Abstract
In studies of sexual mucosal transmission and early stages of simian immunodeficiency virus (SIV) and HIV infections, productive infection predominates in CD4+ T cell populations, in both ostensibly resting and activated cells. The surprising ability of SIV and HIV to replicate in resting cells in vivo, in contrast to propagation of infection in vitro, suggested a model in which during the early stages of infection these viruses exploit the greater availability of resting cells to maintain unbroken chains of transmission from an infected resting cell to another resting cell nearby. Because immune activation in response to infection provides more activated CD4+ T cells, these viruses take advantage of the greater efficiency of virus production and spread in activated cells for propagation and dissemination of infection. In this article, we report the results of experimental tests of this model, including visualization at the light microscopic level and direct analysis of virus production by cells in tissues. Analysis of tissues of rhesus macaques inoculated intravaginally or i.v. with SIV supports the proposed roles of target cell availability, susceptibility, and virus production by infected resting and activated CD4+ T cells in mucosal transmission and early infection, and points to a potential role for topical anti-inflammatory agents in moderating the initial propagation of infection.
In previously reported studies of both the now globally predominant route of sexual transmission of HIV-1 to women (1), and the animal model of intravaginal inoculation of simian immunodeficiency virus (SIV) into rhesus macaques (2), the majority of productively infected cells at the portal of entry and lymphatic tissues (LTs) to which infection spreads were found to be CD4+ T cells. Similarly, in HIV-1 infection, from the acute to late stages of infection, close to 90% of the HIV-1 RNA+ cells in the LT reservoir where virus is produced were typed as CD4+ T cells (3, 4).
In the majority of these infected CD4+ T cells in the early stages of SIV infection, markers of cell activation and proliferation were not detectable, and thus most of the productively infected population in the very early stages of infection was apparently comprised of resting CD4+ T cells (3). This finding was quite surprising, because both in vitro and in vivo HIV-1 was long thought to be incapable of productively infecting quiescent T cells (5–8), and to rely for propagation on productive infection of activated proliferating CD4+ T cells (9–12). Moreover, the rapid depletion of CD4+ T cells in gut-associated LTs and vagina in SIV infection has been thought to be due to preferential replication of SIV in activated CD4+ T cells (13, 14). Nonetheless, recent reports of productive infection of resting cells in ex vivo cultures of LTs and in patients provide additional evidence that in vivo HIV can productively infect resting CD4+ T cells (15–17).
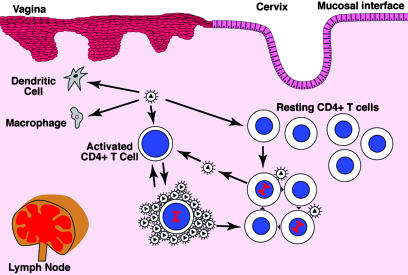
Why would productive SIV and HIV-1 infection be initially found predominantly in resting CD4+ T cells, and what role might infection of these cells play in the early stages of infection? In the target cell substrate availability model shown in Fig. 1, we propose that it is initially mainly resting T cells that are infected after intravaginal inoculation of SIV, simply because of the greater density of resting T cells, compared with other susceptible cell types such as dendritic cells (DCs) at the portal of entry. Whereas DCs may be important in conveying virus across the mucosal barrier (18), we reasoned that the CD4+ T cell would quickly become the major infected cell type because (i) CD4+T cells outnumber other potential target cell types in the female reproductive tract of uninfected rhesus macaques (19) and, (ii) because the spatial proximity of an infected CD4+T cell to other CD4+T cells in small aggregates would facilitate propagation of infection in CD4+ T cells, compared with the more dispersed DCs and macrophages in the submucosa. We further reasoned that more resting CD4+ T cells will be infected in the early stages of infection, because resting CD4+ T cells will greatly outnumber the activated T cell population, with a correspondingly greater likelihood of virus encountering and infecting a resting T cell, until infection induces immune activation both at the portal of entry and in the LTs to which infection disseminates.
Fig. 1.
Target cell substrate availability and virus production by infected resting and activated CD4+ T cells in transmission and acute infection. Virus that crosses the mucosal barrier and spreads to LTs can infect susceptible DCs, macrophages, and both resting and activated CD4+ cells at the portal of entry and in LTs. Because the population of resting CD4+ cells is larger than the other cell types, most of the productively infected cells initially are resting CD4+ T cells. They propagate infection by infecting other nearby resting CD4+ T cells. Infected activated CD4+ T cells produce more virions and transmit infection to cells at greater distances than infected resting cells. Over time, infection-related immune activation leads to an increasing proportion of activated CD4+ T cells in infected cell pool at the portal of entry and LTs, to which infection is disseminated.
In this model, the proposed role of infection of resting CD4+ T cells is to produce sufficient virus to maintain the chain of transmission during the early stages of infection. As infection progresses, the immune activation associated with active viral replication would provide larger numbers of activated CD4+ T cells as potential targets for infection. Whereas HIV and SIV can infect both activated and resting CD4+ T cells, we have shown (3) that activated CD4+ T cells contain >5-fold higher levels of SIV RNA than their resting CD4+ T cell counterparts, which is consistent with higher levels of virus production. We therefore ascribe to infected activated CD4+ T cells the role of amplifiers that propagate and spread infection more efficiently than resting cells, by producing more virus to infect more cells at greater distances.
In this article, we report the results of experimental tests of the predictions of the model: (i) that the proportions of susceptible cell types initially infected should be related to the relative population densities of each susceptible cell type; and, (ii) that changes in the proportions of infected cells of each type should correspond to changes in the population densities. We also describe a method to directly visualize and measure, at the light microscopic level, virus production by cells in tissues, and use this method to examine another major prediction of the model, that infected activated CD4+ T cells produce more virions and could thus infect cells at greater distances than infected resting CD4+ T cells.
Methods
Animals and Experimental SIV Infections. Cervical and LTs that were reexamined and described in this report were from a previous study (3) of female adult Indian rhesus macaques (Macaca mulatta) that had been inoculated intravaginally with SIVmac251 and subsequently killed. Tissues were collected and fixed as described (3). In longitudinal studies of infection of LTs, adult female animals were inoculated i.v. with SIVmac239. Sequential axillary lymph node (LN) biopsies were obtained from anesthetized animals. Plasma SIV RNA levels were determined as described previously (20). All animal housing and care provided and research performed were in conformance with the Guide for the Care and Use of Laboratory Animals (21).
Immunohistochemical Staining (IHCS) and In Situ Hybridization (ISH). IHCS and ISH, single- and double-label IHCS, and combined ISH/IHCS were performed as described (3).
Detection of Virions and Intracellular Viral RNA by ISH/Tyramide Signal Amplification (TSA)/Enzyme-Linked Fluorescence (ELF). To detect virions embedded in agarose and in tissue sections, and viral RNA in cells, sections were cut, deparaffinized, digested with pepsin, washed and refixed in paraformaldehyde, and acetylated. After hybridization to digoxigenin labeled virus-specific RNA probes, washing and digestion with RNase A, bound probe was detected by repeated cycles of TSA and incubation with streptavidin-alkaline phosphatase and ELF 97 substrate (Molecular Probes), followed by extensive washes to reduce or eliminate nonspecific precipitates of the ELF substrate. For combined ISH/IHC, after the ELF 97 substrate step, slides were incubated with rabbit anti-Ki67 antibody, and then anti-rabbit IgG labeled with Cy3.
For further details, see Supporting Methods, which is published as supporting information on the PNAS web site.
Results and Discussion
Target Cell (Substrate) Availability and Proportions of Cell Types Infected in Sexual Mucosal Transmission. The model shown in Fig. 1 predicts that the frequency of infection of different cell types at the portal of entry and LTs to which infection spreads after intravaginal exposure to SIV should be related to the likelihood of virus encountering a susceptible cell and thus to the relative population density of susceptible cell types in the tissues. To test this prediction, we first determined the relationship between target cell availability and proportion of infected cells in cervical and LTs in animals exposed to SIV by intravaginal inoculation. We evaluated target cell availability in archived tissues in which we had already determined at day 7 that SIV RNA+ cells were confined to endocervix and that at day 12 nearly 90% of the SIV RNA+ cells in endocervix and LN were CD4+ Ki67– HLA-DR resting T cells (3). Because the remaining SIV RNA+ cells were CD4+ Ki67+ T cells, DCs, and macrophages, these tissues provided an optimal test of the proposed relationship between the frequency of productive infection of a susceptible cell type and its relative population density.
We found that in endocervix CD4+ T cells were ≈5-fold more abundant than DCs and there were ≈70 times as many Ki67– CD4+ T cells as CD4+ Ki67+ T cells (Table 1). CD4+ T cells were also four times as abundant as macrophages, and both the macrophages and DCs were dispersed through the submucosa, in contrast to the CD4+ T cells that frequently clustered in small follicular aggregates (data not shown). Thus, both the relative density and spatial proximity of target cells would favor infection of predominantly Ki67– CD4+ T cells in these endocervical tissues, which is consistent with our previous findings. Similarly, in the axillary LN to which infection had spread 12 days after inoculation, Ki67–CD4+ T cells outnumbered Ki67+ CD4+ T cells, macrophages, and DCs by an order of magnitude or more (Table 1). Thus, the demonstration in this report that Ki67–CD4+ T cells are the predominant susceptible target cell in these tissues, combined with the previous demonstration that the majority of SIV RNA+ cells in this LN were Ki67–CD4+ T cells, satisfies this prediction of the model that frequency of infection in susceptible cell populations will correlate with the prevalence of target cell types.
Table 1. Target cell availability.
| CD4+T cells
|
||||
|---|---|---|---|---|
| Cells per mm2 | Ki67- | Ki67+ | CD68+ macrophages | S100+ DCs |
| Endocervix* | 413 | 6 | 104 | 90 |
| Axillary LN* | 13,598 | 363 | 1,388 | 1,067 |
| Axillary LN† | 13,699 | 83 | 1,698 | 1,125 |
In rhesus macaque tissues before Intravaginal inoculation.
In rhesus macaque tissues before i.v. inoculation.
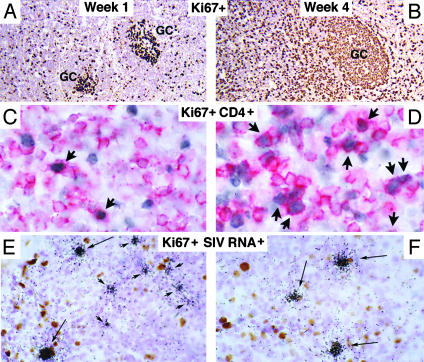
Longitudinal Studies. The second major prediction of the model is that the proportions of infected resting and activated T cells should change as the proportion of activated CD4+ T cells increases with the immune activation that accompanies progressive infection. To test this theory, we examined axillary LN biopsies 1 and 4 weeks after i.v. inoculation of SIV. Consistent with the model, we found that progressive immune activation altered the proportional representation of activated and resting CD4+ T cells in LTs, and thus their availability for infection. In these experiments, we inoculated animals i.v. with a large dose of SIV to maximally expose resting and activated CD4+ T cells, macrophages, and DCs in LTs to virus. At the time of inoculation most of the potential target cells were Ki67–CD4+ T cells (Table 1). Ki67–CD4+ T cells were approximately an order of magnitude more abundant than macrophages or DCs, and more than two orders of magnitude more abundant than activated Ki67+ CD4+ T cells. There was increased immune activation at 4 weeks, compared with 1 week, with dramatic increases in Ki67+ B cells in lymphoid follicles and T cells (Fig. 2 A and B) and specifically, Ki67+CD4+ T cells, in the paracortex (Fig. 2 C and D), which increased 3-fold from 1 to 4 weeks after inoculation (Fig. 3A).
Fig. 2.
Immune activation, availability, and infection of resting and activated CD4+ T cells. (A and B) There are increased numbers of Ki67+ (brown stained) T cells in the paracortex, and B cells in follicles in sections of LN at 4 weeks after i.v. inoculation of SIV, compared with 1 week. (C and D) There are increased numbers of Ki67+ (blue gray nuclei) and CD4+ T cells (red cytoplasm; arrows) at 4 weeks, compared with 1 week. (E and F) There are more Ki67+ (brown nuclei) and SIV RNA+ (black silver grains; long arrows) cells at 4 weeks, compared with 1 week, where most of the SIV RNA+ cells are Ki67– (short arrows).
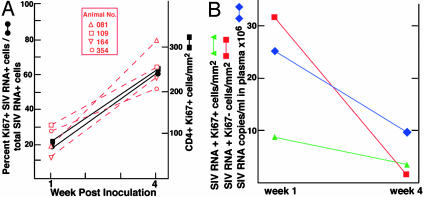
Fig. 3.
(A) Proportions of CD4+ Ki67+ or Ki67– T cells and SIV RNA+ Ki67+ or Ki67– cells. The proportions of SIV RNA+ Ki67+ or Ki67– T cells were determined as described in the text and plotted as averages for the four animals (solid lines and symbols) and individual animals (red dashed lines and open symbols). (B) Changes in frequency of Ki67+- and Ki67–-infected T cells and plasma SIV RNA level from weeks 1 to 4 after i.v. inoculation. Plasma SIV RNA and frequency of SIV RNA+ cells were determined as described in Methods.
To evaluate changes in the size and proportion of Ki67– and Ki67+ SIV viral RNA+ CD4+ T populations, we first determined the proportion of different types of productively infected cells by combining IHCS, using antibodies to identify CD4+ T cells, macrophages, or DCs, with ISH to determine the proportion of each cell type that were SIV viral RNA+. After i.v. inoculation, >95% of the SIV RNA+ cells in the LTs were CD4+ T cells (data not shown), which is equivalent to the proportion found after intravaginal inoculation. To determine the proportion of productively infected resting or activated cells in this population of infected CD4+ cells, we combined IHCS with antibodies to Ki67 and ISH, and then counted the number of singly or doubly labeled cells per mm2 (Fig. 2 E and F). We found that the majority (80%) of SIV RNA+ T cells at 1 week were Ki67–, and 15% were activated CD4+ T cells, which is equivalent to the previously reported proportions of SIV RNA+ resting and activated CD4+ cells in LTs after intravaginal inoculation. The proportion of SIV RNA+ Ki67+ CD4+ cells subsequently increased 3-fold between weeks 1 and 4 (Fig. 3A), which is exactly parallel to the increased proportion of activated T cells. This result thus satisfies the second major prediction of the proposed model of viral propagation in vivo at transmission and early stages of infection.
Substrate Exhaustion and Decreased Plasma Viremia. Two mechanisms have been advanced to explain the decline in plasma levels of HIV-1 and SIV typically observed in the acute stage of infection after the initial peak in plasma viremia. There is evidence that virus-specific cellular immune responses contribute to the decline (22), but there is also theoretical (23) and experimental (13, 14) support for the idea that exhaustion of activated CD4+ T cells as substrates for viral replication plays a major role. In the LTs of the animals we studied, the frequency of activated CD4+ T cells in LTs increased 3-fold between weeks 1 and 4, even as levels of virus in plasma fell (Fig. 3A). This result is inconsistent with the hypothesis that plasma levels decline because of exhaustion of activated CD4+ T cells as substrates for viral replication, at least in peripheral LTs. Because we did not examine gut-associated LTs, it is possible that depletion of activated CD4+ T cells from this compartment could contribute substantially to the decline in plasma viremia. Nonetheless, it is clear that global depletion of activated CD4+ T cells in all LTs cannot completely account for the observed down-regulation of plasma viremia. Moreover, the parallel decreases in plasma SIV RNA and infected CD4+ T cells (Fig. 3B) is consistent with elimination of productively infected cells in LTs by virus-specific CD8+ CTLs or other mechanism as the basis for the decrease in plasma viremia.
Inherent Susceptibilities of Resting and Activated CD4+ T Cells. Although the results shown in Fig. 3A support the target cell availability model, they also suggest that availability alone cannot account for the predominance of infection in the activated CD4+ T cell population at week 4. Even with a 3-fold increase in the relative abundance of activated CD4+ T cells at 4 weeks, there are still many more resting CD4+ T cells that could be infected if they were equally susceptible to infection. Moreover, there are nearly 20 times fewer infected Ki67– CD4+ T cells but only ≈2-fold fewer infected Ki67+ CD4+ T cells at week 4, compared with week 1. We interpret the shift in productive infection to the activated T cell compartment as evidence of a cellular and lymphoid tissue milieu (24) that enhances the susceptibility and ability of activated T cells to replicate virus compared with resting cells.
Speculations That Only a Subset of Resting CD4+ T Cells Can Support Productive Infection and the Origins of Latently Infected Resting Cells. The small fraction of resting CD4+ T cells infected at both 1 and 4 weeks, relative to the sizes of the total resting CD4+ T cell population at these times, and the nearly 20-fold decrease between weeks 1 and 4 in SIV RNA+ Ki67– T cells compared with the 2-fold decrease in SIV RNA+ Ki67+ T cells over this interval, also requires explanation. One speculative possibility we propose here is that only a small fraction of the resting population might actually be capable of supporting productive infection. For example, the infectable population might be recently activated cells returning to a resting state. Although they would be characterized as resting T cells with the reagents currently available for immunophenotyping in tissue sections, they may differ subtly from truly resting cells, e.g., as has been recently shown by Chun et al. (17), in the expression of transcriptional regulatory and other genes, that enable them to support productive infection. With the progressive immune activation that accompanies systemic SIV infection, there would be fewer postactivated cells returning to this state, and thus, a smaller population of resting cells to support productive infection. As the initial population of productively infected resting CD4+ T cells is eliminated by immune or other mechanisms, the decreased availability of an infectable resting CD4+ T cell population, and increased relative frequency of the more susceptible activated CD4+ T cells, would result in the observed greater proportional decrease in the size of the viral RNA+Ki67– T cell population seen at week 4. The decreased numbers of infected resting cells observed at week 4 could also in part reflect conversion of the pool of initially productively infected cells to a state of latent infection, a mechanism in accord with recent ideas about the origins of the latently infected resting CD4+ T cell (25). Additionally or alternatively, the initially infected resting cells could have slowly succumbed to the cumulative cytopathic effects of viral replication, they could have become activated, or they could have been eliminated by host defenses.
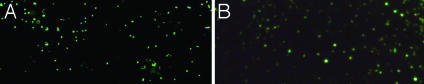
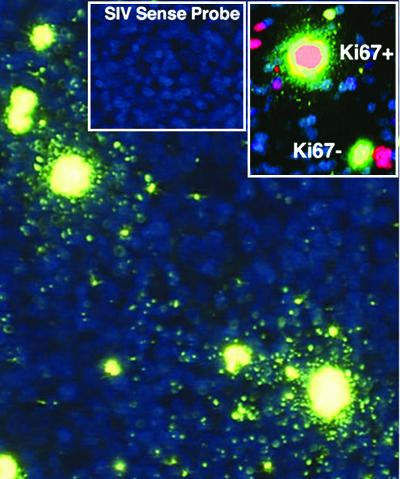
Virus Production and Efficiency of Propagation and Spread of Infection. In the model shown in Fig. 1, infected resting T cells produce fewer virions per cell but sustain infection in its earliest stages by transmitting virus to resting T cells in close proximity. However, infected activated T cells produce more virions per cell and thus may transmit infection more efficiently, through multiplicity effects, and at greater distances. We have previously used relative viral RNA concentrations in infected cells (3) as a surrogate measure of different levels of virus production, and also found higher concentrations of SIV RNA in activated cells, compared with resting cells, in the lymph node biopsies of animals infected i.v. (data not shown). To more rigorously quantify virus production in vivo by resting and activated T cells, we developed an amplification method to visualize virions at the light microscopic level to facilitate counting the number of virions associated with cells in tissues. To visualize virions, we detected viral RNA in them by ISH, and then amplified the signal by TSA, followed by ELF, which generates a strong fluorescent signal that is stable to quenching. We first showed that we that we could visualize virions at the light microscopic level in agarose sections containing predetermined concentrations of virions (Fig. 4), and then applied the method to estimate virus production by resting and activated cells in tissue sections. We first documented the specificity of the method in sections of axillary LN 12 days after intravaginal inoculation. We focused on this tissue because, as described above, we had already shown that 85% of the SIV RNA+ cells in this LN were CD4+ cells, and, of these infected CD4+ T cells, 80% were small Ki67– T cells, whereas 15% were large activated Ki67+ T cells. Fig. 5 illustrates the application of this method and demonstrates several points. There are two large SIV RNA+ cells in the field surrounded by virions concentrated within 2 cell diameters of the infected cell. At the upper left of the field, there are smaller SIV RNA+ cells with relatively few virions visible at the cell surface. The large activated cells are Ki67+, whereas the smaller SIV RNA+ cells with few virions are Ki67– (Inset). Neither viral RNA nor virions were detectable by ISH/TSA/ELF with an SIV sense probe (Inset).
Fig. 4.
Detection of virions by ISH/TSA/ELF. Virions embedded in agarose were detected in sections as described in Methods. The brightest and largest viral particles in the figure are in the plane of focus. [Original magnifications: ×600 (A) and ×2,000 (B).]
Fig. 5.
Detection by ISH/TSA/ELF of SIV RNA in virions and in large Ki67+ and small Ki67– T cells in axillary LN. See text for explanation.
The axillary LN sample also provided an opportunity to test the prediction of the model that activated T cells should transmit infection to more cells, at greater distances, than infected resting T cells. We had previously determined (26) that there was a statistically significant clustering of SIV RNA+ cells within approximately one to two T cell diameters of each other in sections of this LN (26). We now illustrate, in Fig. 5, the clustering of SIV RNA+ cells within a few cell diameters of large activated T cells. The frequent occurrence of small resting SIV RNA+ cells in pairs (see upper left portion of Fig. 5) is consistent with close range transmission of infection between resting cells shown in the model (Fig. 1).
Contributions of Resting and Activated CD4+ T Cells to Virus Production in Acute Infection. By using size to discriminate resting from activated CD4+ T cells, we estimated the numbers of virions associated with small and large SIV RNA+ cells by focusing through several planes at high magnification to clearly identify particles. We counted particles over 100 cells and found that there were, on average, five virions (± 2.9, range 1–17 virions) associated with the small resting cells and 64 virions (± 38.7, range 19–220 virions) associated with large activated T cells. These are minimal estimates, because there may have been some particles that could not be distinguished from the background signal over the cellular interior, mainly from intranuclear viral RNA, and thus would not have been counted. Whereas cumulative virus production over time cannot be determined from this type of analysis, the results nevertheless document in this snap shot of infection a 12-fold greater number of virions associated with activated compared to resting T cells in tissues, which is consistent with the model shown in Fig. 1.
In previous studies (27) of the dynamics of HIV-1 infection in LTs, two populations of mononuclear cells were originally identified that turned over with an apparent t1/2 ≈1–2 days and a t1/2 ≈14 days, respectively, after initiation of antiretroviral therapy. Subsequently, these two dynamic populations were shown to be activated and resting CD4+ T cells (3). Assuming similar t1/2 values in SIV infection (28), we can estimate from the numbers of virions associated with activated CD4+ T cells and resting T cells that cumulative virus production by activated T cells is only ≈1.5-fold greater than resting CD4+ T cells. The relatively high per cell output from resting cells, despite the small numbers of virions associated with resting cells in “snapshots,” reflects the assumption, based on a 14-day half-life, of production of virus over a longer period by resting cells, which, presumably, are better able to survive the cytopathic effects of replication and recognition by immune surveillance because of lower levels of virus gene expression. Because there are so many more infected resting cells, we can calculate virus production from the weighted averages of population size and relative virus production. This calculation leads to the conclusion that the infected resting CD4+ T cell population contributes substantially to virus production in the very earliest stages of infection (C.R., unpublished work).
Virion Production by Resting and Activated Cells and Sexual Mucosal Transmission. We also found that after intravaginal inoculation of SIV there are only a few viral particles at or close to the cell surface of small resting T cells, whereas infected activated CD4+ T cells are surrounded by larger numbers of virions. We had previously shown that almost all of the SIV RNA+ cells detected by ISH with radiolabeled riboprobes in sections of cervix after intravaginal inoculation of SIV were CD4+ T cells (3), reflecting the greater number of resting cells initially available. In reexamining these tissues with ISH/TSA/ELF, we again found that most of the SIV RNA+ T cells in the submucosa (Fig. 6A) were small resting cells. At higher magnification (Fig. 6 B and C), there are a few virions at or near the cell surface of the small resting cells, whereas a cloud of virions envelopes large activated T cells. This image captures the proposed roles in the model of transmission at close distances by resting cells and at greater distances by activated cells.
Fig. 6.
Virus production by resting and activated T cells 12 days after intravaginal inoculation of SIV. (A) Predominance of small over large SIV RNA+ cells in ectocervical submucosa. (B and C) At higher magnification, virions are evident at the surface and surrounding large activated T cells, in addition to the bright yellow staining associated with intranuclear SIV RNA. In C, arrows point to the small number of virions associated with small T cells. [Original magnification: ×600 (B), ×800 (C), and ×1,600 (Inset).]
Mass-Action Model of Transmission and Establishment of Persistent Infection. Expression of CD4 and coreceptors (29–33) define populations of susceptible host cells that HIV and SIV can enter but does not predict per se the relative proportions of susceptible cells that will be productively infected in vivo or define mechanisms of transmission and propagation of infection. We propose here and provide evidence in support of what is essentially a mass-action predator-prey model (34, 35) for the transmission of and initial establishment of persistent SIV infection in LTs that accords to the CD4+ T cell the same preeminent role ascribed to this cell type in HIV-1 infection (36). In this model, transmission and propagation are related to the concentrations of infected cells, susceptible cells, and virus. Continued propagation of infection, relative proportions of productively infected cells of a particular type, and transmission at close or longer range all will depend on the availability of susceptible cellular substrates, the levels of virus production by different cell types, and the proximity of infected cells to susceptible cells; e.g., focal collections of CD4+ T cells in the female reproductive tract and the high density of CD4+ T cells in LTs favor propagation in CD4+ T cells.
This mass-action model predicts that in the earliest stages of infection, productive infection will predominate in the type of susceptible cell at the highest concentration in the tissue. The proportion of productively infected cells of a particular type at later times will correspond to the representation of these cells in the population at that time. The experimental data indicate that this prediction is satisfied for the LTs studied. Within the first week of infection, the majority of infected cells in LTs are resting CD4+ T cells. Between 1 and 4 weeks after i.v. inoculation of SIV, the proportion of activated CD4+ T cells increased 3-fold with a parallel 3-fold increase in the proportion of infected activated CD4+ T cells. These findings are consistent with previous cross sectional studies, that in the early stages of HIV-1 infection, a substantial proportion (≈50%) of the HIV-1 RNA+ cells are resting CD4+ T cells, whereas, at later stages, there is a proportional increase in productively infected activated T cells that parallels chronic immune activation (3). These findings collectively support the idea that the relative availability of susceptible cellular substrates is an important determinant of the relative proportions of particular cell types that are productively infected and the contribution that each cell type makes to the continued propagation of infection at a particular stage of infection. The model can also account for the previously reported finding that SIV RNA was found mainly in macrophages in gut-associated LTs at a time when CD4+ T cells had been depleted (13), and for the shift of infection to macrophages after the essentially quantitative acute depletion of CD4+ T cells in macaques infected with pathogenic simian/HIVs (37).
By visualizing virions, we have been able to directly estimate the relative numbers of viral particles associated with resting and activated cells and show that the surface of activated cells is surrounded by >10 times as many virions as are seen for resting cells. However, because resting cells turn over more slowly and far outnumber infected activated cells in the very earliest stages of infection, they could still contribute importantly to total virus replication levels during early infection. They would thus fulfill their proposed critical role in propagating infection by producing sufficient virus to maintain the chain of transmission to nearby cells. As the pool of activated CD4+ T cells increases with immune activation, and is infected, this population becomes the major source of virus production.
In the mass-action model, the greater production of virus and diffusion of virions away from the productively infected activated T cells should increase, through multiplicity effects, the efficiency and likelihood of infecting another cell at greater distances. Both the greater number of virions associated with infected activated T cells documented here, and previously documented genotypic (38) and spatial clustering of infected cells within two cell diameters (26) of another infected cell, are in accord with and support this model. A high local concentration of virus might also contribute to genetic variability through superinfection and recombination in nearby cells. Thus, the recently documented large number of recombinants and multiple proviruses in cells (39) documented in HIV infection need not arise from high systemic concentrations of virus, but rather, may be generated through local multiplicity effects.
These studies have revealed vital roles for productive infection of both resting and activated CD4+ T cells in intravaginal transmission and establishment of systemic infection. We show in this report that resting cells not only serve to maintain unbroken chains of transmission but also likely contribute substantially to production of the high levels of virus in early infection required for systemic dissemination, a role for this cell population that heretofore has not been appreciated. We document the even higher levels of virus production by infected activated CD4+ T cells that enable infection of susceptible cells at greater distances from virus producing cells both locally and throughout the LTs. The higher levels of virus production by infected activated T cells could be a factor in the increased transmission of HIV-1 in the setting of sexually transmitted diseases, other infections, or other causes of preexisting inflammation, e.g., induced by some microbicides (40–44). In addition to the effects of inflammation on mucosal integrity, the increased numbers of activated cells that accompany inflammation, and the important role that the infected activated T cell plays a role as an amplifier to efficiently propagate and spread infection, suggests that there could be a role for topical anti-inflammatory agents in reducing the availability of these critical target cells, thereby reducing sexual mucosal transmission and/or moderating infection at its earliest stages.
Supplementary Material
Acknowledgments
We thank Tammy Thompson and Colleen O'Neill for preparing the manuscript, and Tim Leonard for the figures. This work was supported in part with Federal funds from the National Institutes of Health under National Cancer Institute Contract NO1-CO-12400 and Grants AI028246 and AI048484.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SIV, simian immunodeficiency virus; LN, lymph node; LT, lymphatic tissue; DC, dendritic cell; ISH, in situ hybridization; IHCS, immunohistochemical staining; TSA, tyramide signal amplification; ELF, enzyme-linked fluorescence.
References
- 1.UNAIDS/WHO (2002) UNAIDS/WHO Report, December 2002 (UNAIDS/WHO, Geneva).
- 2.Miller, C. J., Alexander, N. J., Sutjipto, S., Lackner, A. A., Gettie, A. Hendricks, A. G., Lowenstine, L. J., Jennings, M & Marx, P. A. (1989) J. Virol. 63, 4277–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, Z.-Q., Schuler, T., Zupancic, M., Wietgrefe, S., Reimann, K. A., Reinhart, T. A., Rogan, M., Cavert, W., Miller, C. J., Veazey, D., et al. (1999) Science 286, 1353–1357. [DOI] [PubMed] [Google Scholar]
- 4.Schacker, T., Little, S., Connick, E., Gebhard, K., Zhang, Z.-Q., Krieger, J., Pryor, J., Havlir, D., Wong, J. K., Schooley, R. T., et al. (2001) J. Infect. Dis. 183, 555–562. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson, M., Stanwick, T. L., Dempsey, M. P. & Lamonica, C. A. (1990) EMBO J. 9, 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, C.-S., Ramilo, O. & Vitetta, E. S. (1997) Proc. Natl. Acad. Sci. USA 94, 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang, S., Patterson, B. & Levy, J. A. (1995) J. Virol. 69, 5659–5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spina, C. A., Guatelli, J. C. & Richman, D. D. (1995) J. Virol. 69, 2977–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnittman, S. M., Lane, H. C., Greenhouse, J., Justement, J. S., Baseler, M. & Fauci, A. S. (1990) Proc. Natl. Acad. Sci. USA 87, 6058–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spina, A., Prince, H. E., Douglas D. & Richman J. (1997) Clin. Invest. 99, 1774–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukrinsky, M. I., Stanwick, T. L., Dempsey, M. P. & Stevenson, M. (1991) Science 254, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zack, J. A., Arrigo, S. J., Weitsman, S. R., Go, A. S., Haislip, A. & Chen, I. S. Y. (1990) Cell 61, 213–222. [DOI] [PubMed] [Google Scholar]
- 13.Veazey, R. S., DeMaria. M., Chalifoux, L. V., Shvetz, D. E., Pauley, D. R., Knight, H. L., Rosenzweig, M., Johnson, R. P., Desrosiers, R. C. & Lockner, A. A. (1998) Science 280, 427–431. [DOI] [PubMed] [Google Scholar]
- 14.Veazey, R. S., Marx, P. A. & Lackner, A. A. (2003) J. Infect. Dis. 187, 769–776. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein, D. A., Penn, M. L., Korin, Y. D., Scripture-Adams, D. D., Zack, J. A., Kreisberg, J. F., Roederer, M., Sherman, M. P., Chin, P. S. & Goldsmith, M. A. (2001) Immunity 15, 671–682. [DOI] [PubMed] [Google Scholar]
- 16.Blaak, H., van't Wout, A. B., Brouwer, M., Hooibrink, B., Hovenkamp, E. & Schuitemaker, H. (2000) Proc. Natl. Acad. Sci. USA 97, 1269–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun, T.-W., Justement, J. S., Lempicki, R. A., Yang, J., Dennis, G., Jr., Hallahan, C. W., Sanford, C., Pandya, P., Liu, S., McLaughlin, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pope, M. & Haase, A. T. (2003) Nat. Med. 9, 847–852. [DOI] [PubMed] [Google Scholar]
- 19.Ma., Z., Lü, F. X., Torten, M. & Miller, C. J. (2001) J. Clin. Immunol. 100, 240–249. [DOI] [PubMed] [Google Scholar]
- 20.Suryanarayana, K., Wiltrout, T. A., Vasquez, G. M., Hirsch, V. M. & Lifson, J. D. (1998) AIDS Res. Hum. Retroviruses 14, 183–189. [DOI] [PubMed] [Google Scholar]
- 21.Insitute for Laboratory Animal Research (1996) Guide for the Care and Use of Laboratory Animals (Natl. Acad. Press, Washington, DC).
- 22.Koup, R. A., Safrit, J. T., Cao, Y., Andrews, C. A., McLeod, G., Borkowsky, W., Farthing, C. & Ho, D. D. (1994) J. Virol. 68, 4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips, A. N. (1996) Science 271, 497–499. [DOI] [PubMed] [Google Scholar]
- 24.Fauci, A. S. (1996) Nature 384, 529–534. [DOI] [PubMed] [Google Scholar]
- 25.Persaud, D., Zhou, Y., Siliciano, J. M. & Siliciano, R. F. (2003) J. Virol. 77, 1659–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reilly, C., Schacker, T., Haase, A., Wietgrefe, S. & Krason, D. (2002) J. Am. Stat. Assoc. 97, 943–954. [Google Scholar]
- 27.Cavert, W., Notermans, D. W., Staskus, K., Wietgrefe, S. W., Zupancic, M., Gebhard, K., Henry, K., Zhang, Z.-Q., Mills, R., McDade, H., et al. (1997) Science 276, 960–964. [DOI] [PubMed] [Google Scholar]
- 28.Nowak, M. A., Lloyd, A. L., Vasquez, G. M., Wiltrout, T. A., Wahl, L. M., Bischofberger, N., Williams, J., Kinter, A., Fauci, A. S., Hirsch, V. M. & Lifson, J. D. J. Virol. (1997) 71, 7518–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paxton, W. A., Martin, S. R., Tse, D., O'Brien, T. R., Skurnick, J., VanDevanter, N. L., Padiau, N., Braun, J. F., Kotler, D. P., Wolingky, S. M., et al. (1996) Nat. Med. 2, 412–417. [DOI] [PubMed] [Google Scholar]
- 30.Liu, R., Paxton, W. A., Choe, S., Ceradini, D., Martin, S. R., Horuk, R., MacDonald, M. E., Stuhlmann, H., Koup, R. A. & Landau, N. R. (1996) Cell 86, 367–377. [DOI] [PubMed] [Google Scholar]
- 31.Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Smith, M. W., Allikmets, R., Goedert, J. J., Buchbinder, S. P., Vittinghoff, E., Gomberts, E., et al. (1996) Science 273, 1856–1861. [DOI] [PubMed] [Google Scholar]
- 32.Deng, H. K., Rong, L., Ellmeier, W., Choe, S., Unutmaz, D., Burkhard, M., DiMarzio, P., Marmon, S., Sutton, R. E., Hill, C. M., et al. (1996) Nature 381, 661–666. [DOI] [PubMed] [Google Scholar]
- 33.Alkhatib, G., Combadiere, C., Broder, C. C., Feng, Y., Kennedy, P. E., Murphy, P. M. & Berger, E. A. (1996) Science 272, 1955–1958. [DOI] [PubMed] [Google Scholar]
- 34.DeBoer, R. J. & Perelson, A. S. (1998) J. Theor. Biol. 190, 201–214. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz, E. J., Neumann, A. U., Teixiera, A. V., Bruggeman, L. A., Rappaport, J., Perelson, A. S. & Klotman, P. E. (2002) AIDS 16, 341–345. [DOI] [PubMed] [Google Scholar]
- 36.Haase, A. T. (1999) Annu. Rev. Immunol. 17, 625–656. [DOI] [PubMed] [Google Scholar]
- 37.Igarashi, T., Brown, C. R., Endo, Y., Buckler-White, A., Plishka, R., Bischofberger, N., Hirsch, V. & Martin, M. A. (2001) Proc. Natl. Acad. Sci. USA 98, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinhart, T. A., Rogan, M. J., Amedee, A. M., Murphey-Corb, M., Rausch, D. M., Eiden, L. E. & Haase, A. T. (1998) J. Virol. 72, 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung, A., Maier, R., Vartanian, J. P., Bocharov, G., Jung, V., Fischer, U., Meese, E., Wain-Hobson, S. & Meyerhans, A. (2002) Nature 418, 144. [DOI] [PubMed] [Google Scholar]
- 40.Martin, H. L., Richardson, B. A., Nyange, P. M., Lavreys, L., Hillier, S. L., Chohan, B., Mandaliya, K., Ndinya-Achola, J. O., Bwayo, J., Kreiss, J., et al. (1999) J. Infect. Dis. 180, 1863–1868. [DOI] [PubMed] [Google Scholar]
- 41.Sweankambo, N., Gray, R. H., Wawer, M. J., Paxton, L., McNairn, D., Wabwire-Mangen, F., Serwadda, D., Chuanjun, L., Kiwanuka, N., Hiller, S. L., et al. (1997) Lancet 350, 546–550. [DOI] [PubMed] [Google Scholar]
- 42.Cohn, M. A., Frankel, S. S., Rugpao, S., Young, M. A., Willett, G., Tovanabutra, S., Khamboonruang, C., VanCott, T., Bhoopat, L., Barrick, S., et al. (2001) J. Infect. Dis. 184, 410–417. [DOI] [PubMed] [Google Scholar]
- 43.Moss, G. B., Clemetson, D., D'Costa, L., Plummer, F. A., Ndinya-Achola, J. O., Reilly, M., Holmes, K. K., Piot, P., Maitha, G. M., Hillier, S. L., et al. (1991) J. Infect. Dis. 164, 588–591. [DOI] [PubMed] [Google Scholar]
- 44.Van Damme, L., Ramjee, G., Alary, M., Voylsteke, B., Chandeying, V., Rees, H., Sirivongrangson, P., Mukenge-Tshibaka, L., Ettienge-Traore, V., Uaheowitchai, C., et al. (2002) Lancet 360, 971–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.