15 expression by the parenchyma (Fig. 2B). Furthermore, expression of IL-15 by the parenchyma alone was completely sufficient to allow γδ T cell development (Fig. 2B). Thus, in contrast to splenic CD8 TCRαβ and TCRγδ T cell development, parenchymal cell IL-15Rα and IL-15 expression was critical to TCRγδ IEL generation.
Fig. 2.
TCRγδ IEL development strictly requires parenchymal cell expression of IL-15Rα and IL-15 but not IL-15Rβ. IEL were isolated from various chimeras, and the proportion of TCRγδ T cells was determined. Data are the percentages of donor+CD8α+TCRγδ+ IEL in IL-15Rα–/– chimeras (A), in IL-15–/– chimeras (B), and in IL-15β–/– chimeras (C). These data are representative of four experiments (n = 2–3 mice per group). Numbers represent the average percentage of cells in quadrant ± SD. The percentage of CD8α+TCRγδ+ IEL in the chimeras (IL-15Rα–→ IL-15Rα–/–, Wt→ IL-15Rα–/–, and IL-15Rβ–/–→ Wt) was significantly different from the IL-15Rα–/–→ Wt,Wt→ IL-15Rβ–/–, and control chimeras (Wt→ Wt) (P < 0.05). For all fluorescence-activated cell sorter plots, the log scale is four decades.
One alternative to the theory that IL-15 is transpresented to opposing cells is the possibility that IL-15 acts on an IL-15-responsive cell to induce a secondary factor that acts back on the opposing cell. Thus, we determined whether Wt BM-derived cells could generate TCRγδ IEL in hosts that lack the signaling component IL-15Rβ. In Wt→IL-15Rβ–/– chimeras, TCRγδ IEL were generated in normal percentages (Fig. 2C), indicating that TCRγδ IEL development did not require parenchymal cell expression of IL-15Rβ. BM chimeras were also generated that lack IL-15Rβ in the BM compartment. In these IL-15Rβ–/– →Wt chimeras, TCRγδ IEL failed to develop (Fig. 2C), indicating that this subset required direct IL-15R signaling for their production. Altogether, these data suggested that IL-15 responses by the developing IEL by means of transpresentation of IL-15 by parenchymal cells was the major mechanism of IL-15-mediated IEL development.
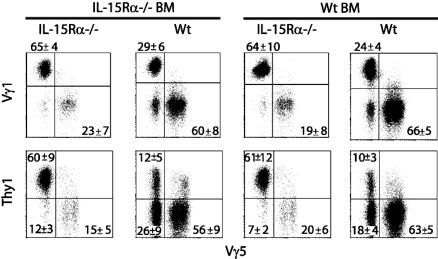
Differences in Vγ Usage and Thy1 Expression Are Dictated by the Pattern of IL-15Rα Expression. TCRγδ T cells in specific tissue locations use distinct TCR V regions. TCRγδ IEL contain T cells that use Vγ5, Vγ1, and Vγ2 (18, 20–22). We wanted to determine whether TCRγδ IEL with different Vγ usage were equally affected by IL-15Rα deficiency. In Wt→Wt chimeras, the predominant IEL subset used Vγ5, followed by Vγ1 (Fig. 3); only a small portion expressed Vγ2 (data not shown). When IL-15Rα expression was absent from the parenchyma, Vγ1+ IEL predominated over Vγ5+ IEL. IEL in IL-15Rα–/–→Wt chimeras were virtually indistinguishable from those in Wt→Wt chimeras, and Wt→IL-15Rα–/– chimeras were similar to those found in IL-15Rα–/–→IL-15Rα–/– chimeras (Fig. 3).
Fig. 3.
Specific TCRγδ IEL subsets are differentially affected by the loss of IL-15Rα expression. Expression of specific Vγ regions and Thy1 by IEL from IL-15Rα–/– chimeras was determined. (Upper) The percentage of Vγ1+ and Vγ5+ cells present in IEL. (Lower) The relationships between Thy1 and Vγ5 expression. All panels demonstrate staining after gating on CD8α+TCRγδ cells. Numbers represent the average percentage of cells in quadrant ± SD (n = 5 mice per group). The differences in the proportions of IEL subsets in the knockout (KO)→ KO and Wt→ KO chimeras were significantly different from those in the KO→ Wt and Wt→ Wt chimeras (P < 0.05). For all fluorescence-activated cell sorter plots, the log scale is four decades.
TCRγδ IEL can be furthered subdivided into Thy1+ and Thy1– populations (23). Thy1 expression by IEL is thought to be a marker of TCR-triggered activation (24). In the control chimeras, most TCRγδ IEL were Thy1–, and this was also true in the IL-15Rα–/–→Wt chimeras (Fig. 3). The few TCRγδ IEL that did develop in Wt→IL-15Rα–/– and IL-15Rα–/–→IL-15Rα–/– chimeras were predominately Thy1+ (Fig. 3). Because most Vγ5+ IEL were Thy1–, this subpopulation was more affected by the absence of IL-15Rα in parenchymal cells (Fig. 3). Therefore, among the TCRγδ IEL, the Thy1– population was more affected by the lack of IL-15Rα expression by parenchymal cells. With respect to the requirements for IL-15 expression and IEL development, the lack of IL-15 expression in parenchymal cells resulted in defective development of Thy1– and Vγ5+ TCRγδ IEL (data not shown). In chimeras with IL-15 expression only in the parenchyma, these IEL subsets were restored to levels similar to the control chimeras (data not shown). Development of the Thy1– and Vγ5+ IEL depended on the expression of IL-15Rβ in the BM-derived compartment but not by the parenchyma (data not shown). Overall, these data suggested that both IL-15Rα and IL-15 expressed by the parenchyma were crucial for the development of specific TCRγδ IEL subsets.
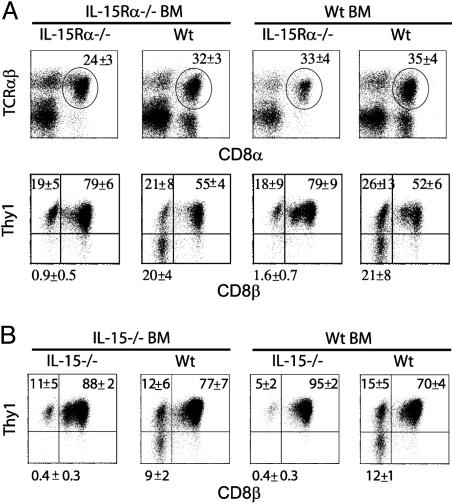
Development of CD8ααTCRαβ IEL Specifically Requires IL-15Rα and IL-15 Expression by Parenchymal Cells. TCRαβ IEL can also be subdivided based on CD8α, CD8β, and Thy1 expression (23), and CD8ααTCRαβ IEL require IL-15/IL-15Rα for development (3, 4). In the IL-15Rα–/– chimeras, analysis of IEL revealed little differences in the development of the overall CD8αTCRαβ population (Fig. 4A). Whereas CD8αβTCRαβ IEL are all Thy1+, the CD8ααTCRαβ IEL are a mixed population of Thy1+ and Thy1– cells. Interestingly, the Thy1– CD8αα population developed similarly in both IL-15Rα–/–→Wt chimeras and Wt→Wt chimeras but failed to develop in Wt→IL-15Rα–/– and IL-15Rα–/–→IL-15Rα–/– chimeras (Fig. 4A). Thus, as with TCRγδ IEL, the expression of IL-15Rα by parenchymal cells alone was sufficient for normal development of CD8ααThy1– IEL.
Fig. 4.
CD8αα TCRαβ IEL development strictly requires IL-15Rα and IL-15 expression by parenchymal cells. IEL were isolated from IL-15Rα and IL-15 chimeras, and the proportion of TCRαβ T cell subsets was determined. (A) The percentage of CD8α+TCRαβ+ IEL in IL-15Rα–/– chimeras. In Upper, the circles indicate the gating used in the secondary analysis depicted in Lower. (Lower) The expression of Thy1 and CD8β after gating on CD8α+TCRαβ+ cells. (B) Analysis of Thy1 and CD8β expression after gating on CD8α+TCRαβ+ IEL in IL-15 chimeras. These data are representative of four experiments (n = 2–3 mice per group). Numbers represent the average percentage of cells in the respective quadrants ± SD. The percentages in the Thy1–CD8α+CD8β– population in the KO→ KO and Wt→ KO chimeras were significantly decreased compared with the KO→ Wt and Wt→ Wt chimeras (P < 0.05). For all fluorescence-activated cell sorter plots, the log scale is four decades.
The requirement for IL-15 expression for the development of CD8αα TCRαβ IEL was also determined in IL-15–/– chimeras (Fig. 4B). In chimeras in which IL-15 was absent from the parenchyma, the percentage of CD8αα TCRαβ cells in the IEL was decreased. This decrease could be accounted for in large part by the absence of the Thy1– population. Therefore, the Thy1– CD8αα TCRαβ IEL had an absolute requirement for both IL-15 and IL-15Rα expression by parenchymal cells. Similar to TCRγδ IEL, the development of CD8αα TCRαβ IEL did not require IL-15Rβ expression by parenchymal cells but did require IL-15Rβ expression by BM-derived cells (data not shown), suggesting that this population also utilizes IL-15 by means of the transpresentation by parenchymal cells.
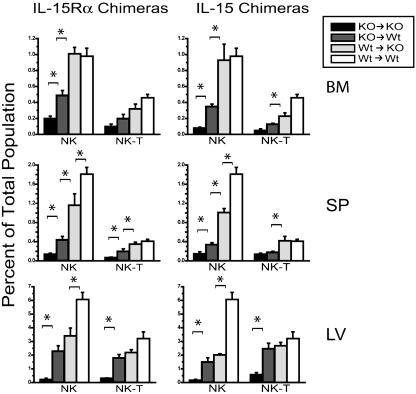
Multiple Cell Types Participate in IL-15-Mediated NK Cell Development. In the absence of IL-15Rα or IL-15 expression, NK and NK/T cells are severely deficient (3, 4). Therefore, we analyzed various tissues in the IL-15Rα and IL-15–/– chimeras for development of NK and NK/T cells. These tissues included BM, spleen, and liver (sites known to have significant numbers of NK cells). When either IL-15Rα or IL-15 was expressed by BM-derived cells or the parenchyma, NK cells were partially or completely restored in the three tissues examined, suggesting that both BM-derived and non-BM-derived cells contributed to NK development (Fig. 5). Within the BM and the spleen, expression of IL-15Rα and IL-15 by the BM-derived cells was more important than expression by parenchyma for NK and NK/T cell development (Fig. 5). This hierarchy did not hold true for the NK and NK/T cells in the liver, where the contribution of BM-derived and parenchymal cells was equivalent. With regard to the role of IL-15Rβ in NK and NK/T cell development, we found that the expression of IL-15Rβ by the parenchyma was dispensable, whereas expression by BM-derived cells was absolutely required (data not shown).
Fig. 5.
NK cell development requires IL-15Rα and IL-15 expression by both BM-derived and non-BM-derived cells. Lymphocytes were isolated from BM, spleen (SP), and liver (LV) of IL-15Rα–/– and IL-15–/– chimeras 10 weeks after irradiation and BM reconstitution. Graphs show the average percentage of CD3-NK1.1+ cells (NK) and CD3+NK1.1+ cells (NK/T) in each tissue as determined by flow cytometry. These data are representative of four experiments (n = 2–3 mice per group). Brackets with asterisks indicate values that are significantly different from each other (P < 0.05).
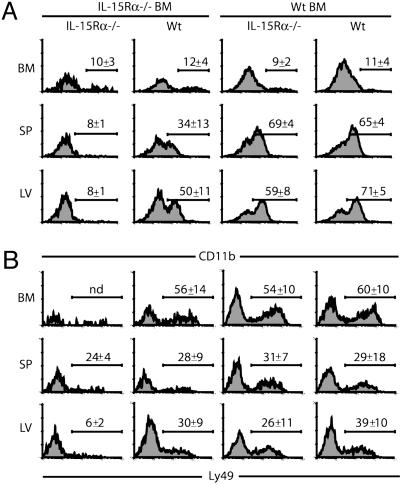
We also examined whether the level of maturation of NK and NK/T cells was differentially affected by IL-15Rα and IL-15 expression by certain cell types. Several cell-surface markers have been proposed as indicators of mature NK cells. In the present study, we analyzed CD11b and Ly49 expression as indicators of conventional NK cell and NK/T cell maturation, respectively (25–27). Few CD11b+ NK cells are normally present in BM (25), and this was reiterated in all of the chimeras (Fig. 6A). For maturation in the spleen, expression of IL-15Rα by BM-derived cells was more important than expression by parenchymal cells, whereas in the liver the contribution of both tissue types was equivalent. Using a second proposed conventional NK cell maturation marker, CD43, we found identical progression in its expression as CD11b (data not shown). For NK/T cells, expression of IL-15Rα by either BM-derived or non-BM-derived cells was equally sufficient to induce some maturation of NK/T cells and demonstrated no preference for expression of IL-15Rα by either BM-derived or parenchymal cells (Fig. 6B). These trends in NK and NK/T cell maturation were also observed in the IL-15 chimeras (data not shown).
Fig. 6.
Maturation of NK and NK/T cells requires IL-15Rα expression. (A) CD11b expression on CD3-NK1.1+ lymphocytes in the BM, spleen (SP), and liver (LV). (B) Ly49 expression on CD3+NK1.1+ lymphocytes in the indicated tissues. Numbers represent the average percentage of CD11b+ or Ly49+ cells ± SD.
Discussion
The traditional view of how cytokines mediate their effects has been challenged by the theory that IL-15 can be transpresented to opposing cells. Evidence is accumulating supporting the idea that transpresentation of IL-15 is a major mechanism of IL-15-mediated actions. IL-15 is widely expressed and has the potential to have deleterious effects if not properly controlled. Sequestration of IL-15 to those cells expressing the high-affinity IL-15Rα may serve to prevent unnecessary responses to IL-15. Moreover, binding of IL-15 to cell-surface IL-15Rα may effectively prolong the half-life of IL-15, thereby allowing continual acquisition of cytokine by lymphoid cells without the necessity for continuous biosynthesis of IL-15, whose half-life in vivo is extremely short (28).
Our study provides further evidence that transpresentation of IL-15 may be used for development of CD8 T cells, NK cells, and IEL subsets in vivo. With respect to IL-15-dependent development of IEL subsets, our data suggest that transpresentation is a major mechanism of IL-15 actions. Whereas IL-15Rα expression by parenchymal tissue alone was completely sufficient to direct IEL development, BM-derived expression of IL-15Rα mediates the proliferation of memory CD8 T cells (Fig. 1B and refs. 10 and 12). Interestingly, naive CD8 T cells and NK cells could use IL-15Rα or IL-15 when expressed by either BM-derived or parenchymal cells. Previous studies have shown that IL-15Rα–/– NK cells exhibit normal survival after transfer into a normal host but fail to survive after transfer into IL-15Rα–/– hosts (11). Together, these studies support the notion that IL-15Rα expression by non-NK hematopoietic cells is important for NK and NK/T cell development and survival. Among the CD8 T cells, NK cells, and IEL subsets where transpresentation of IL-15 is likely the major mechanism of IL-15-mediated development, we show that distinct cell types require expression of IL-15Rα depending on the lymphoid developmental pathway.
It remains possible that the effects of IL-15 on development are mediated through a second factor that drives development after induction by IL-15. For example, IL-15 could act on intestinal epithelial cells to induce another factor (e.g., IL-7) crucial to IEL development. Intestinal epithelial cells express IL-15R components and respond to IL-15 (29). However, our data indicating that parenchymal cells did not require the expression of IL-15Rβ for IEL development was strong evidence that parenchymal cells do not need to respond to IL-15. Furthermore, IEL development required BM-derived cell expression of the IL-15R signaling component IL-2/15Rβ suggesting that IEL or their precursors needed to receive a direct IL-15 signal. In NK transfer studies, survival of NK cells was normal in a γC-deficient background but defective when NK cells lacked γC, suggesting that the NK cells, not host cells, need to respond to IL-15 (30, 31). Thus far, the criteria for transpresentation of IL-15 appear to be expression of IL-15Rα by the presenting cell and IL-15Rβ and γC by the responding cell. One important criterion for this system to operate is the ability of two cell types to interact with each other. IEL are closely apposed to intestinal epithelial cells, thereby promoting an intimate interaction. This is one reason why we theorize that the intestinal epithelial cell is the parenchymal cell type that is transpresenting IL-15 to the IEL, although we cannot currently rule out the possibility that thymic parenchyma is mediating transpresentation of IL-15 to IEL precursors. Which cell types mediate transpresentation of IL-15 to CD8 T cells and NK/NK/T cells is not known. Overall, our demonstration that transpresentation is being used for CD8 T cell, IEL, and NK/NK/T cell development is new evidence that cell–cell interactions are integral for these developmental processes.
Why do particular IEL subsets require IL-15 and IL-15Rα expression by parenchymal cells whereas others appear IL-15-independent? One possibility is that the two IEL subsets affected in our study, the CD8αα TCRγδ and TCRαβ IEL, undergo some level of development directly in the intestine, perhaps in the cryptopatches (32, 33). Thus, CD8αα IEL are touted to be extrathymically derived, although these populations or their precursors can also be thymus-derived (34, 35). Although the evidence of extrathymic development is strongest for TCRγδ IEL (36), it is clear that the CD8αα IEL populations, regardless of TCR expression, represent a distinct lineage that shares some developmental requirements with NK/NK/T cells (37, 38). In contrast, the other major IEL subset is comprised of conventional phenotype TCRαβ CD8αβ T cells, which develop in the thymus and migrate into the IEL compartment in response to activation in the periphery (39, 40). Interestingly, antigen-specific memory CD8 T cells in the IEL would be expected to require IL-15Rα expression by BM-derived cells for their maintenance (10, 12); however, other than T cells, few BM-derived cells are present in the IEL compartment. Therefore, one would suspect either that bona fide memory CD8 T cells in the IEL are short-lived or that these memory cells have distinct requirements for IL-15Rα expression compared with memory cells elsewhere in the body. Whether the TCRαβ CD8αβ IEL are representative of peripheral antigen-specific memory CD8 T cells is not clear. Further studies are needed to examine these issues.
The effects of IL-15 on lymphoid development could be mediated during differentiation, expansion, survival, or a combination of these processes. With respect to IEL, IL-15 could promote IEL precursor differentiation in situ or perhaps in the thymus or BM. Alternatively, or in addition, mature IEL arriving at the epithelium could acquire IL-15 to promote cell expansion and/or survival. In comparison, evidence suggests that generation of naive CD8 T cells does not require IL-15 for thymic development but may help maintain mature, naive CD8 T cell survival (refs. 4, 41, and 42 and our unpublished results). In contrast to naive CD8 T cells, homeostasis of memory CD8 T cells relies on both survival and a continual low level of division, both of which can be mediated by IL-15 (43–47). For NK cells, IL-2/15Rβ is expressed midway through the developmental pathway, and IL-15 has been shown to drive differentiation of NK progenitors to mature NK cells in vitro (48–50). Once NK cells have completed development, NK cells do not undergo slow division as part of a homeostatic mechanism, although mature NK cells will expand in a NK-deficient environment (30, 31). Previous studies have shown that IL-15 is crucial for NK cell survival by maintaining Bcl-2 levels (50, 51). Thus, it is likely that transpresentation of IL-15 can mediate multiple functions, including differentiation, survival, and proliferation.
Overall, our findings emphasized the complexity of the IL-15/IL-15R system and demonstrated a dichotomy in IL-15R-mediated events. Whereas memory CD8 T cells primarily use IL-15Rα expressed by BM-derived cells, IEL development depended strictly on IL-15Rα expression by parenchymal cells. Given the data on transpresentation of IL-15, a plausible scenario would entail production and presentation of IL-15 by IEC, whereas memory cells could acquire IL-15 from dendritic cells as they migrate through secondary lymphoid or tertiary tissues. In contrast, naive CD8 T cells and NK/NK/T cells used IL-15Rα expressed by either hematopoietic or parenchymal cells for development and homeostasis. Future studies will shed light on the mechanism of cell type-specific effects of IL-15 presentation and provide clues as to how cells of distinct lymphoid lineages acquire and respond to IL-15 to mediate development and survival.
Acknowledgments
This work was supported by National Institutes of Health Grants T32-AI07080 (to E.C.N.), HL66963 (to L.P.), AI35917 (to L.L. and L.P.), AI46708 (to H.L.A.), and AI41576, AI051583, and DK45260 (to L.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IEL, intestinal intraepithelial lymphocyte; NK, natural killer; γC, common γ chain; BM, bone marrow; TCR, T cell antigen receptor; Wt, wild-type; IL-15R, IL-15 receptor; KO, knockout.
References
- 1.Tanchot, C., Rosado, M. M., Agenes, F., Freitas, A. A. & Rocha, B. (1997) Semin. Immunol. 9, 331–337. [DOI] [PubMed] [Google Scholar]
- 2.Tough, D. F. & Sprent, J. (1995) Immunol. Res. 14, 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Lodolce, J. P., Boone, D. L., Chai, S., Swain, R. E., Dassopoulos, T., Trettin, S. & Ma, A. (1998) Immunity 9, 669–676. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy, M. K., Glaccum, M., Brown, S. N., Butz, E. A., Viney, J. L., Embers, M., Matsuki, N., Charrier, K., Sedger, L., Willis, C. R., et al. (2000) J. Exp. Med. 191, 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giri, J. G., Kumaki, S., Ahdieh, M., Friend, D. J., Loomis, A., Shanebeck, K., DuBose, R., Cosman, D., Park, L. S. & Anderson, D. M. (1995) EMBO J. 15, 3654–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, D. M., Kumaki, S., Ahdieh, M., Bertles, J., Tometsko, M., Loomis, A., Giri, J., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., et al. (1995) J. Biol. Chem. 270, 29862–29869. [DOI] [PubMed] [Google Scholar]
- 7.Grabstein, K. H., Eisenman, J., Shanebeck, K., Rauch, C., Srinivasan, S., Fung, V., Beers, C., Richardson, J., Schoenborn, M. A. & Ahdieh, M. (1994) Science 264, 965–968. [DOI] [PubMed] [Google Scholar]
- 8.Giri, J. G., Ahdieh, M., Eisenman, J., Shanebeck, K., Grabstein, K., Kumaki, S., Namen, A., Park, L. S., Cosman, D. & Anderson, D. (1994) EMBO J. 13, 2822–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois, S., Mariner, J., Waldmann, T. A. & Tagaya, Y. (2002) Immunity 17, 537–547. [DOI] [PubMed] [Google Scholar]
- 10.Burkett, P. R., Koka, R., Chien, M., Chai, S., Chan, F., Ma, A. & Boone, D. L. (2003) Proc. Natl. Acad. Sci. USA 100, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koka, R., Burkett, P. R., Chien, M., Chai, S., Chan, F., Lodolce, J. P., Boone, D. L. & Ma, A. (2003) J. Exp. Med. 197, 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schluns, K. S., Klonowski, K. D. & Lefrançois, L. (2003) Blood 103, 988–994. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki, H., Kundig, T. M., Furlonger, C., Wakeham, A., Timms, E., Matsuyama, T., Schmits, R., Simard, J. J., Ohashi, P. S., Griesser, H., et al. (1995) Science 268, 1472–1476. [DOI] [PubMed] [Google Scholar]
- 14.Dennert, G., Hyman, R., Lesley, J. & Trowbridge, I. S. (1980) Cell. Immunol. 53, 350–364. [DOI] [PubMed] [Google Scholar]
- 15.Masopust, D., Vezys, V., Marzo, A. L. & Lefrançois, L. (2001) Science 291, 2413–2417. [DOI] [PubMed] [Google Scholar]
- 16.Laky, K., Lefrançois, L. & Puddington, L. (1997) J. Immunol. 158, 1417–1427. [PubMed] [Google Scholar]
- 17.Goodman, T. & Lefrançois, L. (1989) J. Exp. Med. 170, 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira, P., Gerber, D., Huang, S. Y. & Tonegawa, S. (1995) J. Exp. Med. 182, 1921–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, X., Sun, S., Hwang, I., Tough, D. F. & Sprent, J. (1998) Immunity 8, 591–599. [DOI] [PubMed] [Google Scholar]
- 20.Goodman, T. & Lefrançois, L. (1988) Nature 333, 855–858. [DOI] [PubMed] [Google Scholar]
- 21.Asarnow, D. M., Goodman, T., Lefrançois, L. & Allison, J. P. (1989) Nature 341, 60–62. [DOI] [PubMed] [Google Scholar]
- 22.Itohara, S., Farr, A. G., Lafaille, J. J., Bonneville, M., Takagaki, Y., Hass, W. & Tonegawa, S. (1990) Nature 343, 754–757. [DOI] [PubMed] [Google Scholar]
- 23.Parrott, D. M. V., Tait, C., MacKenzie, S., Mowat, A. M., Davies, M. D. J. & Micklem, H. S. (1983) Ann. N.Y. Acad. Sci. 409, 307–320. [DOI] [PubMed] [Google Scholar]
- 24.Lefrançois, L. & Goodman, T. (1989) Science 243, 1716–1718. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S., Iizuka, K., Kang, H. S., Dokun, A., French, A. R., Greco, S. & Yokoyama, W. M. (2002) Nat. Immunol. 3, 523–528. [DOI] [PubMed] [Google Scholar]
- 26.Benlagha, K., Kyin, T., Beavis, A., Teyton, L. & Bendelac, A. (2002) Science 296, 553–555. [DOI] [PubMed] [Google Scholar]
- 27.Gapin, L., Matsuda, J. L., Surh, C. D. & Kronenberg, M. (2001) Nat. Immunol. 2, 971–978. [DOI] [PubMed] [Google Scholar]
- 28.Pettit, D. K., Bonnert, T. P., Eisenman, J., Srinivasan, S., Paxton, R., Beers, C., Lynch, D., Miller, B., Yost, J., Grabstein, K. H., et al. (1997) J. Biol. Chem. 272, 2312–2318. [DOI] [PubMed] [Google Scholar]
- 29.Reinecker, H. C., MacDermott, R. P., Mirau, S., Dignass, A. & Podolsky, D. K. (1996) Gastroenterology 111, 1706–1713. [DOI] [PubMed] [Google Scholar]
- 30.Prlic, M., Blazar, B. R., Farrar, M. A. & Jameson, S. C. (2003) J. Exp. Med. 197, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranson, T., Vosshenrich, C. A., Corcuff, E., Richard, O., Muller, W. & Di Santo, J. P. (2003) Blood 101, 4887–4893. [DOI] [PubMed] [Google Scholar]
- 32.Saito, H., Kanayama, Y., Takemori, T., Nariuchi, H., Kubota, E., Takahashi-Iwanaga, H., Iwanaga, T. & Ishikawa, H. (1998) Science 280, 275–278. [DOI] [PubMed] [Google Scholar]
- 33.Laky, K., Lefrançois, L., Lingenheld, E. G., Ishikawa, H., Lewis, J. M., Olson, S., Suzuki, K., Tigelaar, R. E. & Puddington, L. (2000) J. Exp. Med. 191, 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefrançois, L. & Olson, S. (1994) J. Immunol. 153, 987–995. [PubMed] [Google Scholar]
- 35.Lin, T., Matsuzaki, G., Kenai, H. & Nomoto, K. (1994) Eur. J. Immunol. 24, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 36.Lefrançois, L. & Olson, S. (1997) J. Immunol. 159, 538–541. [PubMed] [Google Scholar]
- 37.Hayday, A., Theodoridis, E., Ramsburg, E. & Shires, J. (2001) Nat. Immunol 2, 997–1003. [DOI] [PubMed] [Google Scholar]
- 38.Shires, J., Theodoridis, E. & Hayday, A. C. (2001) Immunity 15, 419–434. [DOI] [PubMed] [Google Scholar]
- 39.Kim, S. K., Schluns, K. S. & Lefrançois, L. (1999) J. Immunol. 163, 4125–4132. [PubMed] [Google Scholar]
- 40.Lefrançois, L., Parker, C. M., Olson, S., Schon, M. P., Muller, W., Wagner, N. & Puddington, L. (1999) J. Exp. Med. 189, 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, T. S., Lee, J. M., Lai, Y. G., Hsu, J. C., Tsai, C. Y., Lee, Y. H. & Liao, N. S. (2002) J. Immunol. 168, 705–712. [DOI] [PubMed] [Google Scholar]
- 42.Berard, M., Brandt, K., Paus, S. B. & Tough, D. F. (2003) J. Immunol. 170, 5018–5026. [DOI] [PubMed] [Google Scholar]
- 43.Tough, D. F. & Sprent, J. (1994) J. Exp. Med. 179, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Judge, A. D., Zhang, X., Fujii, H., Surh, C. D. & Sprent, J. (2002) J. Exp. Med. 196, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schluns, K. S., Williams, K., Ma, A., Zheng, X. X. & Lefrançois, L. (2002) J. Immunol. 168, 4827–4831. [DOI] [PubMed] [Google Scholar]
- 46.Goldrath, A. W., Sivakumar, P. V., Glaccum, M., Kennedy, M. K., Bevan, M. J., Benoist, C., Mathis, D. & Butz, E. A. (2002) J. Exp. Med. 195, 1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becker, T. C., Wherry, E. J., Boone, D., Murali-Krishna, K., Antia, R., Ma, A. & Ahmed, R. (2002) J. Exp. Med. 195, 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colucci, F., Caligiuri, M. A. & Di Santo, J. P. (2003) Nat. Rev. Immunol. 3, 413–425. [DOI] [PubMed] [Google Scholar]
- 49.Williams, N. S., Moore, T. A., Schatzle, J. D., Puzanov, I. J., Sivakumar, P. V., Zlotnik, A., Bennett, M. & Kumar, V. (1997) J. Exp. Med. 186, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, H., Fehniger, T. A., Fuchshuber, P., Thiel, K. S., Vivier, E., Carson, W. E. & Caligiuri, M. A. (1998) Blood 92, 3647–3657. [PubMed] [Google Scholar]
- 51.Minagawa, M., Watanabe, H., Miyaji, C., Tomiyama, K., Shimura, H., Ito, A., Ito, M., Domen, J., Weissman, I. L. & Kawai, K. (2002) J. Immunol. 169, 4153–4160. [DOI] [PubMed] [Google Scholar]
- 52.Cooper, M. A., Bush, J. E., Fehniger, T. A., VanDeusen, J. B., Waite, R. E., Liu, Y., Aguila, H. L. & Caligiuri, M. A. (2002) Blood 100, 3633–3638. [DOI] [PubMed] [Google Scholar]