Abstract
Skeletal muscle is essential for normal bodily function and the loss of skeletal muscle (i.e. muscle atrophy/wasting) can have a major impact on mobility, whole-body metabolism, disease resistance, and quality of life. Thus, there is a clear need for the development of therapies that can prevent the loss, or increase, of skeletal muscle mass. However, in order to develop such therapies, we will first have to develop a thorough understanding of the molecular mechanisms that regulate muscle mass. Fortunately, our knowledge is rapidly advancing, and in this review, we will summarize recent studies that have expanded our understanding of the roles that Smad signaling and the synthesis of phosphatidic acid play in the regulation of skeletal muscle mass.
The importance of skeletal muscle
Skeletal muscle plays essential roles in the human body, including the generation of limb, trunk, and eye movements and the control of breathing. Furthermore, because skeletal muscle makes up such a large, metabolically active proportion of the body (~45%), it also plays an important role in the regulation of whole-body metabolism (e.g. systemic glucose control) [1,2]. As such, the loss of skeletal muscle mass (i.e. muscle atrophy) can have a major impact on mobility, whole-body metabolism, disease resistance, and quality of life [3-5]. Muscle atrophy occurs in many commonly occurring conditions (e.g. injury-induced immobilization, severe burns, cancer cachexia, aging, heart failure, obesity, diabetes, chronic obstructive pulmonary disease, renal disease, and HIV) and can be induced by changes in a variety of factors, including nutrients, neural activity, cytokines, growth factors, hormones, and mechanical loading [4-9]. Thus, there is clearly a need for therapies that can prevent the loss, or increase, of skeletal muscle mass. However, in order to develop such therapies, we will first have to develop a thorough understanding of the molecular mechanisms that regulate muscle mass.
In recent years, significant progress has been made in identifying potential molecular signaling mechanisms that regulate skeletal muscle mass under a range of different conditions (for recent reviews, see [10-22]). In this review, we will summarize the major advancements that have been made in our understanding of the roles that Smad signaling and the synthesis of phosphatidic acid (PA) play in the regulation of muscle mass (Figure 1). For further background information on these two areas of investigation, the reader is referred to the following reviews: [23-27].
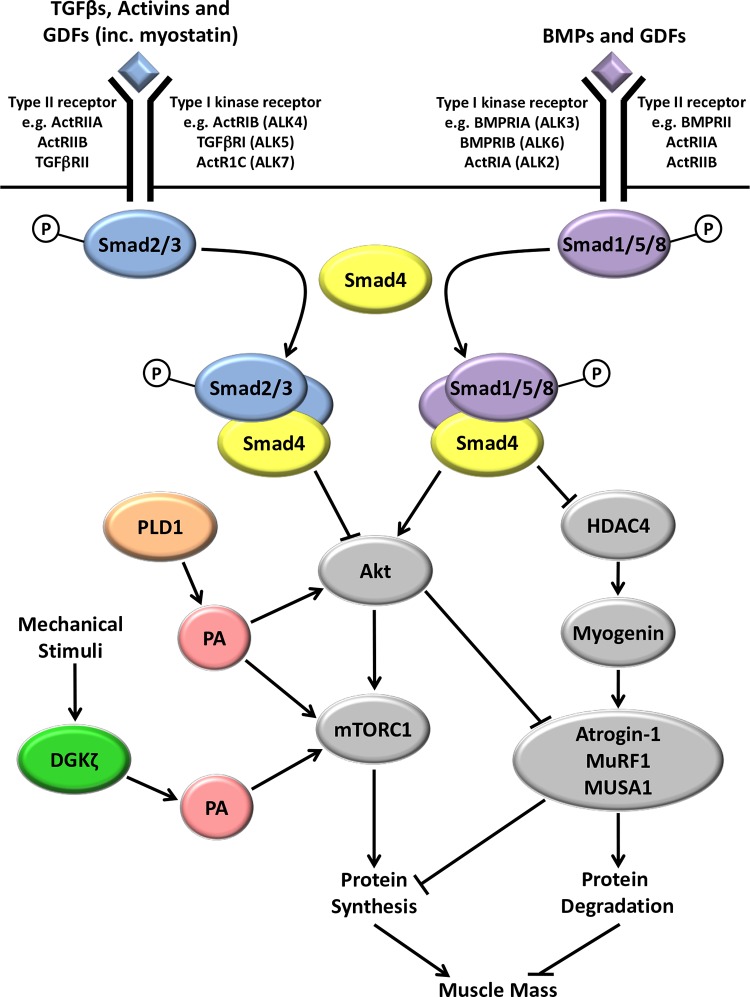
Figure 1. A general overview of recent findings related to Smad signaling and the synthesis of phosphatidic acid in the regulation of skeletal muscle mass.
Increases in transforming growth factor-beta (TGFβ), activins, and some growth and differentiation factors (GDFs), such as myostatin, lead to receptor-mediated phosphorylation of Smads 2 and 3, whereas increases in bone morphogenic proteins (BMPs) and other GDFs result in increased Smad 1, 5, and 8 phosphorylation. Phosphorylated Smads 2/3 compete with phosphorylated Smads 1/5/8 for binding to Smad4 before entering the nucleus to regulate gene transcription. Increased Smad2/3/4 signaling inhibits Akt/mTORC1 signaling and activates the expression of ubiquitin proteasome E3 ligases, atrogin-1 and muscle RING-finger protein-1 (MuRF1), ultimately leading to a decrease in protein synthesis and increased protein degradation. Increased Smad1/5/8/4 signaling activates Akt/mechanistic target of rapamycin complex 1 (mTORC1) signaling and represses the histone deacetylase 4 (HDAC4)/myogenin-mediated activation of E3 ligases (including MUSA1) under atrophic conditions, such as denervation. The net balance between Smad2/3/4 and Smad1/5/8/4 signaling appears to play a major role in determining skeletal muscle mass. The enzyme phospholipase D1 (PLD1) synthesizes PA, and PLD1 overexpression activates mTORC1 signaling and represses the expression of the E3 ligases, atrogin-1 and MuRF1. The zeta isoform of diacylglycerol kinase (DGKζ) also synthesizes PA and is required for a mechanically induced increase in PA and the subsequent activation of mTORC1. See text (“Bone morphogenic protein-induced Smad1/5/8 signaling and the regulation of skeletal muscle mass” section) for a more detailed explanation of these molecular signaling mechanisms.
The role of Smad signaling in the regulation of skeletal muscle mass: recent advances
Myostatin-induced Smad2/3 signaling and the regulation of skeletal muscle mass
Canonical Smad signaling is regulated by members of the transforming growth factor-beta (TGF-β) superfamily of receptor ligand proteins. For example, Smad2/3 signaling is activated by members of the TGF-β and activin subfamilies and by some members of the growth differentiation factor (GDF) subfamily, whereas Smads 1/5/8 are activated by members of the bone morphogenic protein (BMP) and GDF subfamilies [28,29] (Figure 1). Initial interest in the role of Smad signaling in the regulation of skeletal muscle mass largely stemmed from the discovery that the disruption of the GDF-8 (also known as the myostatin) gene, and the inhibition of myostatin receptor binding, produced marked skeletal muscle hypertrophy [30,31]. Furthermore, it has since been shown that an increase in myostatin is often associated with skeletal muscle atrophy [32,33] and an increase in Smad3 phosphorylation [34]. Combined, these studies suggested that myostatin is a negative regulator of skeletal muscle mass and that Smad signaling may, in part, mediate this effect.
Myostatin-mediated Smad signaling is activated by the binding of the mature myostatin peptide to plasma membrane-associated activin type IIB and, to a lesser extent, type IIA receptors (ActRIIB/IIA) (Figure 1) [35]. The activation of ActRIIB/IIA receptors leads to the recruitment and activation of serine/threonine type 1 receptor kinases called activin receptor-like kinase-4 and -5 (ALK4 and ALK5), which phosphorylate the transcription factors Smad2 and 3 [35]. Phosphorylated Smad2 or 3 can then form a heterotrimeric complex with another Smad2 or 3, and a Smad4, and the resulting complex translocates to the nucleus to regulate gene transcription by associating with various transcription factors, co-activators, or co-repressors [36] (Figure 1). Smad2 and 3 are also phosphorylated in response to TGF-β and activin binding to TGF-β type II receptors (TGF-βRII) and ActRIIA/IIB receptors which, in turn, leads to the activation of ALK5 and ALK4/7, respectively [37,38] (Figure 1).
The necessity for Smad signaling in myostatin/activin/TGF-β-induced muscle atrophy in vivo was first demonstrated by Sartori et al. (2009), who showed that the overexpression of a constitutively active mutant of ALK5 (caALK5) increased the promoter activity of the muscle-specific ubiquitin proteasome E3 ligase atrogin-1 gene and induced muscle fiber atrophy via a Smad3-dependent mechanism [39]. Furthermore, Smad3 has been found to be necessary for myostatin-induced increases in atrogin-1 expression and atrophy of cultured myotubes [40,41]. Other studies have also shown that increased myostatin is sufficient to induce a decrease in protein synthesis, possibly by promoting the atrogin-1-mediated degradation of ribosomal proteins and translation initiation factors or via the inhibition of signaling through the protein kinase B (Akt) and mechanistic target of rapamycin complex 1 (mTORC1) pathway [40-49] or both. Importantly, it has been demonstrated that Smad3 signaling is necessary for the myostatin-induced inhibition of the Akt/mTORC1 pathway [40,50]. Combined, these data suggest that myostatin-induced muscle atrophy may be due, in part, to a combination of Smad3-mediated increases in atrogin-1 expression and the inhibition of Akt/mTORC1 signaling [41,51,52] (Figure 1). However, while Smad3 was shown to be necessary for these events, it remained to be determined whether Smad3 is sufficient to induce these events in vivo or whether Smad3 merely plays a permissive role.
To address the issue of whether Smad3 is sufficient to induce many of the reported effects of increased myostatin signaling, we overexpressed Smad3 in vivo and demonstrated that this was sufficient to activate the atrogin-1 promoter, inhibit mTORC1 signaling and protein synthesis, and ultimately induce muscle fiber atrophy [53]. Furthermore, we and others have recently presented evidence that the myostatin/Smad3-induced decrease in Akt/mTORC1 signaling may be mediated by a decrease in the expression of microRNAs (i.e. miR29 or miR486 or both) that normally inhibit the translation of the Akt signaling antagonist, phosphatase and tensin homologue (PTEN) [53,54]. Thus, combined with previous studies, these results demonstrated that Smad3 is both necessary and sufficient to regulate many of the previously reported myostatin-induced events. Furthermore, the results of this study suggest that Smad3 signaling may be a viable target for therapies aimed at ameliorating many of the detrimental effects of increased myostatin, activin, or TGF-β expression (or a combination of these) on skeletal muscle.
Bone morphogenic protein-induced Smad1/5/8 signaling and the regulation of skeletal muscle mass
While there has been a significant amount of research aimed at identifying the factors that regulate the myostatin-Smad3 signaling axis, there has been comparatively less investigation into the potential role(s) of BMP signaling in mature skeletal muscle. BMP signaling is initiated by the binding of BMP ligands to heterodimeric membrane receptor complexes, made up of specific combinations of type II (e.g. BMP type II receptor [BMPRII], ActRIIA, and ActRIIB) and type I receptors (e.g. BMPRIA [ALK3], BMPRIB [ALK6], and ActRIa [ALK2]), which, in turn, leads to the phosphorylation of Smads 1, 5, and 8 [55] (Figure 1). As with Smads 2/3, phosphorylated Smads 1/5/8 form heterotrimeric complexes with Smad4, which then translocates to the nucleus to regulate gene transcription. Importantly, two recent studies have provided compelling evidence that BMPs do indeed play a role in mature skeletal muscle by regulating muscle mass through a Smad1/5/8-dependent mechanism [56,57]. Specifically, it was shown that the inhibition of BMP receptor signaling and the knockdown of Smads 1 and 5 were sufficient to inhibit Smad1/5/8 phosphorylation and induce muscle fiber atrophy [56]. Furthermore, it was found that the overexpression of BMP7 and caALK3 were sufficient to increase Smad1/5/8 phosphorylation and induce substantial muscle fiber hypertrophy [56,57]. Importantly, these hypertrophic responses were repressed by BMP receptor inhibition and knockdown of Smad 1 and 5 [56,57]. Moreover, the overexpression of BMP7 and caALK3 was sufficient to protect against denervation-induced muscle atrophy [56,57]. Together, these data have provided strong evidence that signaling by BMP can positively regulate skeletal muscle mass.
The mechanisms responsible for the effects of BMP signaling on muscle mass appear to be due, at least in part, to increased activity of mTORC1 [57] (Figure 1). Indeed, the overexpression of BMP7 and caALK3 was associated with an increase in mTORC1 signaling (possibly mediated by an increased insulin-like growth factor 1 [IGF-1]-induced Akt-phosphorylation), and BMP7-induced muscle hypertrophy was completely prevented by the mTORC1 inhibitor, rapamycin [57]. Although these results underscore the central role of mTORC1 in regulating skeletal muscle mass, further investigation is required to firmly establish the link between increased BMP signaling, IGF-1 expression, Akt phosphorylation, and mTORC1 activation. Moreover, additional work is needed to determine whether the hypertrophic effects of BMP signaling are associated with an increase in protein synthesis or a decrease in protein degradation or both. Although these two studies provide clear evidence that the overexpression of BMP and caALK3 is sufficient to regulate muscle mass, they also present data showing that endogenous BMP signaling plays a role in limiting the extent of muscle loss that occurs during atrophic conditions, such as denervation. Indeed, it was found that, in denervated muscle, there was an increase in Smad1/5/8 phosphorylation and this was associated with an increase in the expression of BMP13 (GDF6) and 14 (GDF5) [56,57]. Furthermore, denervation-induced muscle fiber atrophy was markedly exacerbated in BMP14 knockout mice and in denervated wildtype mice in which BMP signaling was inhibited with the BMP type I receptor inhibitor, LDN-193189, or overexpression of inhibitory Smad6 [56,57]. These data strongly suggest that endogenous BMP signaling acts to protect skeletal muscle from excessive atrophy. Interestingly, unlike the overexpression of BMP7 and caAKL3, the mechanism behind the endogenous BMP signaling-induced protection against muscle atrophy does not seem to be related to an increase in Akt/mTORC1 signaling [56,57]. Instead, this protective effect appears to be due to the suppression of muscle-specific ubiquitin proteasome E3 ligases, including the recently identified MUSA1 (also known as Fbxo30) [56,57]. For example, inhibition of BMP signaling via overexpression of Smad6 or an extracellular BMP binding protein called Noggin enhanced the increase in MUSA1 expression that occurs in response to denervation [56,57]. Furthermore, the knockdown of MUSA1 was sufficient to protect against denervation-induced atrophy [56,57]. Mechanistically, BMP signaling may protect against the induction of E3 ligases, such as MUSA1, by inhibiting histone deacetylase 4 (HDAC4)-mediated myogenin expression [56,57], a signaling pathway that has also previously been implicated in the expression of atrogin-1 and muscle RING-finger protein-1 (MuRF1) [58] (Figure 1). Combined, the findings from these two recent studies [56,57] highlight the potential for increasing BMP signaling as a possible therapeutic approach for increasing muscle mass or inhibiting muscle atrophy/wasting or both. One question that remains to be answered is why Akt/mTORC1 signaling was not enhanced by the increase in endogenous BMP signaling found in denervated muscles but was enhanced in innervated muscles subjected to the overexpression of BMP7 and caALK3.
Finally, although these two recent BMP studies arrive at different conclusions as to which Smad signaling branch (i.e. myostatin/activin/TGF-β-Smad2/3 or BMP-Smad1/5/8) is dominant under basal conditions (see [56,57] for further details), a major conceptual contribution of these studies is the hypothesis that the net balance between these two pathways plays a major role in determining skeletal muscle mass. Furthermore, it appears that competition for Smad4 plays a central role in this balancing act [56] (Figure 1). For example, under conditions of increased myostatin, activin, or TGF-β signaling (or a combination of these), an increase in phosphorylated Smads 2 or 3 or both would effectively sequester a greater proportion of the Smad4 pool and thus limit the ability of phosphorylated Smads 1, 5, or 8 (or a combination of these) to form Smad4-containing complexes, and vice versa. Therefore, an increase in Smad2/3/4 complexes entering the nucleus, combined with a decrease in nuclear Smad1/5/8/4 complexes, would likely lead to an increase in the Smad2/3-mediated transcriptional events that are reported to occur under various atrophic conditions and a decrease in Smad1/5/8-mediated anabolic events. As a result, the overall effect would be a reduction in skeletal muscle mass. Interestingly, this hypothesis could also explain some of our recently reported findings on the effect of Smad3 overexpression on muscle fiber size (i.e. the overexpression of Smad3 may have decreased BMP/Smad1/5/8 signaling) [53].
In summary, recent studies have significantly advanced our understanding of the role that Smad signaling plays in the regulation of skeletal muscle mass. In particular, the discovery that BMP-Smad1/5/8 signaling plays a significant role in the regulation of muscle mass, and the finding that competition for Smad4 may be fundamental to the balance between catabolic myostatin/activin/TGF-β-Smad2/3 and anabolic BMP-Smad1/5/8 signaling, will hopefully facilitate the development of pharmaceutical or genetic therapies (or both) aimed at regulating Smad signaling and preventing muscle wasting.
The role of phosphatidic acid in the regulation of skeletal muscle mass: recent advances
PA is a glycerophospholipid second messenger that has a diverse range of cellular functions, including the regulation of cell growth (for review, see [59]). Indeed, in 2001, PA was shown to activate mTORC1, a master regulator of protein synthesis and cell growth [60,61]. Since then, numerous studies have shown that the stimulation of cells with exogenous PA, or the overexpression of PA-generating enzymes, can activate mTORC1 signaling [62-66], whereas blocking the generation of PA has been reported to inhibit the activation of mTORC1 that occurs in response to various types of stimuli [60,67-70]. Mechanistically, it has been reported that PA can directly activate mTORC1 kinase activity in vitro and this appears to be due to its ability to bind to the FKBP12-rapamycin binding (FRB) domain of mTOR [60,65,71,72].
PA can be synthesized by a number of different classes of enzymes including the following: phospholipase D (PLD), which synthesizes PA from phosphatidylcholine (PC); lysophosphatidic acid acyltransferases (LPAAT), which synthesize PA from lysophosphatidic acid (LPA); and the diacylglycerol kinases (DAGKs), which synthesize PA from diacylglycerol (DAG) [59,66]. To date, the number of studies that have examined the role of PA-synthesizing enzymes in skeletal muscle is quite limited (for reviews, see [25,26]); however, two recent studies have significantly expanded our understanding of the role of the PA-synthesizing enzymes, PLD1 and the zeta isoform of DGK (DGKζ), in the regulation of mTORC1 signaling and skeletal muscle mass [73,74] (Figure 1).
The synthesis of phosphatidic acid by phospholipase D1 as a regulator of skeletal muscle mass
In the first of these studies, the pharmacological inhibition, and short interfering RNA (siRNA)-mediated knockdown, of PLD1 was shown to induce atrophy in cultured myotubes, suggesting that PA produced by PLD1 is necessary for maintaining muscle mass [73]. Conversely, the overexpression of PLD1 was sufficient to induce hypertrophy of myotubes and of muscle fibers in vivo [73]. PLD1 overexpression was also able to inhibit dexamethasone-induced myotube atrophy, whereas the addition of exogenous PA inhibited myotube atrophy induced by dexamethasone and tumor necrosis factor-alpha (TNFα) [73]. Combined, these data provide strong support for the hypotheses that PA derived from PLD1 contributes to the regulation of skeletal muscle mass and that the activation of PLD1 has the potential to prevent muscle atrophy. Mechanistically, it appears that PLD1 exerts effects on both protein synthesis and protein degradation (Figure 1). For instance, the overexpression of PLD1 was associated with an increase in mTORC1 signaling, whereas the siRNA-mediated knockdown of PLD1 had the opposite effect [73]. As the activation of mTORC1 signaling is known to be sufficient to increase protein synthesis and induce muscle fiber hypertrophy [75], these data suggest that PLD1-derived PA controls skeletal muscle mass, in part, via the regulation of mTORC1 and protein synthesis. Interestingly, PLD1 overexpression was also associated with a decrease in the expression of FoxO3 and the E3 ligase genes, atrogin-1 and MuRF1 [73]. Moreover, the expression of these genes was increased by pharmacological inhibition and knockdown of PLD1, whereas the addition of exogenous PA attenuated the activation of these genes by dexamethasone [73]. The PLD1/PA-induced inhibition of these atrophy genes appears to be due to the activation of Akt, possibly via the activation of the rapamycin-insensitive mTOR complex 2 (mTORC2) [76-78]. For example, PLD1 overexpression was associated with increased Akt phosphorylation, whereas the knockdown of PLD1 had the opposite effect [73]. Combined, these data suggest that PLD1-derived PA regulates skeletal muscle mass via the activation of mTORC1 signaling and protein synthesis and possibly by an mTORC2/Akt-mediated inhibition of ubiquitin proteasome-mediated protein degradation (Figure 1). Given that most of these data were obtained from cultured myotubes, more research is now required to confirm these findings in vivo. Nevertheless, the results of this study highlight that the activation of PLD1/PA signaling may be a potential therapeutic strategy for inducing muscle hypertrophy or preventing muscle wasting or both.
The role of diacylglycerol kinase in the mechanical activation of mechanistic target of rapamycin complex 1 signaling
Not only is PLD/PA signaling implicated in the regulation of basal skeletal muscle mass, but previous studies have also suggested that this signaling pathway plays a role in the activation of mTORC1 and protein synthesis by mechanical stimuli [68,79]. For example, ex vivo passive stretch, which activates mTORC1 signaling, has been shown to increase PLD activity and the synthesis of PA [68]. Furthermore, the stretch-induced increases in PA and mTORC1 signaling were prevented when muscles were incubated with the PLD inhibitor 1-butanol [68]. In a separate study, in vivo eccentric contractions were also shown to increase PA and mTORC1 signaling and 1-butanol prevented the activation of mTORC1 signaling [79]. Combined, the data obtained with 1-butanol suggested that PLD-derived PA plays a significant role in the mechanical activation of mTORC1 signaling.
Although 1-butanol has been used as a PLD inhibitor for many years, more recent studies have questioned its specificity by showing that several of the biological effects of 1-butanol cannot be attributed to its effects on the synthesis of PA by PLD [80-82]. Thus, we re-examined whether the mechanically induced increase in PA requires PLD by using a small-molecule inhibitor of PLD called 5-fluoro-2-indolyl des-chlorohalopemide (FIPI) [80]. To our surprise, we found that FIPI did not inhibit the passive stretch-induced increases in PA or mTORC1 signaling, suggesting that changes in PLD activity were not necessary for these events [74]. Instead, it was shown that passive stretch induces an increase in DAG, suggesting that there could be corresponding increases in DGK activity. Indeed, a passive stretch was sufficient to increase membrane DGK activity, but further experiments showed that the α, β, γ, and θ isoforms of DGK were not necessary for the mechanically induced increase in PA and mTORC1 signaling [74]. These findings prompted us to explore the potential role of DGKζ, and using DGKζ knockout mice, it was found that the passive stretch-induced increase in PA was almost completely abolished and that the activation of mTORC1 signaling was markedly impaired [74]. These data suggested that DGKζ is predominantly responsible for the increase in PA and contributes to the mechanical activation of mTORC1 signaling (Figure 1). Additional experiments also showed that the in vivo overexpression of DGKζ was sufficient to induce muscle fiber hypertrophy in a kinase-dependent manner (i.e. via the synthesis of PA) and that this hypertrophic response was largely inhibited by rapamycin [74]. Combined, the results of this study highlight a novel role for DGKζ (but not PLD as previously thought) in the mechanical activation of PA-mTORC1 signaling and provide new insights into the potential mechanism(s) through which mechanical stimuli regulate muscle mass. Further research is now required to confirm the role of DGKζ in other models of mechanical stimulation (e.g. eccentric contractions and synergist ablation-induced mechanical overload) and to determine whether DGKζ-derived PA is sufficient to increase protein synthesis or decrease protein degradation or both.
Conclusions
Significant progress continues to be made in our understanding of the diverse range of molecular mechanisms that can regulate skeletal muscle mass under various physiological and pathophysiological conditions. In this review, we have specifically focused on recent studies that have significantly advanced our understanding of the roles that Smad signaling and the synthesis of PA play in the regulation of muscle mass. It is expected that these advancements will facilitate the development of therapies for conditions that are associated with muscle atrophy/wasting, metabolic disease, and reduced mobility and ultimately help to improve quality of life.
Acknowledgments
This work was supported by the National Institutes of Health grants AR057347 (TAH) and AR063256 (CAG and TAH).
Abbreviations
- ALK
activin receptor-like kinase
- BMP
bone morphogenic protein
- caALK
constitutively active ALK
- DAG
diacylglycerol
- DGKζ
diacylglycerol kinase zeta isoform
- FIPI
5-fluoro-2-indolyl des-chlorohalopemide
- GDF
growth differentiation factor
- IGF-1
insulin-like growth factor 1
- mTORC
mechanistic target of rapamycin complex
- MuRF1
muscle RING-finger protein-1
- PA
phosphatidic acid
- PLD
phospholipase D
- PLD1
phospholipase D1
- siRNA
short interfering RNA
- TGF-β
transforming growth factor-beta
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/6/20
References
- 1.Lee RC, Wang Z, Heo M, Ross R, Janssen I, Heymsfield SB. Total-body skeletal muscle mass: development and cross-validation of anthropometric prediction models. Am J Clin Nutr. 2000;72:796–803. doi: 10.1093/ajcn/72.3.796. [DOI] [PubMed] [Google Scholar]
- 2.Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–72. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seguin R, Nelson ME. The benefits of strength training for older adults. Am J Prev Med. 2003;25:141–9. doi: 10.1016/S0749-3797(03)00177-6. [DOI] [PubMed] [Google Scholar]
- 4.Srikanthan P, Karlamangla AS. Relative muscle mass is inversely associated with insulin resistance and prediabetes. Findings from the third National Health and Nutrition Examination Survey. J Clin Endocrinol Metab. 2011;96:2898–903. doi: 10.1210/jc.2011-0435. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307005
- 5.Lynch GS. Tackling Australia's future health problems: developing strategies to combat sarcopenia--age-related muscle wasting and weakness. Intern Med J. 2004;34:294–6. doi: 10.1111/j.1444-0903.2004.00568.x. [DOI] [PubMed] [Google Scholar]
- 6.Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008;23:160–70. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 8.Frost RA, Lang CH. Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J Appl Physiol. 2007;103:378–87. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- 9.Bodine SC. mTOR signaling and the molecular adaptation to resistance exercise. Med Sci Sports Exerc. 2006;38:1950–7. doi: 10.1249/01.mss.0000233797.24035.35. [DOI] [PubMed] [Google Scholar]
- 10.Zacharewicz E, Lamon S, Russell AP. MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front Physiol. 2013;4:266. doi: 10.3389/fphys.2013.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhees KJP, Pansters NAM, Schols AMWJ, Langen RCJ. Regulation of skeletal muscle plasticity by glycogen synthase kinase-3β: a potential target for the treatment of muscle wasting. Curr Pharm Des. 2013;19:3276–98. doi: 10.2174/1381612811319180011. [DOI] [PubMed] [Google Scholar]
- 12.Tabebordbar M, Wang ET, Wagers AJ. Skeletal muscle degenerative diseases and strategies for therapeutic muscle repair. Annu Rev Pathol. 2013;8:441–75. doi: 10.1146/annurev-pathol-011811-132450. [DOI] [PubMed] [Google Scholar]
- 13.Senf SM. Skeletal muscle heat shock protein 70: diverse functions and therapeutic potential for wasting disorders. Front Physiol. 2013;4:330. doi: 10.3389/fphys.2013.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schakman O, Kalista S, Barbé C, Loumaye A, Thissen JP. Glucocorticoid-induced skeletal muscle atrophy. Int J Biochem Cell Biol. 2013;45:2163–72. doi: 10.1016/j.biocel.2013.05.036. [DOI] [PubMed] [Google Scholar]
- 15.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol. 2013;45:2121–9. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ljubicic V, Jasmin BJ. AMP-activated protein kinase at the nexus of therapeutic skeletal muscle plasticity in Duchenne muscular dystrophy. Trends Mol Med. 2013;19:614–24. doi: 10.1016/j.molmed.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49:59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace MA, Lamon S, Russell AP. The regulation and function of the striated muscle activator of rho signaling (STARS) protein. Front Physiol. 2012;3:469. doi: 10.3389/fphys.2012.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Bhatnagar S, Paul PK. TWEAK and TRAF6 regulate skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2012;15:233–9. doi: 10.1097/MCO.0b013e328351c3fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman CA, Mayhew DL, Hornberger TA. Recent progress toward understanding the molecular mechanisms that regulate skeletal muscle mass. Cell Signal. 2011;23:1896–906. doi: 10.1016/j.cellsig.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch GS, Ryall JG. Role of beta-adrenoceptor signaling in skeletal muscle: implications for muscle wasting and disease. Physiol Rev. 2008;88:729–67. doi: 10.1152/physrev.00028.2007. [DOI] [PubMed] [Google Scholar]
- 22.Goodman CA. The Role of mTORC1 in Regulating Protein Synthesis and Skeletal Muscle Mass in Response to Various Mechanical Stimuli. Rev Physiol Biochem Pharmacol. 2014 doi: 10.1007/112_2013_17. [DOI] [PubMed] [Google Scholar]
- 23.Han HQ, Zhou X, Mitch WE, Goldberg AL. Myostatin/activin pathway antagonism: molecular basis and therapeutic potential. Int J Biochem Cell Biol. 2013;45:2333–47. doi: 10.1016/j.biocel.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchida K. The role of myostatin and bone morphogenetic proteins in muscular disorders. Expert Opin Biol Ther. 2006;6:147–54. doi: 10.1517/14712598.6.2.147. [DOI] [PubMed] [Google Scholar]
- 25.Hornberger TA. Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle. Int J Biochem Cell Biol. 2011;43:1267–76. doi: 10.1016/j.biocel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs BL, Goodman CA, Hornberger TA. The mechanical activation of mTOR signaling: an emerging role for late endosome/lysosomal targeting. J Muscle Res Cell Motil. 2013 doi: 10.1007/s10974-013-9367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams GR, Bamman MM. Characterization and regulation of mechanical loading-induced compensatory muscle hypertrophy. Compr Physiol. 2012;2:2829–70. doi: 10.1002/cphy.c110066. [DOI] [PubMed] [Google Scholar]
- 28.Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005;16:251–63. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Moustakas A, Heldin C. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 30.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307010
- 31.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–11. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Cadavid NF, Taylor WE, Yarasheski K, Sinha-Hikim I, Ma K, Ezzat S, Shen R, Lalani R, Asa S, Mamita M, Nair G, Arver S, Bhasin S. Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proc Natl Acad Sci USA. 1998;95:14938–43. doi: 10.1073/pnas.95.25.14938. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718307015
- 33.Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee S. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–8. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1006668
- 34.Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, Kambadur R. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J Biol Chem. 2002;277:49831–40. doi: 10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1012804
- 35.Elkina Y, von Haehling S, Anker SD, Springer J. The role of myostatin in muscle wasting: an overview. J Cachexia Sarcopenia Muscle. 2011;2:143–51. doi: 10.1007/s13539-011-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Walton KL, Makanji Y, Harrison CA. New insights into the mechanisms of activin action and inhibition. Mol Cell Endocrinol. 2012;359:2–12. doi: 10.1016/j.mce.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 38.Tsuchida K, Nakatani M, Uezumi A, Murakami T, Cui X. Signal transduction pathway through activin receptors as a therapeutic target of musculoskeletal diseases and cancer. Endocr J. 2008;55:11–21. doi: 10.1507/endocrj.KR-110. [DOI] [PubMed] [Google Scholar]
- 39.Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:C1248–57. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- 40.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol. 2009;296:C1258–70. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307022
- 41.Lokireddy S, McFarlane C, Ge X, Zhang H, Sze SK, Sharma M, Kambadur R. Myostatin induces degradation of sarcomeric proteins through a Smad3 signaling mechanism during skeletal muscle wasting. Mol Endocrinol. 2011;25:1936–49. doi: 10.1210/me.2011-1124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; http://f1000.com/prime/718307029
- 42.Rodriguez J, Vernus B, Toubiana M, Jublanc E, Tintignac L, Leibovitch S, Bonnieu A. Myostatin inactivation increases myotube size through regulation of translational initiation machinery. J Cell Biochem. 2011;112:3531–42. doi: 10.1002/jcb.23280. [DOI] [PubMed] [Google Scholar]
- 43.Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27:1266–76. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1120440
- 44.Amirouche A, Durieux A, Banzet S, Koulmann N, Bonnefoy R, Mouret C, Bigard X, Peinnequin A, Freyssenet D. Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology. 2009;150:286–94. doi: 10.1210/en.2008-0959. [DOI] [PubMed] [Google Scholar]
- 45.Taylor WE, Bhasin S, Artaza J, Byhower F, Azam M, Willard DH, Kull FC, Gonzalez-Cadavid N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab. 2001;280:E221–8. doi: 10.1152/ajpendo.2001.280.2.E221. [DOI] [PubMed] [Google Scholar]
- 46.Welle S, Bhatt K, Pinkert CA. Myofibrillar protein synthesis in myostatin-deficient mice. Am J Physiol Endocrinol Metab. 2006;290:E409–15. doi: 10.1152/ajpendo.00433.2005. [DOI] [PubMed] [Google Scholar]
- 47.Welle S, Burgess K, Mehta S. Stimulation of skeletal muscle myofibrillar protein synthesis, p70 S6 kinase phosphorylation, and ribosomal protein S6 phosphorylation by inhibition of myostatin in mature mice. Am J Physiol Endocrinol Metab. 2009;296:E567–72. doi: 10.1152/ajpendo.90862.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welle S, Mehta S, Burgess K. Effect of postdevelopmental myostatin depletion on myofibrillar protein metabolism. Am J Physiol Endocrinol Metab. 2011;300:E993–E1001. doi: 10.1152/ajpendo.00509.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hulmi JJ, Oliveira BM, Silvennoinen M, Hoogaars WMH, Ma H, Pierre P, Pasternack A, Kainulainen H, Ritvos O. Muscle protein synthesis, mTORC1/MAPK/Hippo signaling, and capillary density are altered by blocking of myostatin and activins. Am J Physiol Endocrinol Metab. 2013;304:E41–50. doi: 10.1152/ajpendo.00389.2012. [DOI] [PubMed] [Google Scholar]
- 50.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 51.Lokireddy S, Mouly V, Butler-Browne G, Gluckman PD, Sharma M, Kambadur R, McFarlane C. Myostatin promotes the wasting of human myoblast cultures through promoting ubiquitin-proteasome pathway-mediated loss of sarcomeric proteins. Am J Physiol Cell Physiol. 2011;301:C1316–24. doi: 10.1152/ajpcell.00114.2011. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307035
- 52.Lokireddy S, Wijesoma IW, Sze SK, McFarlane C, Kambadur R, Sharma M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am J Physiol Cell Physiol. 2012;303:C512–29. doi: 10.1152/ajpcell.00402.2011. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307057
- 53.Goodman CA, McNally RM, Hoffmann FM, Hornberger TA. Smad3 induces atrogin-1, inhibits mTOR and protein synthesis, and promotes muscle atrophy in vivo. Mol Endocrinol. 2013;27:1946–57. doi: 10.1210/me.2013-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hitachi K, Nakatani M, Tsuchida K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int J Biochem Cell Biol. 2014;47:93–103. doi: 10.1016/j.biocel.2013.12.003. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718213299
- 55.Mueller TD, Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012;586:1846–59. doi: 10.1016/j.febslet.2012.02.043. [DOI] [PubMed] [Google Scholar]
- 56.Sartori R, Schirwis E, Blaauw B, Bortolanza S, Zhao J, Enzo E, Stantzou A, Mouisel E, Toniolo L, Ferry A, Stricker S, Goldberg AL, Dupont S, Piccolo S, Amthor H, Sandri M. BMP signaling controls muscle mass. Nat Genet. 2013;45:1309–18. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718122888
- 57.Winbanks CE, Chen JL, Qian H, Liu Y, Bernardo BC, Beyer C, Watt KI, Thomson RE, Connor T, Turner BJ, McMullen JR, Larsson L, McGee SL, Harrison CA, Gregorevic P. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J Cell Biol. 2013;203:345–57. doi: 10.1083/jcb.201211134. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718151542
- 58.Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/5481962
- 59.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45:250–78. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–5. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1003461
- 61.Park I, Bachmann R, Shirazi H, Chen J. Regulation of ribosomal S6 kinase 2 by mammalian target of rapamycin. J Biol Chem. 2002;277:31423–9. doi: 10.1074/jbc.M204080200. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307095
- 62.Avila-Flores A, Santos T, Rincón E, Mérida I. Modulation of the mammalian target of rapamycin pathway by diacylglycerol kinase-produced phosphatidic acid. J Biol Chem. 2005;280:10091–9. doi: 10.1074/jbc.M412296200. [DOI] [PubMed] [Google Scholar]
- 63.Tang W, Yuan J, Chen X, Gu X, Luo K, Li J, Wan B, Wang Y, Yu L. Identification of a novel human lysophosphatidic acid acyltransferase, LPAAT-theta, which activates mTOR pathway. J Biochem Mol Biol. 2006;39:626–35. doi: 10.5483/BMBRep.2006.39.5.626. [DOI] [PubMed] [Google Scholar]
- 64.O'Neil TK, Duffy LR, Frey JW, Hornberger TA. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J Physiol (Lond) 2009;587:3691–701. doi: 10.1113/jphysiol.2009.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You JS, Frey JW, Hornberger TA. Mechanical stimulation induces mTOR signaling via an ERK-independent mechanism: implications for a direct activation of mTOR by phosphatidic acid. PLoS ONE. 2012;7:e47258. doi: 10.1371/journal.pone.0047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 67.Ballou LM, Jiang Y, Du G, Frohman MA, Lin RZ. Ca(2+)- and phospholipase D-dependent and -independent pathways activate mTOR signaling. FEBS Lett. 2003;550:51–6. doi: 10.1016/S0014-5793(03)00816-0. [DOI] [PubMed] [Google Scholar]
- 68.Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci USA. 2006;103:4741–6. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takahara T, Hara K, Yonezawa K, Sorimachi H, Maeda T. Nutrient-dependent multimerization of the mammalian target of rapamycin through the N-terminal HEAT repeat region. J Biol Chem. 2006;281:28605–14. doi: 10.1074/jbc.M606087200. [DOI] [PubMed] [Google Scholar]
- 70.Ha SH, Kim D, Kim I, Kim JH, Lee MN, Lee HJ, Kim JH, Jang SK, Suh P, Ryu SH. PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals. Cell Signal. 2006;18:2283–91. doi: 10.1016/j.cellsig.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 71.Yoon M, Sun Y, Arauz E, Jiang Y, Chen J. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. J Biol Chem. 2011;286:29568–74. doi: 10.1074/jbc.M111.262816. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718307100
- 72.Veverka V, Crabbe T, Bird I, Lennie G, Muskett FW, Taylor RJ, Carr MD. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene. 2008;27:585–95. doi: 10.1038/sj.onc.1210693. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307125
- 73.Jaafar R, de Larichaudy J, Chanon S, Euthine V, Durand C, Naro F, Bertolino P, Vidal H, Lefai E, Némoz G. Phospholipase D regulates the size of skeletal muscle cells through the activation of mTOR signaling. Cell Commun Signal. 2013;11:55. doi: 10.1186/1478-811X-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718307142
- 74.You J, Lincoln HC, Kim C, Frey JW, Goodman CA, Zhong X, Hornberger TA. The Role of Diacylglycerol Kinase ζ and Phosphatidic Acid in the Mechanical Activation of Mammalian Target of Rapamycin (mTOR) Signaling and Skeletal Muscle Hypertrophy. J Biol Chem. 2014;289:1551–63. doi: 10.1074/jbc.M113.531392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–39. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1019081
- 77.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/S1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718307368
- 78.Jaafar R, Zeiller C, Pirola L, Di Grazia A, Naro F, Vidal H, Lefai E, Némoz G. Phospholipase D regulates myogenic differentiation through the activation of both mTORC1 and mTORC2 complexes. J Biol Chem. 2011;286:22609–21. doi: 10.1074/jbc.M110.203885. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718307375
- 79.O'Neil TK, Duffy LR, Frey JW, Hornberger TA. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J Physiol (Lond) 2009;587:3691–701. doi: 10.1113/jphysiol.2009.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su W, Yeku O, Olepu S, Genna A, Park J, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2009;75:437–46. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718307388
- 81.Yanase Y, Carvou N, Frohman MA, Cockcroft S. Reversible bleb formation in mast cells stimulated with antigen is Ca2+/calmodulin-dependent and bleb size is regulated by ARF6. Biochem J. 2010;425:179–93. doi: 10.1042/BJ20091122. [DOI] [PubMed] [Google Scholar]
- 82.Sato T, Hongu T, Sakamoto M, Funakoshi Y, Kanaho Y. Molecular mechanisms of N-formyl-methionyl-leucyl-phenylalanine-induced superoxide generation and degranulation in mouse neutrophils: phospholipase D is dispensable. Mol Cell Biol. 2013;33:136–45. doi: 10.1128/MCB.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]